1. INTRODUCTION
Lignocellulosic waste, a primary component of renewable biomass, is broken down by enzymes such as laccase, cellulase, hemicellulase, and pectinase. These enzymes catalyze the hydrolysis of complex phenols and carbohydrates in plant cell walls into simpler molecules such as sugars and aromatic compounds [1]. This process has significant implications in various fields, including bioremediation, wastewater treatment plants, bioethanol production, anaerobic digestion, the paper and pulp industry, animal feed manufacturing, and the textile industry. Lignocellulolytic enzymes, such as hydrocarbons and pesticides, are essential for breaking down organic contaminants in polluted environments, contributing to ecosystem recovery. They also facilitate the breakdown of organic materials, reducing biological oxygen demand (BOD) and chemical oxygen demand (COD) in effluents before release into water bodies [2]. They also produce bioethanol from LCB, decomposing cellulose and hemicellulose into sugars that microbes can ferment. In the paper and pulp industry, they expedite delignification and bleaching procedures, enhancing the effectiveness of pulp bleaching and reducing environmental consequences linked to chemical bleaching agents. They also improve nutrient availability, animal growth, and health by breaking down complex carbohydrates such as maize stover and wheat straw in feedstocks. In the textile industry, they aid in bioscouring and denim bleaching, eliminating contaminants and natural waxes from textile fibers, and improving dye absorption and overall textile quality. Lignocellulolytic enzymes offer sustainable solutions for biomass usage, waste management, and the generation of renewable energy and bioproducts [3].
Jute and banana fibers are crucial cash crops in South Asian countries, with lignocellulosic biomasses (LCBs) being the primary sources of these fibers. However, some plants are wasted due to their low-quality fiber content. Various physical, chemical, or physicochemical treatments cannot provide desirable modifications, but biological processes can transform these low-grade fibers into valuable fibers. Lignocellulolytic enzymes, such as cellulases and hemicellulases, can breakdown the cellulose and hemicellulose parts of jute and banana fibers, making them softer and more flexible [4-6]. This process enhances their flexibility and facilitates their use in various applications. Enzymatic treatment can also modify the surface characteristics of fibers by eliminating pollutants, increasing surface roughness, and improving adherence to matrix materials. This leads to enhanced adhesion and harmonization between different materials in composite structures. Enzymatic treatment can decrease the energy needed for mechanical processing, such as carding, spinning, and weaving, and enhance dyeability by expanding the fiber structure and increasing exposure to additional dye-binding sites. Enzymatic alteration of jute and banana fibers can create high-value products such as textiles, nonwoven fabrics, paper goods, environmentally friendly packaging materials, and composite materials. Enzymes can also introduce functional groups onto fiber surfaces, resulting in antibacterial activity, flame retardancy, and water repellency. Enzymatic modification is an environmentally friendly and sustainable method that limits the use of harsh chemicals and reduces environmental contamination. Enzymatically altered jute and banana fibers are eco-friendly substitutes for synthetic materials. Optimizing enzymatic treatment methods can lower processing costs and increase economic viability in various sectors [7-9].
The bacterial system is the most acceptable among microbial systems because of its fast growth under different broad-range environmental conditions. The Sundarban mangrove isolate Stenotrophomonas maltophilia GD1 showed extensive laccase activity and little cellulase and pectinase activity during its screening in this research study. Stenotrophomonas maltophilia GD1 is a gram-negative bacterial system that is highly adaptable and is found in various environmental habitats, including soil, water, plants, and hospitals. It thrives at high or low nutrient levels and under stressors such as heavy metals, antibiotics, and organic pollutants. This microorganism is known for its adaptable metabolism and ability to break down complex organic substances, making it crucial for bioremediation and environmental remediation. It has potential applications in biotechnological procedures such as bioremediation, biodegradation, and bioproduction. It has also been researched for its ability to break down pollutants and contaminants in soil and water, such as aromatic hydrocarbons, insecticides, industrial colors, and industrial effluents, making it valuable for remediating polluted areas. Certain strains of S. maltophilia possess enzymes of biotechnological significance, such as laccases, proteases, lipases, cellulases, and pectinases, which are used in industries such as textiles, food, pharmaceuticals, and biofuel production. Overall, S. maltophilia has a wide range of applications in various industries. Characterization studies have shown that laccases from S. maltophilia possess the characteristic traits of both fungal and bacterial laccases, including a blue copper center and conserved amino acid residues. These laccases can oxidize phenolic and nonphenolic substances, including phenolic acids and phenolics from lignin, aromatic amines, and dyes. They have demonstrated potential for removing color from textile dyes and bleaching denim, making them environmentally safe substitutes for chemical bleaching agents. However, there is limited research on the application of fiber modification to S. maltophilia, particularly for jute and banana fibers. Studies on bacterial alterations in natural fibers have primarily focused on cellulose-rich materials such as cotton, hemp, and flax. Although specific strains of Bacillus and Pseudomonas have been extensively studied for their ability to modify cellulose fibers, studies on the potential of S. maltophilia are limited.
Microbial modification of high-root-content jute waste fiber and pseudostem waste fiber of the banana plant has not been explored exhaustively. Hence, much research is needed to amplify the grading of these low-quality fibers for their valorization. The treatment of these fibers with the high-potential laccase producer S. maltophilia GD1 is an innovative and novel approach that significantly enhances the quality of the fiber in this research study, as validated by different procedures, such as FTIR, SEM, Young’s modulus, statistical analysis, and the like, and S. maltophilia GD1-based jute and banana waste fiber modification is the first reported method among all the literature to date.
2. MATERIALS AND METHODS
2.1. Sample Collection
Liquid and soil samples were collected from the Dobanki region of the Sundarban mangrove forest in West Bengal, India (latitude, DMS: 21°59′19.99′′N; longitude, DMS: 88°45′18.72′′E). Within 24 h of collection, the samples were delivered to the laboratory in sterile sample bottles and zipper packets. After that, the samples were kept at 4°C for additional analysis.
2.2. Chemicals
Carboxymethyl cellulose, sodium nitrate, dipotassium phosphate, magnesium sulfate, ferrous sulfate, yeast extract, agar, and potassium chloride were used in this study. Various colors, such as methylene blue (MB), malachite green (MG), Congo red (CR), and peptone, glucose, malt extract, ABTS (2,2′-azino-bis [3-ethylbenzothiazoline-6-sulfonic acid]), galacturonic acid, citrus pectin, (NH4)2SO4, K2HPO4, KH2PO4, DNS (3,5 dinitro salicylic acid), commercially available pure pectinase, cellulase, and laccase were purchased from Sigma-Aldrich, Loba Chemie, Himedia, Thermo Fisher Scientific India, etc.
Luria–Bertani (LB) agar consisted of tryptone (1%), yeast extract (0.5%), NaCl (1%), and agar (1.5%).
Laccase media consisted of glucose (2%), malt extract (3%), peptone (0.1%), and agar (1%).
Cellulase media consisted of carboxymethyl cellulose (0.2%), sodium nitrate (0.1%), dipotassium phosphate (0.1%), potassium chloride (0.1%), magnesium sulfate (0.05%), ferrous sulfate (0.001%), yeast extract (0.5%), and agar (1.5%).
Pectinase media consisted of wheat bran (2%), (NH4)2SO4 (1%), K2HPO4 (0.1%), KH2PO4 (0.1%), glucose (1%), yeast extract or peptone or urea (1%), and agar (1.5%).
2.3. Instruments
UV-Vis spectrophotometer (SHIMADZU, Model No: UV-1800), light microscope (400×, 1000× magnification in Magnus MLXi Plus, Magcam DC3), cold centrifuge (10,000 rpm) CM-8 PLUS, REMI, water bath (Sarada Labs, India), incubator (Sarada Labs, India), vacuum freeze-drying apparatus (EYELA, FDU-1200), PerkinElmer Frontier FTIR spectrophotometer, Denovix DS-11 spectrophotometer, ZEISS scanning electron microscope, Traveling Microscope-Model-TVM-02-SES Instruments Pvt. Ltd., and laminar flow (HEPA filter, Sarada Labs, India).
2.4. Isolation of Lignocellulolytic Enzyme-Producing Bacteria
Serial dilutions of soil and water samples were used to isolate lignocellulolytic bacteria. The sample underwent a series of dilutions (10−1–10−9). The spread plate method was used to plate 0.1 mL of the 10−3 dilution with a dilution factor of 103 onto nutrient agar plates and incubating them for 24 h at 37°C to count the number of bacterial colony present. The colonies on the plates were manually counted. Then, using the streak plate method, a single colony was plated on several selective media, such as laccase, cellulase, and pectinase media, as described by Pundi et al., Atri et al., and Tekere et al. [11-13] and incubated at 37°C for 24 h. The media compositions are provided in Section 2.2. This process was performed primarily to screen for microorganisms able to digest lignocellulosic components such as lignin, cellulose, and pectin. Most of the screened bacterial isolates were stored at 4°C on selective media. Due to high growth in the laccase media plates and negligible growth in the cellulase and pectinase media plates, we continued our research based on laccase production with this natural isolate.
2.5. Qualitative Analysis and Screening of Laccase-Producing Mangrove Isolates
The screening for lignin breakdown by the natural isolate was determined using a plate assay. LB agar is a widely employed growth medium in microbiology, specifically for the propagation of bacteria. The formulation of LB agar typically comprises tryptone, a product derived from the digestion of casein in the pancreas. It serves as a source of amino acids and peptides. Yeast extract is a liquid extract derived from yeast cells undergoing autolysis. It is abundant in vitamins, minerals, and amino acids. Sodium chloride (NaCl) supplies vital ions necessary for bacterial development and maintenance of osmotic equilibrium. Agar is used to make the medium semisolid, which helps in the proliferation of bacteria. The precise ratios may differ based on the particular formulation or producer. However, a standard recipe for LB agar could include 1% tryptone, 0.5% yeast extract, 1% sodium chloride, and 1.5–2.0% agar. Once the components are combined with water, the resulting mixture is subjected to autoclaving for sterilization. In the present study, the sterilized liquid was mixed with 0.1% ABTS and placed into petri dishes, where it solidified. Then, the microbial culture was streaked over the agar surface, and the plates were incubated at 30°C–37°C for 24 h [14]. The production of green coloration on the agar surface indicates the oxidation of ABTS by the enzyme laccase. Laccase-positive microbial colonies were preserved as glycerol stocks at –80°C.
2.6. Quantitative Analysis of Dye Degradation and Validation by FTIR Spectroscopy
A pure culture of the laccase-producing microbial system was used to estimate dye degradation quantitatively. A standard culture medium, laccase-selective media broth mixed with a specific dye (CR/MB/MG), was used for the experiment. CR (1%), MG, and MB were prepared. A total of 20 mL of sterile media were placed in a 100 mL conical tube. Next, 200 μL of dye and 200 μL of a 24-h culture mixture (GD1) were mixed. After the culture was added, a 2 mL sample of the mixture was collected and centrifuged at a speed of 10,000 revolutions per minute for a duration of 8 min. The optical density of the supernatant was then determined at wavelengths of 497 nm, 617 nm, and 668 nm for the CR, MG, and MB dyes, respectively. The identical method was repeated at intervals of 2 h for up to 6 h. The media was used as the blank. The percentage of dye degradation was determined using the following standard formula:
Decolorization (%) = (zero-hour absorbance—final hour absorbance)/zero-hour absorbance × 100. The change in the UV-Vis spectral shift has been considered for standardizing dye degradation [15].
Research and observation of the chemical bond changes and degradation of CR, MB, and MG dyes were conducted using Fourier transform infrared (FTIR) spectroscopy. After 6 h of incubation in nutrient broth, the samples were removed and centrifuged at 12,000 rpm for 10 min to precipitate all the bacterial cells. Then, using a vacuum freeze-drying apparatus (EYELA, FDU-1200), some of the supernatant was dried to a uniform weight for FTIR analysis. Samples of lyophilized dried powder were used for FTIR analysis. The FTIR spectra in the 4000–400 cm−1 region were measured using a PerkinElmer Frontier FTIR spectrometer [15].
2.7. Laccase Assay for Substrate Specificity and Kinetic Study
ABTS oxidation was used to evaluate laccase enzyme activity. The enzyme laccase oxidizes a nonphenolic dye called ABTS to a more stable cation radical. The blue-green color, which is closely related to enzyme activity and can be measured with a spectrophotometer at 420 nm (420 = 3.6 × 10−4 M−1 cm−1), can be attributed to the cation radicals produced following oxidation. The assay mixture included 0.1 mL of crude enzyme incubated for 10 min at 40°C with 0.2 mL of 0.5 mM ABTS and 1.7 mL of 0.1 M sodium acetate buffer (pH 4.5). All the reaction mixtures were reciprocally agitated at 120 rpm. The quantity of laccase that oxidized 1 mol of ABTS per minute was considered one unit of the enzyme.
The relative oxidation rates for ABTS were calculated from the enzyme activity (at their optimal pH). To explore the kinetic characteristics of the crude laccase, the standard substrate ABTS was generated at concentrations ranging from 50 to 500 M and used for routine enzyme assays. A Lineweaver-Burk plot was used to determine the Michaelis–Menten constant (km) and maximum velocity (Vmax) [16].
Laccase enzyme activity (U/L) = (ΔE × Vt) × 106/(t × ε × d × Vs)
where ΔE = Change in the extinction of light [min−1] at 420 nm,
ε = Molar absorption coefficient of ABTS [M−1 cm−1],
d = Thickness [cm] of the cuvette,
Vt = Total volume measured,
Vs = Volume of the enzyme added,
t = Reaction time (min).
2.8. Jute and Banana Waste Fiber Treatment and Validation by DNS Assay
This assay aims to use a novel strain to treat the waste portion of jute and banana fibers and calculate the chemical degradation of biopolymer components by taking into account the amount of reducing sugars released at various time points, namely, 0, 7, 14, and 21 days [17]. First, 100 mL of nutrient broth was autoclaved and divided into 50 mL Falcon tubes (two Falcon tubes for banana and two Falcon tubes for jute), each containing 10 mL of broth. Then, 0.1 g of banana and jute fiber waste was added to each Falcon tube. After the addition of fiber, 100 µL of laccase-producing culture was added. A total of 21 days of treatment were given at 37°C incubation with a reciprocal agitation of 120 rpm for both jute and banana fiber waste. Microbial biomass on days 0, 7, 14, and 21 was determined by measuring the dry weight of the biomass. To measure the dry weight, 2 mL of the treated sample was collected in an Eppendorf tube and centrifuged at 8000 rpm for 10 min to sediment the microbial biomass. After centrifugation, the supernatant was discarded, and the biomass was left at the incubation temperature to dry. After drying, a vacant Eppendorf tube was weighed, and the dry biomass was weighed with an Eppendorf tube. The value of the vacant Eppendorf tube was deducted from the value of the dry biomass containing the Eppendorf tube to obtain the weight of the dry biomass. The amount of reducing sugar released on different days (0, 7, 14, and 21) against the glucose standard curve was calculated using the DNS assay procedure outlined by Kriger et al. [10]. Using IBM SPSS 26 software, we performed a statistical analysis to compare the microbial biomass with the substantial amount of released reducing sugar against the number of days for their validation [18]. Positive (PC) and negative (NC) controls were used to standardize the experimental procedure. In PC, 100 µL of 0.1% commercial laccase was used to treat the waste fibers, and in NC, the whole setup was the same without any microbial culture or commercial enzyme.
2.9. Statistical Analysis
The statistical analysis was performed using IBM SPSS software (version 26.0). Using the correlation coefficient test, the day, biomass concentration, and released reducing sugars of pure bacterial cultures seeded in sterile laccase-selective medium were correlated at the 0.05 significance level (two-tailed) (Pearson correlation).
2.10. Scanning Electron Microscopic Analysis
A ZEISS scanning electron microscope was used to scan the surface of the treated jute and banana waste fibers at an accelerating voltage of 5.00 kV to determine the fiber modification by the ligninolytic microbial isolate.
2.11. FTIR Analysis of Treated Jute and Banana Waste Fibers
The modified chemical bonds of the treated high-root-content jute and banana waste fibers were observed using FTIR spectroscopy. After 21 days of incubation in nutrient broth, the samples were removed and centrifuged at 12,000 rpm for 10 min to precipitate all the bacterial cells. Then, using a vacuum freeze-drying apparatus (EYELA, FDU-1200), some of the supernatant was dried to a uniform weight for FTIR analysis. The lyophilized dried powder samples were used to form a pellet with KBr for the FTIR analysis. FTIR spectra in the 4000–400 cm−1 range were measured using a PerkinElmer Frontier FTIR spectrometer [16].
2.12. Pathogenicity Test
The biosafety of the microbial isolate was assessed using a blood agar test. Autoclaved blood agar was made using a high-media blood agar media protocol. Two wells were prepared on the agar plates. 100 µL of 20% hemolysin buffer (ammonium chloride potassium or ACK buffer) was added to a single well, and 100 µL of the overnight-grown test sample was added to a different well. In the broth media experiment, 10 mL of PBS, 5 mL of sheep blood, and other required substances were thoroughly mixed (pH 7.2). The supernatant was collected after 10 min of centrifugation at 2000 rpm at 4°C. For the erythrocyte suspension stock, 2 mL of PBS (pH 7.2) was combined homogeneously with the pellet after centrifugation. The positive control was then made by combining 0.5 mL of erythrocyte suspension with 1.5 mL of hemolysis buffer (ACK buffer). Using 1.5 mL of overnight culture solution, the same amount of microbial crude extract (sonicated in PBS buffer) was mixed with 0.5 mL of erythrocyte suspension separately. The testing vials were prepared and kept at 30°C for 2 h. Following incubation, the samples were centrifuged at 8000 rpm for 8 min. The supernatant was then meticulously and aseptically removed from each vial, and the absorbance was calculated at 412 nm [19]. Here, the criterion for assessing pathogenicity was the hemolysis capability of the isolated strain, as hemolysis refers to the red blood cell disintegration process. The capacity of bacterial colonies to cause hemolysis when cultured on blood agar is employed to categorize specific microorganisms as pathogenic.
2.13. Molecular Characterization and Phylogenetic Study of a Laccase-Producing Mangrove Bacterial Isolate
A bacterial DNA isolation kit was used to process the laccase-producing microbial isolate for genomic DNA isolation, and the purity and concentration were analyzed using a Denovix DS-11 spectrophotometer. The 16S rDNA region of the isolated DNA was amplified using a Bacterial 16S rDNA PCR Kit Fast (800), Catalog number-# RR310A. The kit details include Lot ID-#AL41019N and an expiration date of April 2023. The amplification of an 800 bp amplicon was obtained using 16S rDNA Primer Mix and TaKaRa Taq™ HS Fast Detect Premix in the sample provided and in the PC used (Escherichia coli). No amplicon was obtained for the non-template control (NTC). The 800 bp test amplicon was purified via column purification, and bidirectional cycle sequencing was carried out with forward primers and reverse primers using a BDT V3.1 cycle sequencing kit on an ABI 3730 Genetic Analyzer according to the sequencing protocol.
Laccase enzyme FASTA sequences for typical laccase-producing bacteria and a fungal laccase source were obtained from the NCBI database [https://www.ncbi.nlm.nih.gov/]. Multiple sequence alignments of the laccase enzyme sequence were performed with the muscle algorithm, and MEGA 11 was used to conduct phylogenetic analysis. The neighbor-joining method was used to construct the tree. A specific outgroup with a fungal origin was chosen for the laccase enzyme. Using the outgroup as a guide, the root was located. The bootstrap method was used to examine the consistency of the phylogenetic tree [20,21].
2.14. Determination of the Mechanical Properties of the Fibers Using Traveling Microscopy
Vernier microscopes or traveling microscopes are portable microscopes with brass steel scales running horizontally and vertically. It is employed to measure small lengths. A metal carriage glides over the top of the properly machined cast iron foundation, which is equipped with leveling screws. A clamping screw allows the microscope slide to be clamped in any position. The horizontal carriage is a similar-functioning vertical slide. This vertical carriage has a rack and pinion microscope, which may be clamped in either a vertical or horizontal position. A perspex plate is attached to the stage where the measurement objects are set up. There are achromatic lenses used. The slides were rotated 160 mm vertically and 210 mm horizontally.
Young’s modulus (E) is a material feature that indicates how easily a material can stretch and flex. The Young’s modulus of jute and banana fiber was determined by the stress and strain ratio obtained using the maximum weight carried by the biologically treated fibers of a fixed diameter and length. The stress was determined by calculating the ratio of force to area by hanging the maximum weight on the fiber using a holder. On the other hand, the strain was calculated using the ratio of the change in length to the length. After stress and strain were measured, Young’s modulus of both fibers was determined to ascertain their increased stretchability. All the measurements were performed using a traveling microscope [22,23].
3. RESULTS AND DISCUSSION
3.1. Qualitative Screening of Ligninolytic Mangrove Isolates
An encouraging sign of laccase was the development of a solid green color on the ABTS-LB agar plates. This justified the current study’s objective of assessing the potential ligninolytic microbial system isolated from the Sundarban mangrove area. This ability will also satisfy the modification concept of high-root-content jute and banana waste lignocellulose fiber as a target that can significantly improve fiber quality by degrading the phenolic polymer lignin and enhancing the economic value of the modified waste fiber in the overseas marketplace [24] [Figure 1A]. Reducing the organic load of different industrial effluents used for irrigation can also play an essential role in bioremediation, resulting in sustainable, toxin-free agricultural products.
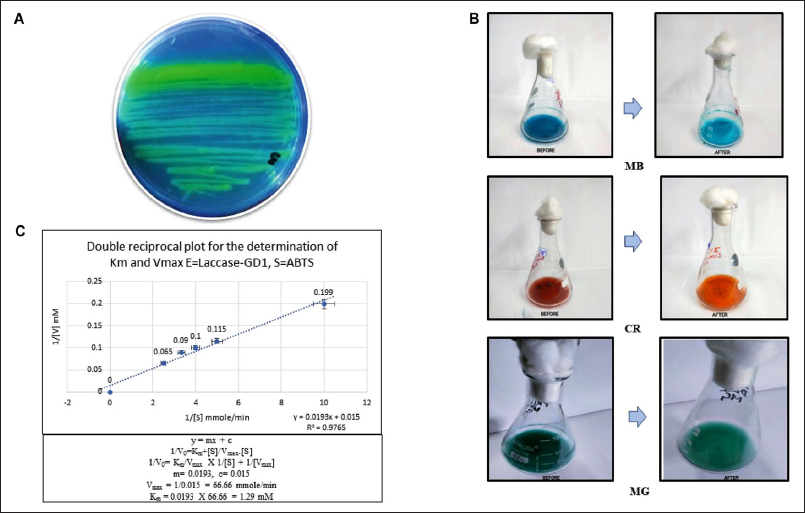 | Figure 1: (A) Qualitative analysis of the laccase-producing mangrove isolate Stenotrophomonas maltophilia GD1: oxidation of ABTS and production of green color in LB-Agar-ABTS plates. (B) Quantitative analysis by dye decolorization. (C) Lineweaver–Burk or double reciprocal plot for the determination of the enzyme kinetics of laccase produced by S. maltophilia GD1.
[Click here to view] |
3.2. Quantitative Analysis by Dye Decolorization
Three distinct dyes—CR, MG, and MB—were used to quantitatively evaluate the dye decolorization potential of isolate GD1. The enzyme laccase degraded these colors in suitable selective media. The dye degradation rate was monitored at specific wavelengths every 2, 4, and 6 h (CR–497 nm, MG –617 nm, and MB–668 nm). The decolorizing percentages of three distinct dyes used by the isolate are represented in Table 1, which provides information on the decolorizing percentages of the dyes used by the isolate. The greatest degradation was observed for MG, followed by CR and MB. Here, laccase oxidizes dye molecules in the presence of oxygen. The enzyme adheres to the surface of the dye molecule and binds to specific chemical groups. This process produces reactive oxygen species (ROS), damaging the chromophore groups of dye molecules. These ROS fragment the dye into smaller, less pigmented molecules that are more soluble in water and less environmentally damaging [25] [Figure 1B].
Table 1: Dye degradation by the natural mangrove isolate GD1.
| Strain Name | Dye | ?max | Time | % Degrade |
|---|
GD1
Isolated from Dobanki, Sundarban, West Bengal, India | CR | 497 | 6 h | 59.15 |
| MG | 617 | 6 h | 60.2 |
| MB | 668 | 6 h | 31.3 |
3.3. Laccase Enzyme Activity and Enzyme Kinetic Study
According to the laccase enzyme assay results, the GD1 strain has more significant laccase activity. This finding suggests that this mangrove isolate has tightly regulated genes responsible for this action, which explains its high expression level (9.72 U/L) and potential for breaking the lignin part of the fiber and destroying the chemical structure of the dyes, leading to a reduction in the organic load.
The Lineweaver-Burke plot was used to determine the enzyme kinetic action of laccase, that is, km and Vmax, for oxidizing ABTS. Laccase produced by the test organism has a high catalytic efficiency for ABTS, with a Vmax of 1.29 mM and 66.66 mmol/min, respectively. The nature of the substrate and phenolic ring substitution significantly affect the ability of laccases to oxidize compounds and their affinity for those chemicals [Figure 1C].
These results on the kinetic parameters of the isolated laccase enzyme are significant in industry. Enzyme kinetic studies help researchers understand how laccases facilitate reactions. Laccase activity can also serve as a biomarker for environmental surveillance, indicating contaminants or conditional changes. Establishing connections between laccase activity and environmental factors aids in environmental health evaluation.
3.4. FTIR Analysis of GD1-Treated Dye, Jute, and Banana Waste Fibers
The CR treated with GD1 displayed spectral peaks for the stretching vibrations of C-N between 1100 and 1200 cm−1 and the asymmetric vibrations of the CH3 group between 2900 and 3000 cm−1. The peak between 2300 and 2400 cm−1 appeared due to the symmetric and asymmetric stretching of the tertiary amine salt. The C-N stretch peak of an aromatic tertiary amine was observed at 1300–1400 cm−1. Following biodegradation by GD1, aromatic ring degradation in the 1600–1700 cm−1 range was observed. This peak results from the stretching of C=C [26] [Figure 2A].
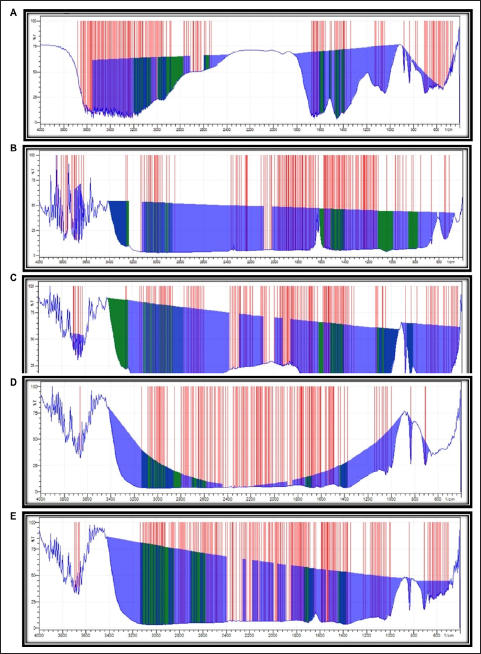 | Figure 2: Fourier transform infrared spectroscopy (FTIR) spectral analysis of the treated dye and waste fiber: (A) Congo red, (B) methylene blue, (C) malachite green, (D) jute waste fiber, and (E) banana waste fiber.
[Click here to view] |
The FTIR spectrum of untreated MB exhibited distinct peaks at 3400 and 3500 cm−1, 1400 and 1500 cm−1, and 1100 and 1200 cm−1, which were attributed to NH, N=NO, and CS stretching, respectively. The CH2 group was present, as evidenced by the peak between 600 and 700 cm−1. After treatment with GD1, a CS stretching peak was observed at 1100–1200 cm−1, NH stretching at 3500–3600 cm−1, and OH stretching at 3399 cm–1, and C-N and OH bending peaks were observed between 1400 and 1500 cm−1 and between 600 and 700 cm−1, respectively. The peak at approximately 3000 cm−1 was an indication of N=N=N antisymmetric stretching. The findings revealed that numerous new peaks emerged, while some completely vanished. This might be due to the laccase activity of the GD1 bacterial isolate, which modifies and degrades the molecules of the MB dye [27] [Figure 2B].
The spectrum of MG dye treated with GD1 showed mono- and para-disubstituted benzene ring peaks between 1500 and 500 cm−1, supporting the C=C stretching characteristic of the benzene ring. The spectra also showed peaks for the C-N stretching vibrations between 1100 and 1200 cm−1. Two bands between 3200 and 3500 cm−1 in the FTIR spectra of the dyes were attributed to the N-H stretching vibrations of primary amines. These findings demonstrate the breakdown of MG dye by GD1 bacteria [28-31] [Figure 2C].
High-root-content jute and banana waste fibers treated with GD1 showed significant functional group removal from the fibers listed in the supplementary files. The results confirmed that lignin removal decreases fiber quality and increases the economic value of fibers. In both cases, C-H and CH2/CH3 bending vibrations and C=O stretching vibrations were observed between 1380 and 1450 cm−1 and between 1680 and 1730 cm−1, respectively. Another stretching peak attributed to C-H was detected in the wavenumber range between 2855 and 3120 cm−1 [Figures 2D and 2E].
Details of the functional groups involved in dye degradation and fiber modification can be found in the supplementary data file.
3.5. SEM Analysis
Scanning electron microscopy (SEM) was used to examine the surface morphologies of the high-root-content jute fibers and banana waste fibers before and after treatment with laccase. The control surface had a smooth texture. The distorted lookup of both waste fibers after laccase treatment is probably a result of the enzymatic removal, relocation, and loosening of the bulky lignin on the surface. The laccase enzyme facilitated the oxidation of the phenolic hydroxyl groups present in the lignin on the surface of both fibers. This process results in the formation of phenoxyl radicals, which subsequently bond together to form ether structures. Nevertheless, the involvement of lignin in laccase-mediated interactions enhanced the hydrophobicity of the fiber surface and improved its tensile properties. Furthermore, the laccase-treated fiber membranes exhibited more combinations of short fibers derived from jute and banana fibers, which suggested an enhancement in the mechanical properties [29-35] [Figures 3A and 3B].
 | Figure 3: Scanning electron microscopy analysis: (A) jute waste fiber, (B) banana waste fiber—(a) 300×, (b) 1000×, and (c, d) 5000× magnification.
[Click here to view] |
3.6. Determination of the Release of Reducing Sugars from Jute and Banana Fiber Waste via DNS Assay
According to the observation, a constant stable phase and a modest drop in sugar level were noted on day 21 in the case of the jute fiber treatment. However, the rate of sugar removal from the banana fiber increased along with the microbial biomass until day 21. These observations indicate the utilization of the nutrient broth by the laccase-producing microbial strain for the first 24 h, as proven in our previous study by analyzing the growth curve in nutrient broth [15]. After the exhaustion of the nutrient broth, the microbial system starts breaking the lignin portion for utilization of the sugar, depending upon which they survive, and ends up retting the waste fibers, which ultimately modifies the fiber quality [Figures 4A and 4C].
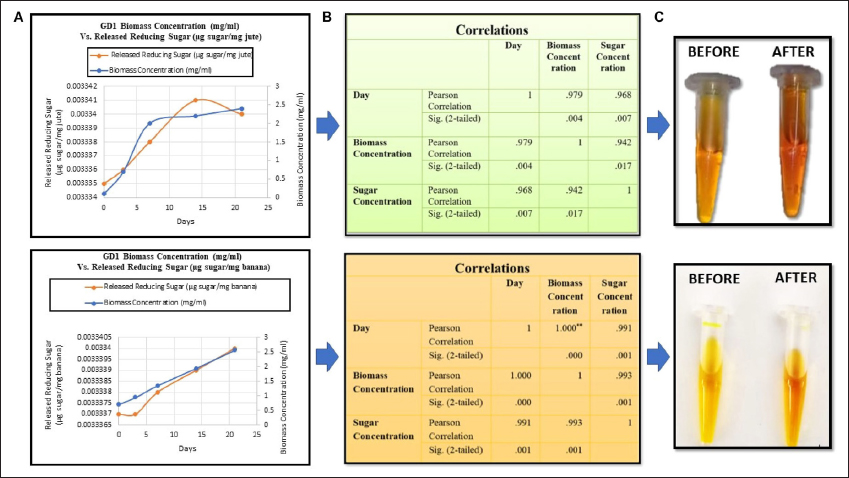 | Figure 4: (A) Comparison between the amount of released reducing sugars and the biomass of the strain, (B) statistical analysis of the day, biomass, and amount of released reducing sugars, and (C) DNS assay confirming the release of reducing sugars.
[Click here to view] |
3.7. Statistical Analysis
The correlation coefficient test, carried out by IBM SPSS 26 statistical software at the 0.05 significance level, confirmed a significant relationship between the day, biomass concentration of the laccase-producing strain GD1, and the released reducing sugars in particular, demonstrating that there was a significant relationship among all three parameters [Figure 4B]. This result indicates that there is a direct proportional relationship among all three parameters; that is, the microbial biomass is directly proportional to the amount of reducing sugar released during the 21-day period. After 21 days of treatment, the biomass and the amount of released reducing sugars declined rapidly and proportionately, which indicated the end of the retting process. Again, the t-value and p-value statistically correlate with all the parameters supporting the alternative hypothesis and reject the null hypothesis.
3.8. Pathogenicity Test
The GD1 isolate developed significantly fewer halo zones on the blood agar media than did the positive control. Beta-hemolysis was expected to act as a positive control (clear zones). The activity of the bacterial hemolysin in this test was lower than that of the control hemolysin in the ACK lysis buffer. The ACK lysis buffer absorbance in the liquid medium was given a value of 100% hemolysis. The pure culture and crude extract had 8.66% and 3.14% hemolysis activity, respectively. In the broth medium experiment, the hemolysis activity of the test organism was minimal. This suggests that the bacteria may not be very hazardous to the environment after being implanted. This work has allowed for the expansion of this isolate, which could be valuable for commercialization in the jute and banana fiber processing industries all over fiber-producing countries for reducing waste fiber loss and can sustainably increase the economic growth of fiber industries as well as the textile industry and dye industry for reducing organic loads and recycling wastewater dye industry effluents for its utilization in water-scarce agricultural fields [Figures 5A and 5B].
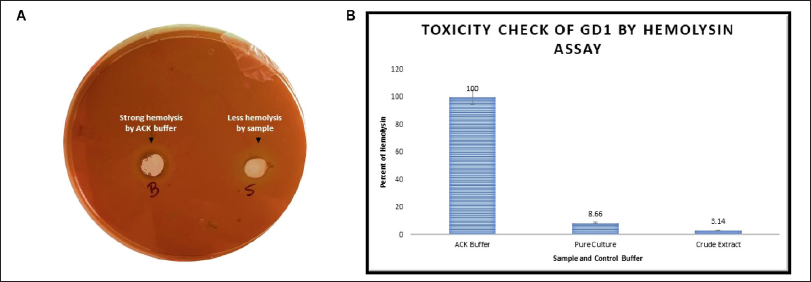 | Figure 5: Hemolysis assay. (A) Blood agar media containing ACK buffer in a well (B) showing high hemolysis and an overnight sample in a well (S) showing less hemolysis. (B) Hemolysis in liquid blood media showing the quantitative hemolysis data obtained with ACK buffer, pure culture, and crude extract.
[Click here to view] |
3.9. Molecular Characterization and Analysis of the Phylogenetic Study
The sequences obtained by sequencing with forward and reverse primers were assembled using Gene-Tool software to generate a consensus sequence of the 16S rRNA gene. BLAST analysis was carried out to compare the sequences with those in the NCBI GenBank database. The first 10 sequences in the database that showed the highest similarity were selected based on the maximum identity score and phylogenetic tree results. The GD1 isolate was more similar (100%) to S. maltophilia. The resulting GD1 isolate was referred to as S. maltophilia GD1. Furthermore, the 16S rRNA nucleotide sequences of S. maltophilia GD1 were used to generate a phylogenetic tree by comparison with other necessary laccase-producing bacterial sequences in the databases via multiple sequence alignment. A total of nine laccase sequences from the bacterial species and one laccase sequence from the fungal origin as an outgroup were chosen for the experiment based on their laccase activity, ABTS oxidation capacity, and capacity to release reducing sugars. A total of 1000 bootstrapping operations were performed using the MEGA 11 program, following numerous sequence alignments using the MUSCLE algorithm. The neighbor-joining (NJ) process was used to construct the phylogenetic tree. Multiple sequence analyses revealed different locations for conserved amino acids, suggesting that these residues must have had a significant role in the evolution of the bacterial laccase sequences. Based on the knowledge of sequence alignment and the phylogenetic tree, PCR (polymerase chain reaction) primers may be utilized for laccase gene isolation. The bootstrap method, a statistical analysis approach that resamples the data and evaluates the strength of the tree’s nodes, was used to examine the consistency of the phylogenetic tree. The highest bootstrap value is 100%, and up to 90% support is vital. The reliable groupings correspond to 50% and 70%, respectively [Figures 6A–E].
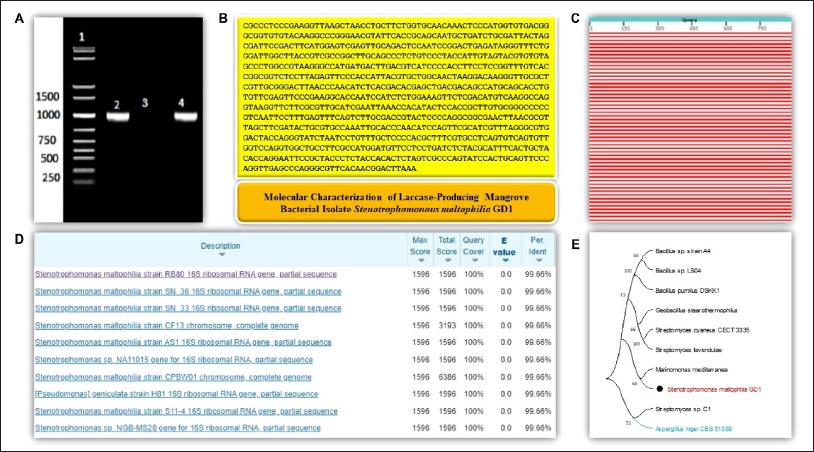 | Figure 6: Molecular characterization of the Sundarban mangrove isolate Stenotrophomonas maltophilia GD1. (A) A single 1-kb PCR product of the 16S rRNA gene on a 2% agarose gel. Lane 1: 1 KB DNA ladder, Lane 2: 16S amplicon (positive control), Lane 3: negative control, Lane 4: sample. (B) Consensus sequence of S. maltophilia GD1. (C) Graphical summary of BLAST analysis. (D) Sequences producing significant alignments in the NCBI database: Max Score: the highest alignment score from that database sequence; Total Score: the total alignment scores from all alignment segments; Query Coverage: the percentage of query covered by alignment to the database sequence; E-Value: the best (lowest) expect value (E-value) of all alignments from that database sequence; Identity: the highest percent identity of all query–subject alignments. (E) Phylogenetic tree analysis: The evolutionary history was inferred using the neighbor-joining method. A bootstrap consensus tree inferred from 1000 replicates was constructed to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% of the bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The evolutionary distances were computed using the maximum composite likelihood method and are in units of the number of base substitutions per site. This analysis involved 10 nucleotide sequences. The codon positions included were 1st+2nd+3rd+noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There were a total of 2856 positions in the final dataset. Evolutionary analyses were conducted in MEGA11. Laccase-producing strains from the NCBI database that were compared with the isolated strain S. maltophilia GD1 in the phylogenetic tree are as follows: >KX674736.1 Bacillus sp. strain A4, >GU972589.1 Bacillus sp. LS04, >KC010297.2 Bacillus pumilus strain DSKK1, >AF184209.1 Marinomonas mediterranea, >HQ857207.1 Streptomyces cyaneus strain CECT 3335, >MW344649.1 Geobacillus stearothermophilus, >AB092576.1 Streptomyces lavendulae, >JF821185.1 Streptomyces sp. C1, and >XM_001389488.2 Aspergillus niger CBS 513.88.
[Click here to view] |
3.10. Determination of the Young’s Modulus of the Fibers Using a Traveling Microscope
Microbiologically treated fibers showed excellent results. A traveling microscope revealed an improvement in the stretchability, which was determined using Young’s modulus; the results are shown in Table 2. The test jute fiber showed a 38.58% increase in elasticity, whereas the banana test fiber showed an increase of 38.21%. This positive enhancement in mechanical properties will increase the economic value of waste fibers and open the door for new commercial products made from these raw materials with sustainability and zero toxicity.
Table 2: Determination of the Young’s modulus of natural fibers using a traveling microscope.
| Biologically Treated Natural Fiber | Microbial System Used for the Retting | Treated Fiber Diameter (mm) | Treated Fiber Length (mm) | Young’s Modulus (MPa) |
|---|
| Jute (control) | GD1 | 0.002 | 90 | 6.02 |
| Jute (test fiber) | 0.002 | 90 | 9.802 |
| Banana (control) | 0.01 | 90 | 1.135 |
| Banana (test fiber) | 0.01 | 90 | 1.837 |
REFERENCES
1. Ghosh D, Das S. Genetic and metabolic engineering approaches for improving accessibilities of lignocellulosic biomass toward biofuels generations. In: Kuila A, Sharma V, editors. Genetic and metabolic engineering for improved biofuel production from lignocellulosic biomass. Elsevier; 2020. p. 13–35. [CrossRef]
2. Pradeep K. Microbial fermentation and enzyme technology. In: Mohapatra S, Mondal KC, editors. Boca Raton (FL): CRC Press; 2020. [CrossRef]
3. Hossain MM, Siddiquee S, Kumar V. Critical factors for optimum biodegradation of bast fiber’s gums in bacterial retting. Fibers (Basel) [Internet]. 2021;9(8):52. [CrossRef]
4. Ghosh D, Das S. Engineering of microbial cellulases for value-added product generations. In: Tuli KD, Kuila A, editors. Current status and future scope of microbial cellulases. Elsevier; 2021. p. 171–87. [CrossRef]
5. Das S, Ghosh D. Potential of lignocellulolytic biocatalysts of native and proposed genetically engineered microbial cell factories on jute fiber modification and jute waste recycling: a review. J Exp Biol Agric Sci [Internet]. 2022;10(5):932–52. [CrossRef]
6. Sfiligoj M, Hribernik S, Stana K, Kree T. Plant fibers for textile and technical applications. In: Grundas S, Stepniewski A, editors. Advances in agrophysical research. InTech; 2013. [CrossRef]
7. Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev [Internet]. 2012;25(1): 2–41. [CrossRef]
8. Khan A, Iftikhar K, Mohsin M, Ubaidullah M, Ali M, Mueen A. Banana agro-waste as an alternative to cotton fiber in textile applications. Yarn to fabric: an ecofriendly approach. Ind Crops Prod [Internet]. 2022;189(115687):115687. [CrossRef]
9. Priyadarshana RWIB, Kaliyadasa PE, Ranawana SRWMCJK, Senarathna KGC. Biowaste management: banana fiber utilization for product development. J Nat Fibers [Internet]. 2022;19(4):1461–71. [CrossRef]
10. Abdelgalil SA, Attia AM, Reyed RM, Soliman NA. Response surface methodology for optimization laccase production by Alcaligenes faecalis NYSO using Agro-industrial wastes as cosubstrate. J Bioremediat Biodegrad [Internet]. 2019;10(2):1–10.
11. Pundir RK, Mishra VK, Rana S, Lakhani M. Screening of laccase producing fungi from soil samples-an in vitro study. Electronic J Biol. 2016;12(3):254–7.
12. Atri N, Kumari B, Upadhyay RC, Sharma SK. Nutritional and sociobiological aspects of culinary-medicinal Termitophilous mushrooms from North India. Int J Med Mushrooms [Internet]. 2012;14(5):471–9. [CrossRef]
13. Tekere M, Zvauya R, Read JS. Ligninolytic enzyme production in selected subtropical white rot fungi under different culture conditions. J Basic Microbiol [Internet]. 2001;41(2):115–29. [CrossRef]
14. Pinar O, Ct B. Heterologous expression and characterization of a high redox potential laccase from Coriolopsis polyzona MUCL 38443. Turkish J Biol. 2017;41(2):278–91. [CrossRef]
15. Das S, Sinha N, Sen M, Ghosh D. Isolation and screening of dye degrading lignocellulolytic bacteria from Sundarban Mangrove Ecosystem, West Bengal, India. J Pure Appl Microbiol. 2023;17(1):609–26. [CrossRef]
16. Falade AO, Nwodo UU, Iweriebor BC, Green E, Mabinya LV, Okoh AI. Lignin peroxidase functionalities and prospective applications. Microbiologyopen [Internet]. 2017;6(1):e00394. [CrossRef]
17. Devulder G, de Montclos MP, Flandrois JP. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int J Syst Evol Microbiol [Internet]. 2005;55(1):293–302. [CrossRef]
18. Abdelgalil SA, Attia AM, Reyed RM, Soliman NA, Enshasy E. Application of experimental designs for optimization the production of Alcaligenes faecalis NYSO laccase. J Sci Ind Res. 2018;77(12):713–22.
19. Huang B, Liang Y, Pan H, Xie L, Jiang T, Jiang T. Hemolytic and cytotoxic activity from cultures of Aureococcus anophagefferens-a causative species of brown tides in the North-Western Bohai Sea, China. Chemosphere. 2020;247:125819. [CrossRef]
20. Satpathy R. Computational phylogenetic study and data mining approach to laccase enzyme sequences. J Phylogenetics Evol Biol [Internet]. 2013;01(02):108–114. [CrossRef]
21. Evans BC, Nelson CE, Yu SS, Beavers KR, Kim AJ, Li H, et al. Ex vivo red blood cell hemolysis assay for the evaluation of pH-responsive endosomolytic agents for cytosolic delivery of biomacromolecular drugs. J Vis Exp [Internet]. 2013;(73):e50166. [CrossRef]
22. Ferrante A, Santulli C, Summerscales J. Evaluation of tensile strength of fibers extracted from banana peels. J Nat Fibers. 2020;17(10):1519–31. [CrossRef]
23. Chanda D. An approach for measurement of young’s modulus of glass in the form of a slab. Indian J Sci Technol. 2017;10(18):1–9. [CrossRef]
24. Islam T, Repon M, Islam T, Sarwar Z, Rahman MM. Impact of textile dyes on health and ecosystem: a review of structure, causes, and potential solutions. Environ Sci Pollut Res. 2023;30:9207–42. [CrossRef]
25. Satheesh Babu S, Mohandass C, Vijayaraj AS, Dhale MA. Detoxification and color removal of Congo red by a novel Dietzia sp. (DTS26) - a microcosm approach. Ecotoxicol Environ Saf [Internet]. 2015;114:52–60. [CrossRef]
26. D’Souza E, Fulke AB, Mulani N, Ram A, Asodekar M, Narkhede N, et al. Decolorization of Congo red mediated by marine Alcaligenes species isolated from Indian West coast sediments. Environ Earth Sci [Internet]. 2017;76(20):721. [CrossRef]
27. Islam, MT, Islam T, Islam T, Repon MR. Synthetic dyes for textile colouration: process, factors and environmental impact. Textile Leather Rev. 2022;5:327–73. [CrossRef]
28. Repon MR, Dev B, Rahman MA, Jurkoniene S, Haji A, Alim MA, et al. Textile dyeing using natural mordants and dyes: a review. Environ Chem Lett. 2024;22:1473–520. [CrossRef]
29. Dong A, Fan X, Wang Q, Yu Y, Cavaco-Paulo A. Enzymatic treatments to improve mechanical properties and surface hydrophobicity of jute fiber membranes. Bioresources [Internet]. 2016;11(2):3289–302. [CrossRef]
30. Akter N, Akter N, Repon MR, Islam T, Al Mamun MA, Shukhratov S. Evaluation and optimization of pretreatment process for lyocell knitted fabric dyeing with reactive dyestuff. Cellulose. 2024;31:3205–19. [CrossRef]
31. Ghosh D, Talukdar P. Relevance of microbial enzymes in textile industries emphasizing metabolic engineering panorama. In: Thatoi H, Mohapatra PKD, Mohapatra S, Mondal KC, editors. Microbial fermentation and enzyme technology. 1st ed. CRC Press. 2020. p. 195–205.
32. Das S, Rudra S, Khatun I, Sinha N, Sen M, Ghosh D. Concise review on lignocellulolytic microbial consortia for lignocellulosic waste biomass utilization: a way forward? Microbiology. 2023;92:(3):301–17. [CrossRef]
33. Das S, Gole A, Chakraborty A, Mal S, Rudra S, Ghosh D. Lignocellulolytic microbial systems and its importance in dye decolourization: a review. J Pure Appl Microbiol. 2023;17(2):705–21. [CrossRef]
34. Repon MR, Islam MT, Mamun MAA. Ecological risk assessment and health safety speculation during color fastness properties enhancement of natural dyed cotton through metallic mordants. Fash Text. 2017;4:24. [CrossRef]
35. Repon, MR, Islam T, Sadia HT, Mikucioniene D, Hossain S, Kibria G, et al. Development of antimicrobial cotton fabric impregnating AgNPs utilizing contemporary practice. Coatings. 2021;11:1413. [CrossRef]





