1. INTRODUCTION
Sweet corn (Zea may L. saccharata) is a valuable crop that is widely cultivated worldwide and is known for its high sugar in the endosperm due to the allelic mutant gene shrunken 2 (sh2sh2), which controls sugar accumulation content [1]. As a result, sweet corn is extensively grown and consumed extensively worldwide. In the United States, it is the second–largest fresh crop in the fresh market and industry [2]. In Thailand, sweet corn is a domestic crop that makes approximately US$165 million annually from by-products, including canned, frozen, and processed corn, which are exported to markets such as Japan, Korea, Taiwan, the US, the Philippines, and so on [3]. Planting sweet corn to achieve desirable high yield and ear quality traits requires an excellent hybrid variety and field practice. However, sweet corn yields are often reduced by various factors, such as weather, poor management, pests, and diseases. Corn disease is one of the problems, especially northern corn leaf blight (NCLB) and downy mildew (DM), which are severe in many areas where corn is planted. These diseases can cause yield losses from 20% to 100%, depending on the severity and the variety of corn [4–8]. NCLB is caused by Exserohilum turcicum (Pass) Leonard and Suggs [syn. Helminthosporium turcicum (Pass)]. Early symptoms include blisters on the lower leaves, which later develop into greenish-gray lesions that expand to 2 to 30 cm in a long, elliptical shape. The lesions eventually turn brown, causing blight [9]. This disease can be severe under optimal conditions, with temperatures between 18°C and 27°C and relative humidity of over 90% [6], and can cause yield losses of up to 90% [10]. DM is an obligate oomycete pathogen belonging to the family Peronosporaceae. In Thailand, P. sorghi and P. maydis are common corn-attacking species [11]. This systemic disease begins with an infection at the base of the leaves during the seedling stage and extends to the entire plant. The initial symptom is chlorosis and affected leaves typically die within 4 weeks of infection [12]. Under disease-favorable conditions, with a relative humidity of over 89% and temperatures ranging from 15°C to 32°C, susceptible varieties can suffer complete yield loss [13]. However, resistant and tolerant varieties of corn can effectively control these diseases, significantly reducing economic damage [12,14]. The lack of DM-resistant sweet corn varieties in Thailand makes it difficult to manage the crop effectively. Thai farmers rely heavily on chemical treatments, which are costly and environmentally harmful. The presence of DM can affect the marketability of sweet corn, particularly for export, because many international markets have strict phytosanitary regulations. This can result in farmers and exporters losing revenue. Although resistance is present in some waxy corn varieties, sweet corn has not been widely studied for DM resistance. Therefore, in this study, sweet corn parental lines were improved to resist NCLB and DM. The Chai Nat Field Crops Research Center (CNFCRC) at the Department of Agriculture of Thailand developed two sweet corn populations, PopHX75C1 and PopCH66C1, which could resist NCLB from E. turcicum but were susceptible to DM. Additionally, waxy corn inbred lines, F4305 and AGWX001, were developed at the same time and could be resistant to DM from P. sorghi and P. maydis. These waxy corns were used to cross with sweet corn, facilitating the transfer of DM resistance genes to sweet corn. Therefore, the objective of this study was to improve the resistance to both NCLB and DM in sweet corn lines and hybrids.
2. MATERIALS AND METHODS
2.1. Plant Materials
Two sweet corn lines, PopHX75C1 and PopCH66C1, which are resistant to NCLB but susceptible to DM, and two waxy corn inbred lines, F4305 and AGWX001, which are resistant to DM, from the CNFCRC, were used in this study. Two groups of S6 populations were then developed through crosses between sweet corn and waxy corn inbred lines to transfer DM resistance genes from waxy to sweet corn. The specific crosses were PopX75C1 × F4305 and PopCH66C1 × AGWX001, as illustrated in Figure 1.
2.2 Breeding Process for NCLB and DM Resistance
Five hundred of S1 wrinkled seeds in each cross, which indicated the presence of the sweet corn gene, were planted and evaluated for NCLB resistance in an artificial disease field at the National Corn and Sorghum Research Center (NCSRC), Nakhon Ratchasima province (14°38'32.6"N, 101°18'54.6"E) during the dry season of 2020. Plants that exhibited resistance or fewer symptoms were self-pollinated to produce S2 generated by pedigree selection. Fifty S2 lines with wrinkled seeds were then planted, with approximately 42 plants in each S2 line, and evaluated for DM resistance during the rainy season of 2020, also in an artificial disease field at the NCSRC. The selected resistant plants were self-pollinated to produce the S3 generation. Twenty-four lines from PopX75C1 × F4305 and 21 lines from PopCH66C1 × AGWX001 were planted around 84 plants/line in the late rainy season of 2020 and underwent natural selection for both NCLB and DM resistance at the NCSRC. The pollens of resistant plants were tested for the waxy gene (wxwx) using I2–KI staining [15], where plants showing a deep blue color in pollen grains were self–pollinated to produce the S4 generation. During the dry season of 2021, S4 seeds were planted following the same procedure used for the S3 generation. Plants with desirable agronomic characteristics, such as a strong root system, lodging resistance, proper timing of anthesis and silking, and absence of disease lesions, particularly NCLB and DM, were selected for advanced generation in S5 lines. The S5 lines were planted and evaluated for DM resistance in an artificial disease field at the NCSRC during the 2021 rainy season. Furthermore, eight S5 lines (X1–X8) were selected from the cross of PopX75C1 × F4305, while seven S5 lines (C1–C7) were selected from the cross of PopCH66C1 × AGWX001. The selected S5 lines were then self-pollinated to develop the S6 line. Additionally, these S5 lines from groups X and C were crossed using an 8 × 7 method (line × tester), resulting in fifty–six F1 hybrids, which were evaluated in a preliminary yield trial during the dry season of 2022 to assess the general combining ability (GCA) and specific combining ability (SCA) of ear yield, together with some agronomic traits and eating qualities. Therefore, 14 promising F1 crosses were selected and repeatedly crossed in the S6 generation for further yield trials.
 | Figure 1. Breeding scheme for the improvement of sweet corn lines resistant to NCLB and DM from 2019 to 2022. [Click here to view] |
In the hybrid yield trials, four commercial single-cross hybrids, Hibrix 3-1 and Hibrix 3-2 from Pacific Seeds (Thai) Ltd., Song Kla 84–1 (SK84–1), and Chai Nat 2 (CN2) from CNFCRC, were included as check varieties. Furthermore, 14 selected hybrids along with their parental S6 lines and 3 commercial hybrids were evaluated for resistance to NCLB and DM in artificial disease fields at the NCSRC and yield trials during the rainy season in 2022.
2.3. Artificial NCLB Disease Field
NCLB evaluation was performed using the spreader-row technique. Hibrix 3 was used as an infected line to spread the disease [16]. The culture and inoculation methods were modified based on the work of Asea et al. [17]. Briefly, E. turcicum was isolated from diseased leaves in the field, and pure cultures of E. turcicum were grown on a PDA (potato dextrose agar) medium. Colonies were then inoculated into sterilized sorghum grain, which was used to produce large inoculum quantities. The inoculum was incubated at room temperature for 2 weeks. After the spreader row (Hibrix 3) had grown for 3–4 weeks, sorghum grain colonized by E. turcicum was inoculated into the leaf whorl of each plant. After inoculation, disease lesions began to appear on leaves within 2–3 weeks. Once this occurred, the tested sweet corn lines were planted in blocks adjacent to the spreader row. The NCLB disease severity was evaluated 65 days after planting using a five-category rating system modified from Min et al. [5]. Briefly, based on the percentage of leaf area infected, we assessed the resistance level, separating into 0%–3% = highly resistant (HR), 4%–10% = resistant (R), 11%–30% = moderately resistant (MR), 31%–70% = moderately susceptible (MS), and >70% = highly susceptible (HS).
2.4. Artificial DM Disease Field
The spreader-row technique was used to screen sweet corn lines for resistance and susceptibility to DM [18]. The Tuxpeño variety was used as the infected row and was planted 2 weeks before inoculation with a DM spore suspension. Tested sweet corn lines were planted 4 weeks after the Tuxpeño variety using one spreader row for 10 rows of tested lines, each row 5 m in length, with a spacing of 25 cm between plants and 75 cm between rows. For a DM spore preparation, corn leaves infected with P. sorghi and P. maydis were collected from the DM nursery field in the morning when the plants were 4 weeks old and rinsed with water to prepare them for spore production. The leaves were then incubated under high humidity at room temperature for 8 hours to stimulate the formation of new conidia and spores. The spore suspension containing 5 × 104 spores ml−1 was sprayed onto the seedlings in the spreader row.
After 2–3 weeks, the DM symptoms appeared. Then, the sweet corn lines were planted in blocks adjacent to the spreader rows. Disease severity data were collected 35 days after planting based on the number of infected plants. The DM score was evaluated using the six–category rating system described by Craig et al. [19] based on the percentage of infected plants: 0% = HR, 1%–10% = R, 11%–25% = MR, 26%–50% = MS, 51%–75% = S, and 76%–100% = HS.
2.5. I2-KI Testing for sh2sh2WxWx Selection
Corn pollen grains can be identified for the presence of the waxy gene using an iodine–potassium iodide (I2-KI) solution, which comprises 1% potassium iodide (w/v) and 0.3% iodine (w/v) [15]. After solution treatment, the pollen grains exhibit distinct colors. The reddish–brown color indicates the presence of the waxy gene (wxwx), whereas the deep blue color indicates regular corn. A microscope with 10× magnification was used to observe and distinguish the colors of the stained pollen grains (Fig. 2).
2.6. Disease Evaluation and Hybrid Yield
An alpha–lattice design was used to carry out the preliminary yield trial of fifty–six F1 hybrids, comparing them with four commercial hybrid varieties, Hibrix 3-1, Hibrix 3-2, Song Khla 84–1, and Chai Nat 2, at CNFCRC (15°09'17.7"N, 100°11'02.3"E) during the dry season 2022. In each experimental unit, sweet corn seeds were planted in two rows, each 5 m in length, with a spacing of 25 cm between plants and 75 cm between rows. A total of 42 plants were obtained from each plot. All plants in each plot were harvested for ear yield, which was then converted to tons ha−1 GCA and SCA were analyzed to estimate the ear yield potential of parental lines and hybrids.
In the rainy season of 2022, 14 sweet corn hybrids that were selected for good eating quality, high yield, and good agronomic traits were evaluated for yield. Hybrids were produced from selected S6 parental lines. Fourteen F1 hybrids were tested for yield against three commercial check varieties using a randomized complete block design (RCBD) with three replications at two locations, namely, NCSRC and CNFCRC. The experimental plot comprised four rows, each 5 m in length, with a spacing of 25 cm between plants and 75 cm between rows. The data on yield and agronomic traits were collected from the two middle rows in each plot. Therefore, 42 plants were harvested for ear yield in each plot and converted to tons ha−1. A combined analysis was performed for yield and agronomic characteristics at the two sites.
During the same season, 14 F1 hybrids, along with 2 susceptible varieties, were evaluated for NCLB and DM resistance under the artificial field at NCSRC and conducted in RCBD with 3 replications. Each plot comprised one 5-m-long row with a plant spacing of 75 × 25 cm. The severity scores and resistance levels for NCLB and DM were assessed using the rating systems modified from Min et al. [5] and Craig et al. [19], respectively. Severity scores were measured for individual plants in each plot, and the average score in each plot was used for analysis of variance (ANOVA) of RCBD.
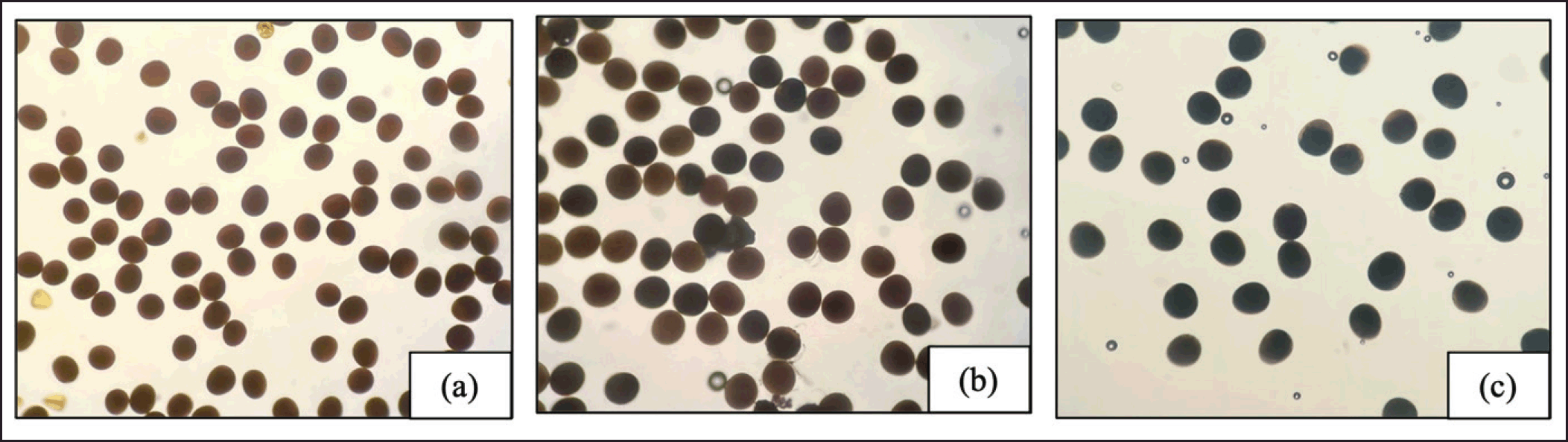 | Figure 2. Microscopic images at 10× magnification showed pollen grains stained with I2–KI in the pollen of sweet corn lines: (a) reddishbrown, (b) mixed reddish–brown and deep blue, and (c) deep blue. [Click here to view] |
2.7. Data Collection of Yield Trials
The agronomic characteristics were collected, including anthesis and silking time (from the first day of watering until 50% of anthesis and silking occurs in each plot), plant and ear heights, husk cover score 1–5 (1 representing the best cover and 5 the poorest), ear diameter (D), ear length (L1), and tip length (L2). Ten random samples were measured in each experiment plot, and the mean was calculated. Yield was measured from fresh ear weight (with and without husk) per plot 20 days after silking and converted to tons per hectare. The sweetness was determined in brix by measuring the squeezed corn kernel liquid using a hand-held refractometer. Quality bite tests were conducted on corn ears cooked for 15 minutes in boiling water. Eight panelists evaluated the samples using a five-point rating scale adapted from Dermail et al. [20]. The evaluation criteria included: sweetness (S): 1–5 (not sweet-highly sweet), tenderness (T), 1–5 (no creamy texture–highly creamy texture), and overall liking (F); 1–5 (most unfavorable–most favorable).
2.8. Statistical Analysis
All the recorded data were analyzed using R statistical software (version 4.2.2) [21]. ANOVA was conducted according to alpha lattice and RCBD. A combined ANOVA was performed for the ear yield and agronomic traits across two sites. Before conducting the combined analysis of variance, we assessed the homogeneity of variance for ear yield using the method described by Snedecor and Cochran [22]. Because the variances were homogeneous, all agronomic traits were combined across the two sites. The least significant difference (LSD) test at a significance level of 0.05 was used to compare the treatment means. The combining ability parameters were analyzed using the line × tester method [23].
3. RESULTS AND DISCUSSION
3.1. I2-KI Testing for sh2sh2WxWx Selection
Considering only the two genes of sweet corn (sh2sh2) and waxy corn (wxwx) in the crossing between them, theoretically, the genotypes in the F2 or S1 generations should have the ratio of 9Sh2_Wx_: 3 Sh2Sh2Wx_: 3 sh2sh2Wx_: 1 sh2sh2wxwx in the segregating population. In the S1 generation, only wrinkled seeds were used to retain the sweet corn genotype. However, there were three genotypes—sh2sh2WxWx, sh2sh2Wxwx, and sh2sh2wxwx—that exhibited a wrinkled seed phenotype, indicating that the shrunken–2 gene (sh2sh2) had an epistatic effect over the waxy gene (wxwx) in wrinkled seeds [24,25]. Therefore, waxy must be eliminated to restore regular sweet corn in the selected line without waxy in the advanced generation. The I2-KI reaction is commonly used to detect starch in endosperm and pollen grains, distinguishing waxy and regular corn seeds [26,27]. In the S3 and S4 generations, we utilized the I2-KI technique to eliminate the sh2sh2Wxwx and sh2sh2wxwx genotypes. When we added I2-KI to the pollen of each plant, we saw a deep blue color (WxWx), a mix of reddish-brown and deep blue colors (Wxwx) or reddish–brown color (wxwx) (Fig. 2). These colors indicate that sh2sh2WxWx, sh2sh2Wxwx, and sh2sh2wxwx were separated, as explained by Talukder et al. [28]. Therefore, we selected plants with only deep-blue pollen grains representing the sh2sh2WxWx genotype for further studies.
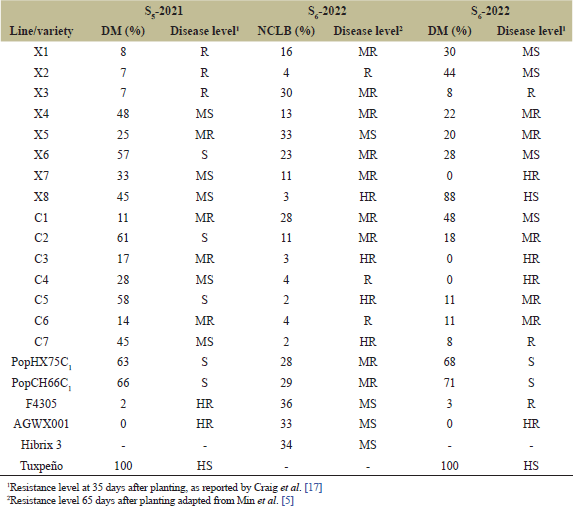 | Table 1. Percentages and resistance levels to NCLB and DM in the S5 and S6 lines tested in the artificial field at NCSRC from 2021 to 2022. [Click here to view] |
3.2. Evaluation of NCLB and DM and Selection of Inbred Lines
As described above, in the early generations (S1–S4), selected plants were evaluated for NCLB and DM resistance in artificial disease fields and natural disease selection at the NCSRC (data not shown). In the S5 generation, the results showed that the selected line X1–X8, obtained from crossing PopHX75C1 × F4305, had a range of 7%–57% DM disease incidence, indicating disease levels from R to S (Table 1). Similarly, the selected line C1–C7, resulted from crossing PopCH66C1 × AGWX001, exhibited a range of 11%–61% DM disease incidence, with disease levels ranging from MR to S (Table 1). The Tuxpeño variety (a susceptible variety) had a 100% DM disease incidence (Table 1). Therefore, only asymptomatic or less symptomatic plants of each line were selected and self-pollinated to produce seed until S6 lines.
The NCLB evaluation of the S6 generation found that X1–X8 exhibited HR to MR levels, ranging from 3% to 30% NCLB disease incidence, except for X5, which showed a MS level of 33% (Table 1). Additionally, C1–C7 showed HR to MR, ranging from 2% to 28%. Meanwhile, the check variety Hibrix 3 for comparison in the NCLB trials demonstrated a MS level of 34%. As for the DM evaluation, X1–X8 demonstrated HR to MR levels, ranging from 0% to 22%, except for X1, X2, X6, and X8, which showed MS to HS levels of 28%–88%. The results for C1–C7 showed HR to MR levels, ranging from 0% to 18%, except for C1, which showed a MS level of 48% NCLB disease incidence (Table 1).
In comparison to the sweet corn parent lines PopHX75C1 and PopCH66C1, which had a high susceptibility to DM at 68% and 71%, respectively (Table 1), 10 elite sweet corn lines (X3, X4, X5, X7, C2, C3, C4, C5, C6, and C7) showed HR (HR, 0% DM disease incidence) to MR (MR, 18% DM disease incidence) to DM. These lines exhibited higher DM resistance than the parent lines, indicating that the DM resistance genes were transferred from the resistance source of the inbred waxy corn lines F4305 and AGWX001. Similarly, in a previous study, Sukto et al. [29] demonstrated that crosses between resistant varieties (Takpha1 or TF and Nei9008) and susceptible varieties (Tein Leang Khon Kaen or TY and Tein Ban Kao or BK) yielded MR (Nei9008/BK–24–9–B) and R (TY/TF–33–1–B) to DM. Additionally, nine elite lines (X3, X4, X7, C2, C3, C4, C5, C6, and C7) remained resistant to NCLB, showing HR to MR.
Moreover, the levels of DM resistance increased when only asymptomatic or less symptomatic plants of each line were self–pollinated from the S5 to the S6 generations, except for X1, X2, X3, X8, and C1. Notably, each generation showed plants resistant to artificial disease fields, demonstrating a clear distinction between resistance and susceptibility. Resistant plants consistently exhibited no or fewer symptoms of DM in controlled environments, whereas susceptible plants displayed apparent disease symptoms (Table 1). This divergence in response under identical conditions highlights the distinction. However, DM resistance is controlled by polygenic genes [4,29]. Therefore, crossing between resistant lines can increase DM resistance in progeny only under suitable selection conditions, such as methods of DM inoculation, favorable temperature, and high humidity. Without these conditions, the breeding program may fail to identify resistant progeny, resulting in reduced effectiveness and slower progress in enhancing DM resistance in advanced generations. Similarly, NCLB resistance breeding can be performed in the same manner as DM resistance breeding because it is also controlled by polygenic genes [30,31]. The consistent results observed across multiple generations (from S1 to S5) are a promising sign of the success of breeding programs in selecting for disease resistance. The use of artificial and natural disease selection combined with the pedigree selection method ensures that these results are not only consistent within the breeding program but also reproducible under different environmental conditions. Variability in resistance levels within the S5 generation reflects natural genetic diversity in disease resistance traits, which is expected and manageable. As the program progresses, further validation across multiple growing seasons and locations will be essential to confirm the long-term stability and global applicability of these resistant sweet corn lines.
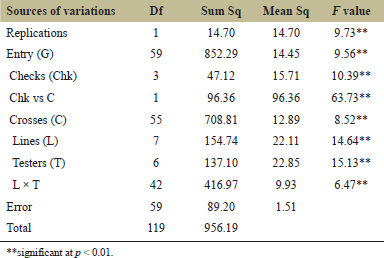 | Table 2. The ANOVA of line × tester of the preliminary yield trial planted at the CNFCRC in the dry season, 2022. [Click here to view] |
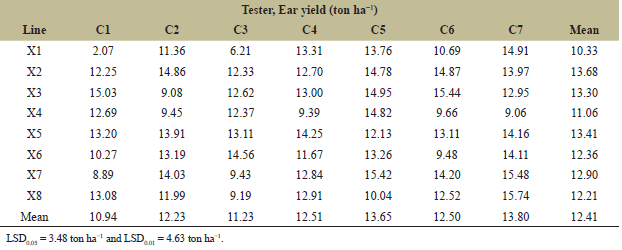 | Table 3. Mean ear yield (ton ha−1) from 8 lines × 7 testers planted at the CNFCRC in the dry season of 2022. [Click here to view] |
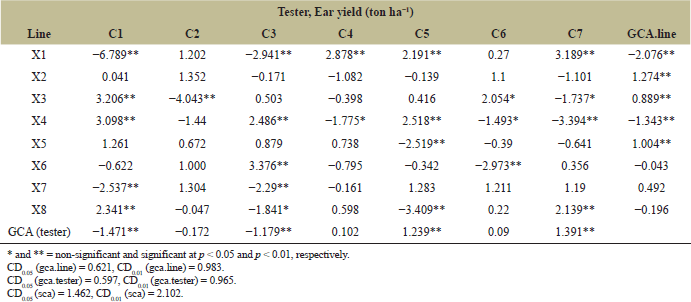 | Table 4. GCA and SCA of ear yield from 8 lines, 7 testers, and fifty-six crosses planted at CNFCRC in the dry season, 2022. [Click here to view] |
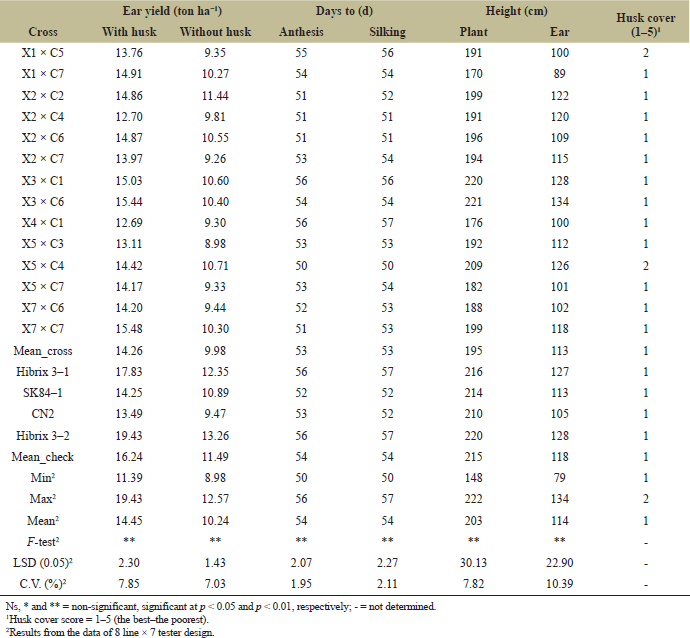 | Table 5. Ear yield with and without husk, days to anthesis and silking, plant and ear height, and husk cover score of 14 sweet corn hybrids selected and evaluated at the CNFCRC in the dry season, 2022. [Click here to view] |
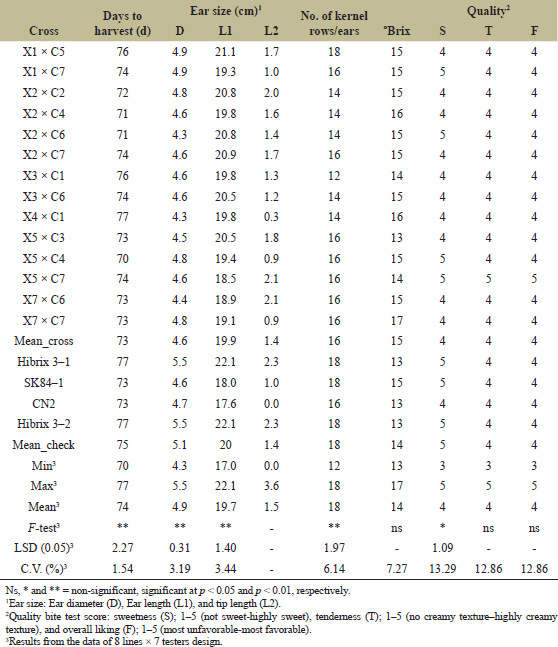 | Table 6. Days to harvest, ear size, number of kernel rows/ear, degree of brix, and eating qualities of 14 sweet corn hybrids evaluated at the CNFCRC in the dry season, 2022. [Click here to view] |
3.3 GCA and SCA Effects From S5 Lines and Their F1 Hybrids
The variance analyses of ear yield were significantly different for crosses, line, tester, and line × tester (Table 2). The ear yield of 56 crosses from line × tester was presented in Table 3. The ear yield of the tested crosses ranged from 2.07 to 15.74 tons ha−1, with an average yield of 12.41 tons ha−1. Moreover, the significant effects of GCA and SCA revealed the presence of additive and non-additive gene effects on ear yield (Table 4). GCA effects are useful for identifying superior parents for direct use in breeding programs. The selected inbred lines should have high GCA values that are significantly different from zero and high mean values to predict the best progeny based on GCA [32]. Tables 4 and 5 present the ear yield and GCA effects from 8 lines and 7 testers, respectively. The X2, X3, X5, C5, and C7 lines exhibited good GCA for ear yield, indicating an additive gene effect. Therefore, these lines were selected for further breeding programs. Moreover, SCA described the performance of crosses relative to the average performance of hybrids in an experiment related to nonadditive gene effects, including dominance and epistasis [32,33]. The SCA effect is presented in Table 4, and then some crosses with positive significance were selected, such as X1 × C5, X1 × C7, X3 × C1, X3 × C6, and X4 × C1. However, some crosses showing higher yield averages with a significant positive SCA effect were not selected, such as X4 × C5, X6 × C3, and X8 × C7. Because a quality bite test of overall liking (F) was from 1 to 2 (data not shown). Therefore, 6 lines (X1, X2, X3, X5, and X7) from group X and 7 lines (C1, C2, C3, C4, C5, C6, and C7) from group C were selected and made into a combination of 14 hybrids with good combining ability of ear yield and eating quality for further evaluation of ear yield and disease resistance (Table 5). In specialty corn, eating quality is the major trait for selecting superior sweet corn hybrids [20,34]. Hence, the goal of a sweet corn hybrid breeding program is to improve not only the ear yield but also the eating quality and excellent agronomic characteristics. Sweet corn was studied for its eating quality using a bite test score, which included sweetness (S), tenderness (T), and overall liking (F). In the taste tests, all crosses were rated with a bite test score from 4 to 5, which was the same quality as that of the commercial varieties. At the same time, the degree of brix ranged from 13 to 17, and the agronomic characteristics were in the acceptable range (Tables 5 and 6). The ear yield of selected crosses ranged from 12.69 to 15.48 tons ha−1 with an average of 14.26 tons ha−1 and they had a good husk cover (Table 5). Therefore, based on our results, 14 superior crosses were selected for the yield trials and disease evaluations at two sites.
 | Table 7. NCLB and DM evaluation of the parental lines (S6) and their hybrids tested at the NCSRC in the rainy season of 2022. [Click here to view] |
 | Table 8. Ear yield with and without husk, days to anthesis and silking, and plant and ear height of 14 hybrid crosses and 3 check varieties in yield trials at CNFCRC and NCSRC in the rainy season, 2022. [Click here to view] |
3.4 Evaluation of NCLB and DM in S6 Lines and Their F1 Hybrids
For the previous generation, 14 selected F1 hybrids were produced from the S5 lines, including six lines from group X and seven lines from group C. Therefore, the 14 selected crosses were made of F1 hybrids from the S6 lines with desirable traits, such as high yield and disease resistance, to evaluate NCLB and DM, including their parental lines in the artificial disease fields at NCSRC in the rainy season of 2022. The results showed that the parental lines were rated as HR to MS for NCLB and DM (Table 7), whereas the 14 crosses were selected based on ear yield by GCA and SCA effects, good agronomic characteristics, and eating quality by the bite test. Comparing the selected S6 lines with the original parents with NCLB resistance (PopHX75C1 and PopCH66C1) and DM resistance (F4305 and AGWX001), these selected S6 lines combined the NCLB and DM resistance in their lines (Table 7). Moreover, the 14 F1 hybrids were between HR to MR to NCLB, whereas they were R to S to DM. However, there were four crosses with a favorable combination of NCLB and DM resistances, including crosses of X3 × C1, X3 × C6, X4 × C1, and X5 × C4. Most corn breeders favor the use of quantitative NCLB resistance testing in their cultivar development programs [35]. In the present study, we did not directly test the effects of genes; however, the progenies showed varying resistance levels, which appear to be controlled by multiple genes within the plant lines. To obtain the plants desired by breeders, classical breeding involves crossing two plants with desired traits, such as high yield and/or disease resistance [36]. In addition, when crosses were carried out between lines with moderate resistance (X1) and high resistance (C7), the hybrid (X1 × C7) showed good resistance to NCLB, and in the case of another line with resistance (X2) crossed with a line with high resistance (C7), the hybrid (X2 × C7) also displayed good resistance. Consequently, hybridization outcomes led to improvements in yield and yield-related traits, which were supported by other authors’ findings [37,38]. For the DM evaluation, in some crosses between a resistant line (X3) and a MR line (C6), the hybrid (X3 × C6) showed resistance to DM. However, this was not true in all crosses because nine crosses were S to DM. This result may not be in accordance with those previously reported in other studies, which illustrated that the additive gene effect was predominant and of higher magnitude than the non-additive gene effect for the inheritance of resistance to DM in maize [39,40]. However, at least four of our crosses did demonstrate R to MR for both NCLB and DM diseases, such as X3 × C1, X3 × C6, X4 × C1, and X5 × C4. The breeding strategy described in this study has far-reaching implications for sweet corn breeding in Thailand and globally. By developing varieties with improved resistance to NCLB and DM, breeders can enhance food security, reduce pesticide use, and increase the sustainability of corn production. This study could serve as a foundation for breeding programs worldwide, particularly in tropical and subtropical regions with high disease pressures, ultimately benefiting farmers and consumers.
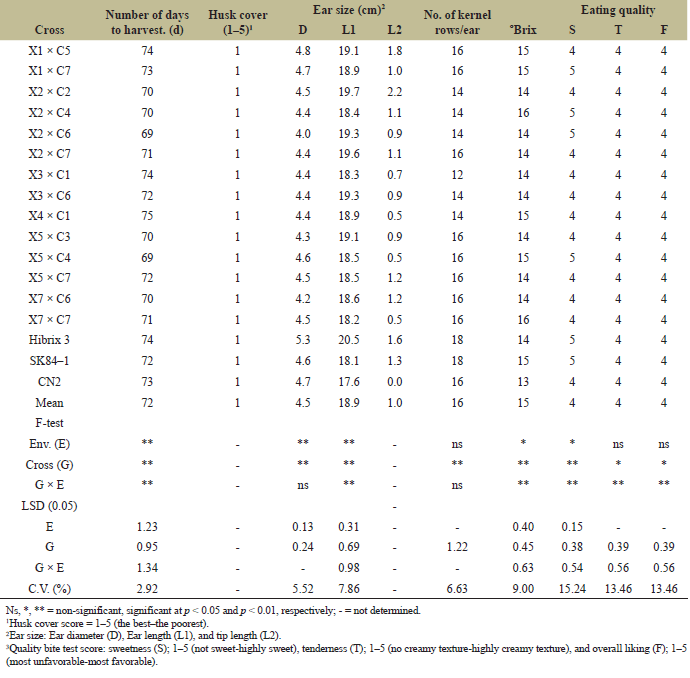 | Table 9. Days to harvest, husk cover score, ear size, number of kernel rows/ear, degree brix, and eating quality of 14 hybrid crosses and 3 check varieties in yield trials at CNFCRC and NCSRC in the rainy season, 2022. [Click here to view] |
3.5 Combined ANOVA Over Environments
The cross variance was homogeneous in terms of ear yield (ear yield with husk). Therefore, a combined ANOVA was conducted for ear yield and agronomic traits. The results showed that the environment (E) had significant effects on all traits except for plant and ear height, number of kernel rows/ear, tenderness, and overall eating quality (Tables 8 and 9). Likewise, there were significant differences in crosses (G) for all the studied traits. Moreover, significant interactions between genotype and environment (G × E) for ear yield and ear yield without husks indicated that hybrid responses differed between environments. The relative ranking or performance of the hybrids varies depending on the environment in which they are grown. For example, X1 × C5, X2 × C6, X5 × C4, and X5 × C7 at CNFCRC had ear yields with husks higher than in NCSRC environments, while CN2 had ear yields at CNFCRC lower than at NCSRC (Table 8). Therefore, multienvironment experiments must be conducted to select superior hybrids and a widely adapted variety [41]. There was a strong correlation between ear yield and ear yield without husks (r = 0.88**) (data not shown). Therefore, a large ear yield would be considered a high yield or good yield variety. From the combined environment, the ear yields of crosses ranged between 11.39 and 14.88 tons ha−1, while the ear yields of check hybrids ranged between 14.15 and 18.79 tons ha−1. Together with NCLB and DM evaluations, four crosses had a good combination of ear yield and disease resistances, namely, X3 × C1, X3 × C6, X4 × C1, and X5 × C4, giving ear yields of approximately 12.68, 14.67, 12.68, and 11.39 tons ha−1, respectively (Tables 7 and 8). The crosses of X3 × C6 had higher ear yields than the other three crosses. The X3 × C6 was infected with NCLB at about 19% and DM at about 7%, and this hybrid came from good parental lines that had high disease resistance to NCLB and DM, showing that the resistance in the inbred parents can be transferred to the hybrids (Table 7). In addition, X3 had a high GCA (0.889*), and X3 × C6 had a high SCA (2.054*) for ear yield in the S5 (Table 4). This yield was in agreement with the findings of Khamphasan et al. [32], indicating that selected inbred lines should have a high GCA and be useful for identifying superior parents for direct use in breeding programs [42]. In addition, hybrids showed high and significant SCA effects that may be valuable for breeding programs [33,43,44]. Superior sweet corn has good eating quality, including sweetness (S), tenderness (T), and overall liking (F). The interesting hybrids, X3 × C6, also had good eating qualities and a high Brix degree (Table 9). They had good husk cover, a total of days to anthesis and silking, and a total of days to harvest, and their ear size was the same as that of the favorable commercial hybrid Hibrix 3 (Tables 8 and 9). However, the best–yield hybrid with NCLB and DM resistances from this study still had a lower ear yield than Hibrix 3, meaning that we have to do other crosses for ear yield and try to incorporate that trait together with NCLB and DM resistances. We can utilize this line of study as a germplasm source for resistance to DM and NCLB in the backcross breeding program, serving as a donor parent to enhance ear yield while improving resistance to these diseases. We have identified some sweet corn hybrids that exhibit high ear yield, comparable to Hybrix 3, yet lack resistance to DM and NCLB. Consequently, X3, which demonstrates good GCA, can be employed as a donor parent for a parental line within the same heterotic group as one of the parent lines. Additionally, C1 (from the cross X3 × C1, which shows good SCA) can serve as the donor parent for a different parental line. The selected lines from 2 to 3 generations of backcrossing from each heterotic pattern can then be crossed to assess ear yield and disease resistance. These strategies could facilitate the development of superior hybrids that deliver higher yields and improved disease resistance compared to older or commercially available hybrids. However, this study contributes by emphasizing the importance of considering marketable yield, eating quality, and NCLB and DM resistances in breeding. This broad scope offers valuable insights into the development of disease-resistant, high-quality sweet corn varieties that can perform well under diverse environmental conditions. In addition, introducing disease-resistant sweet corn lines could have significant environmental and economic impacts. Environmentally, the reduced need for chemical pesticides due to enhanced resistance to NCLB and DM could lead to lower environmental pollution and healthier ecosystems. Economically, higher ear yields and improved disease resistance could enhance farmers' profitability by reducing crop loss and input costs while also ensuring more sustainable and reliable sweet corn production in Thailand.
4. CONCLUSION
This study successfully enhanced the resistance of sweet corn lines and their hybrids to NCLB and DM diseases by incorporating DM resistance from waxy corn. In the S6 generation, which involved 14 hybrids, three parental lines (X3, X4, and X7) from group X and six lines (C2, C3, C4, C5, C6, and C7) from group C exhibited resistance to both NCLB and DM, ranging from HR to MR. This confirms the successful transfer of DM-resistant genes from the source of resistance. The best hybrid, X3 × C6, had a high ear yield of 14.67 tons ha−1 and showed MR to NCLB and R to DM diseases. It also demonstrates excellent eating quality, and its agronomic characteristics are similar to those of commercial varieties. However, its yield was lower than that of the best commercial hybrid, Hibrix 3 (18.79 tons ha−1). Despite this, the selected sweet corn lines with combined resistance to NCLB and DM can be used as sources of resistance for future sweet corn breeding programs as a donor parent in backcross breeding programs to improve ear yield and these diseases' resistance.
5. ACKNOWLEDGMENTS
We would like to thank the National Corn and Sorghum Research Center, Nakhon Ratchasima Province, Nuclear Technology Research Center, Faculty of Science, Kasetsart University, and Chai Nat Field Crops Research Center, Chai Nat Province, Department of Agriculture (DOA), Thailand, for providing the plant materials, experimental fields, and research facilities.
6. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
8. CONFLICTS OF INTEREST
The authors report no financial or other conflicts of interest related to this study.
9. FUNDING
This study was supported by the Agricultural Research Development Agency (public organization) (ARDA).
10. DATA AVAILABILITY
The data that support the findings of this study are available upon request from the corresponding author.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Ruanjaichon V, Khammona K, Thunnom B, Suriharn K, Kerdsri C, Aesomnuk W, et al. Identification of gene associated with sweetness in corn (Zea mays L.) by genome-wide association study (GWAS) and development of a functional SNP marker for predicting sweet corn. Plants 2021;10(6):1239; CrossRef
2. Agricultural marketing resource center [Internet]. Sweet corn; c2022 [cited 2022 Feb 1]. Available via https://www.agmrc.org/commodities-products/vegetables/sweet-corn
3. The Customs Department [Internet]. Sweet corn (Zea mays var. saccharata): exports of Thailand; 2021 [cited 2022 Feb 1]. Available via http://www2.ops3.moc.go.th
4. Kim HC, Kim KH, Song K, Kim JY, Lee BM. Identification and validation of candidate genes conferring resistance to downy mildew in maize (Zea mays L.). Genes 2020;1:191.
5. Min J, Chunyu Z, Khalid H, Suwen W, Feng L. Pyramiding resistance genes to northern leaf blight and head smut in maize. Int J Agric Boil 2012;14:430–4.
6. Ogliari JB, Guimarães MA, Geraldi IO, Camargo LEA. New resistance genes in the Zea mays -Exserohilum turcicum pathosystem. Genetic Mol 2005;28:435–9.
7. Bonde MR, Peterson GL, Duck NB. Effect of temperature on sporulation, conidial germination and infection of maize by Peronosclerospora sorghi from different geographical areas. Phytopathology 1985;75(1):122–6.
8. Rifin A. Downy mildew resistance of single cross progenies between Indonesia and Philippine corn inbred lines. Penelitian Pertanian 1983;3:81–3.
9. Abebe D, Singburaudom N. Morphology, culture, and pathogenicity variation of Exserohilum turcicum (Pass) Leonard and Suggs isolates in maize (Zea mays L.). Kasetsart J 2006;40:341–52.
10. Cox RS. Control of the Helminthosporium blight disease on sweet corn in south Florida. Phytopathology 1956;46:112–5.
11. Janruang P, Unartngam J. Morphological and molecular based identification of corn downy mildew distributed in Thailand. Int J Agric Technol 2018;14(6):845–60.
12. Ajala SO, Kling JG, Kim SK, Obajimi AO. Improvement of maize populations for resistance to downy mildew. Plant Breed 2003;122:328–33.
13. George MLC, Prasanna BM, Rathore RS, Setty TAS, Kasim F, Azrai M, et al. Identification of QTLs conferring resistance to downy mildews of maize in Asia. Theor Appl Genet 2003;107(3):544–51.
14. Dermail A, Suriharn B, Lertrat K, Chankaew S, Sanitchon J. Reciprocal cross effects on agronomic traits and heterosis in sweet and waxy corn. SABRAO J Breed Genet 2018;50(4):444–60.
15. Nelson OE. The waxy locus in maize. I. Intralocus recombination frequency estimates by pollen and by conventional analyses. Genetics 1962;47:737–42.
16. Wongwarat T, Kerdsri C, Phruetthithep C. Identification of SSR markers associated with northern corn leaf blight resistance in sweet corns. Thai Agri Res J 2024;42(2):180–91.
17. Asea G, Vivek BS, Bigirwa G, Lipps PE, Pratt RC. Validation of consensus quantitative trait loci associated with resistance to multiple foliar pathogens of maize. Phytopathology 2009;99(5):540–7.
18. Anahosur KH, Hegde RK. Assessment of the techniques used for screening sorghum genotypes to downy mildew. Mysore J Agric Sci 1979;13:449–51.
19. Craig J, Bockholt AJ, Frederiksen RA, Zuber MS. Reaction of important corn inbred lines to Sclerospora sorghi. Plant Dis Rep 1977;61:563–4.
20. Dermail A, Fuengtee A, Lertrat K, Suwarno WB, Lübberstedt T, Suriharn K. Simultaneous selection of sweet-waxy corn ideotypes appealing to hybrid seed producers, growers, and consumers in Thailand. Agronomy 2022;12:87; CrossRef
21. R core team. R project for statistical computing. Version 4.2.2 [software]. 2022 Oct 31 [cited 2022 Dec 1]. Available via https://www.r-project.org
22. Snedecor GW, Cochran WG. Statistical methods. 7th edition. Ames, IA: Iowa State University Press; 1980.
23. Kempthorne O. An introduction to genetics. New York, NY: John Willey and Sons. Inc.; 1957.
24. Dong L, Qi X, Zhu J, Liu C, Zhang X, Cheng B, et al. Supersweet and waxy: meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol J 2019;17(10):1853–5.
25. Simla S, Lertrat K, Suriharn B. Gene effects of sugar compositions in waxy corn. Asian J Plant Sci 2009;8:417–24.
26. Neuffer MG, Coe EH, Wessler SR. Mutants of maize. New York, NY: Cold Spring Harbor Laboratory Press; 1997.
27. Nakamura T, Yamamori M, Hirano H, Hidaka S, Nagamine T. Production of waxy (amylose-free) wheats. Molec Gen Genet 1995;248:253–9.
28. Talukder ZA, Muthusamy V, Zunjare RU, Chhabra R, Reddappa SB, Mishra SJ, et al. Pollen staining is a rapid and cost-effective alternative to marker-assisted selection for recessive waxy1 gene governing high amylopectin in maize. Physiol Mol Biol Plants 2022;28(9):1753–64.
29. Sukto S, Lomthaisong K, Sanitchon J, Chankaew S, Falab S, Lübberstedt T, et al. Breeding for prolificacy, total carotenoids and resistance to downy mildew in small-ear waxy corn by modified mass selection. Agronomy 2021;11(9):1793; CrossRef
30. Rashid Z, Sofi M, Harlapur SI, Kachapur RM, Dar ZA, Singh PK, et al. Genome-wide association studies in tropical maize germplasm reveal novel and known genomic regions for resistance to northern corn leaf blight. Sci Rep 2020;10:21949; CrossRef
31. Poland JA, Peter J, Bradburya B, Edward S, Bucklera B, Nelsona RJ. Genome-wide nested association mapping of quantitative resistance to northern leaf blight in maize. PNAS 2011;108(17):6893–8.
32. Khamphasan P, Lomthaisong K, Harakotr B, Scott MP, Lertrat K, Suriharn B. Combining ability and heterosis for agronomic traits, husk and cob pigment concentration of maize. Agriculture 2020;10:510; CrossRef
33. Sun J, Gao J, Yu X, Liu J, Su Z, Feng Y, et al. Combining ability of sixteen USA maize inbred lines and their outbreeding prospects in China. Agronomy 2018;8(12):281; CrossRef
34. Lertrat K, Pulam T. Breeding for increased sweetness in sweet corn. Int J Plant Breed 2007;1(1):27–30.
35. Abdelsalam NR, Balbaa MG, Osman HT, Ghareeb RY, Desoky EM, Elshehawi AM, et al. Inheritance of resistance against northern leaf blight of maize using conventional breeding methods. Saudi J Biol Sci 2021;29(3):1747–59.
36. Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science 2010;327(5967):818–22; CrossRef
37. Ohunakin AO, Odiyi A, Akinyele B. Genetic variance components and GGE interaction of tropical maize genotypes under northern leaf blight disease infection. Cereal Res Commun 2020;49:277–83.
38. Razzaq T, Khan MF, Awan SI. Study of northern corn leaf blight (NCLB) on maize (Zea mays L.) genotypes and its effect on yield. Sarhad J Agri 2019;35:1166–74.
39. Maruthi RT, Jhansi RK. Genetic variability, heritability and genetic advance estimates in maize (Zea mays L.) inbred lines. J Appl Nat Sci 2015;7(1):149–54.
40. Motawei AA. Diallel analysis for grain yield and resistance to downy mildew disease in maize, Egypt J plant Breed 2011;14(4):39–50.
41. Worrajinda J, Lertrat K, Suriharn K. Combining ability of super sweet corn inbred lines with different ear sizes for ear number and whole ear weight. SABRAO J Breed Genet 2013;45(3):468–77.
42. Mahan AL, Murray SC, Rooney LW, Crosby KM. Combining ability for total phenols and secondary traits in a diverse set of colored (red, blue, and purple) maize. Crop Sci 2013;53(4):1248–55.
43. Abera W, Hussein S, Derera J, Worku M. Heterosis and combining ability of elite maize inbred lines under northern corn leaf blight disease prone environments of the mid-altitude tropics. Euphytica 2016;208:291–400.
44. Ketthaisong D, Suriharn B, Tangwongchai R, Lertrat K. Combining ability analysis in complete diallel cross of waxy corn (Zea mays var. ceratina) for characteristics starch pasting viscosity. Sci Hort 2014;175:229–35.