1. INTRODUCTION
Azo dyes constitute around 60%–70% of dyes synthesized globally [1,2]. These are characterized by the presence of one or more azo groups, −N=N− [3]. These further consist of phenyl and naphthyl groups modified with various functional groups [3]. Such complex modifications in their structure makes them xenobiotic in nature and highly resistive to breakdown [4]. They are resistant to light, washing, chemical and microbial attack, easy to synthesis with low energy, and low-cost consumption [4–6]. They are extensively used in textile, paper, food, leather, cosmetics, and pharmaceutical industries [7]. Almost 10%–15% of dye is released in wastewater during its manufacture and application in various industries [8]. Contaminants generated by azo dyes constitute of dye particles and their breakdown products; primarily amines that are proven to be toxic and mutagenic [9,10]. They also degrade the water quality by increasing its biological oxygen demand and chemical oxygen demand [11,12]. Many reports indicate that textile dyes and effluents have toxic effect on the germination rates and biomass of several plant species [11,13]. Azo dyes like tartrazine and carmoisine affect the functioning of vital organs like kidney and liver in rats and also induce the formation of free radicals leading to oxidative stress [14].
Various physiochemical methods like membrane filtration, adsorption on activated carbon, flocculation, electro coagulation, ozonation, froth floatation, reverse osmosis, and ion exchange are currently implied for azo dye decolorization. However, these methods have limitations like high cost, high energy input, and sludge generation [15]. Recently microorganisms have been shown to decolorize azo dyes in a cost effective and environment friendly manner [16]. Bacteria, algae, fungi, and yeast have shown cost effective and eco-friendly degradation of textile dyes [11]. Furthermore, versatility in microbial structure makes their exploitation easy for decolorization of most of the dyes [17]. Dye decolorization by higher order organisms like fungi is a slow process due to the slow growth of the organism [18]. Bacteria on the other hand have emerged as a very promising category for dye decolorization [19]. They proliferate rapidly under both aerobic and anaerobic condition thus can achieve dye degradation at a faster rate [20]. They primarily decolorize dyes under anaerobic conditions [21]. Few bacteria have been reported to decolorize azo dyes in aerobic conditions as well [21]. It has also been shown that under a mixture of anaerobic and aerobic conditions, complete degradation of some dyes can be achieved [22]. Often textile waste waters have high metal content and salinity; hence, it is important to utilize microorganisms tolerant to extreme environment conditions. Bacterial species like Enterococcus faecalis and C. bufermentas have been identified as highly halotolerant azo dye degrading microorganisms [23].
This study was done to isolate and identify a bacterial strain capable of decolorizing three azo dyes: methyl orange (MO), congo red (CR), and Eriochrome Black T. MO is an acid class single azo dye and was found to be mutagenic in a Salmonella/microsome assay [24]. CR is a benzidine-based anionic diazo dye and has been shown to metabolize to benzidine, a known human carcinogen [25]. Eriochrome Black T (EBT) is a single azo naphthol derived azo dye and is known to be recalcitrant to oxidative biodegradation [26]. Various physiochemical parameters like temperature and pH were also optimized for efficient decolorization of dyes.
2. MATERIALS AND METHODS
2.1. Isolation, Screening, and Identification
Soil sample was collected in a sterile glass container from dumping site of textile industry located in Bada Bagh Industrial area, New Delhi. Samples were serially diluted 105 times and were enriched in Luria broth (LB) amended separately with 100 mg/l of MO, CR, and Eriochrome Black Tat 37°C for 48 hours. Subsequent transfers of 1 ml of the culture were carried out in fresh media amended with dye (100 mg/l) till decolorization of the media was observed. Decolorized samples were serially diluted 107 times and were spread on petri plates with 100 mg/l dye concentration. Plates were incubated at 37°C for 48 hours. Morphologically distinct colonies with zone of decolorization were screened for their decolorizing ability in media supplied with dye (100 mg/l). Out of all the colonies observed on plate, EBT-2 strain showed the best decolorization zone and hence was picked up for further experiments. EBT-2 strain was identified using 16s rDNA sequencing and similarity search was conducted against database through BLASTn for 16s rDNA. MEGA 4.0 software was used to conduct evolutionary analysis by Neighbour Joining method [27]. Biochemical characteristics of EBT-2 strain were also assessed according to Bergey’s Manual of Determinative Bacteriology [28].
 | Diagrammatic representation of Methodology [Click here to view] |
2.2. Effect of Initial Dye Concentration on Dye Decolorization
The effect of initial dye concentration on dye decolorizing potential of EBT-2 strain was measured by studying decolorization at different initial concentrations of MO, CR and Eriochrome Black T (10–500 mg/l). LB media was amended with dye concentrations ranging from 10 to 500 mg/l and inoculated with 1% EBT-2 isolate (OD 0.8–1). It was incubated at 37° for 16 hours under static condition. Samples were centrifuged at every 24-hour interval at 10,000 rpm for 10 minutes. Supernatant was collected and absorbance was measured at absorbance peak of each dye i.e., 480, 492, and 535 nm for MO, CR, and EBT respectively. Decolorization efficiency was measured for a period of 120 hours in terms of percentage decolorization [29].
% Decolorization = [(Initial absorbance – Observed absorbance)/ Initial absorbance] × 100
The experiment was done thrice, and data was represented as the mean ± Standard error of mean. Abiotic controls were always included.
2.3. Effect of pH on Dye Decolorization
Decolorization efficiency of EBT-2 strain was studied at different pH values. LB containing MO, CR and EBT at the concentration of 100 mg/l was inoculated with 1% inoculum and pH was set from 5 to 11 (with increment of 1 pH unit). pH was adjusted with 0.1 N HCL or 0.1 N NaOH. The test was conducted under static condition at 37°C for a period of 96 hours and decolorization was measured for every 24-hour interval.
2.4. Effect of Agitation on Dye Decolorization
Effect of agitation on decolorization was checked at 37°C under static and agitated state (200 rpm) for MO, CR, and EBT. Dye concentration was 100 mg/l, and the study was carried out for 96 hours with decolorization percentage measured for every24-hour interval.
2.5. Effect of Temperature on Dye Decolorization
100 mg/l dye containing LB media was incubated at 25°C, 35°C, 45°C, and 55°C at pH 7 to study the effect of temperature on dye decolourization. The test was conducted under static condition and decolourization was measured at every 24-hour interval for 96-hour duration.
3. RESULT AND DISCUSSION
3.1. Isolation, Screening, and Identification
EBT-2 strain was identified as Citrobacter sp. on basis of morphological, biochemical (Table 1), and 16s rDNA sequencing (Fig. 1). It matched 86% identity with Citrobacter sp. and fell in the cluster of Klebsiella sp.
3.2. Effect of Initial Dye Concentration on Dye Decolorization
The isolate was able to decolorize MO up to 300 mg/l (Fig. 2a–c) and both CR and EBT up to 500 mg/l dye concentration (Figs. 3a–c and 4a–c). Contrary to results shown by previous works [18,30], dye decolorization did not show a gradual decreasing pattern with increase in its initial concentration. It remained independent of dye concentration and drastically fell to ~50% for 300–500 mg/l dye concentration. It has been also shown that Acid Red dye decolorization by Acinetobacter radioresistens is independent of its concentration [31]. A probable reason for this observation can be extracellular reduction of dye, the rate of which is independent of its concentration [32]. The decrease in decolorization efficiency beyond 100 mg/l may occur due to several reasons like the toxicity of the dye to bacteria and/or insufficient biomass concentration for the uptake of higher concentrations of dye [33].
 | Table 1: Phenotypic and Biochemical characters of EBT-2 strain. [Click here to view] |
3.3. Effect of Agitation on Dye Decolorization
MO showed 98% decolorization in 96 hours but could only achieve 10% decolorization under agitated condition for the same time period (Fig. 5a). Similar trend was observed for CR, it showed 95% decolorization in static and 18% decolorization in shaking condition in 96 hours (Fig. 5b). Eriochrome Black T showed 90% decolorization in static and 35% decolorization in shaking condition for the same time period (Fig. 5c). Dye decolorization was reduced significantly in shaking condition as compared to static condition. This agrees with the results previously demonstrated studies [34,35]. A possible cause for this trend could be that in many bacteria, degradation of azo dyes to their corresponding amines occurs due to reduction of azo linkage with the aid of cytoplasm azoreductase enzyme. Azoreductase mediated degradation of azo dyes is inhibited by the presence of oxygen because oxygen is a preferable terminal electron acceptor over the azo groups for the oxidation of reduced electron carriers such as nicotinamide adenine dinucleotide - hydrogen (reduced) [22,36].
 | Figure 1: Phylogenetic tree of the Citrobacter sp. EBT-2 strain based on 16S rDNA partialsequences. [Click here to view] |
 | Figure 2: (a) Effect of dye concentration on dye colorization (MO: 10–50 mg/l). (b) Effect of dye concentration on dye decolorization (MO: 60–100 mg/l). (c) Effect of dye concentration on dye decolorization (MO: 200, 300 mg/l). [Click here to view] |
 | Figure 3: (a): Effect of dye concentration on dye decolorization (CR; 10–50 mg/l). (b): Effect of dye concentration on dye decolorization (CR; 60–100 mg/l). (c): Effect of dye concentration on dye decolorization (CR: 200–500 mg/l). [Click here to view] |
3.4. Effect of pH on Decolorization
MO and CR showed maximum decolorization at pH 7 in 96 hours (97.24% and 95.23%, respectively) (Fig. 6a,b). Maximum decolorization was observed in a range of pH 7–9 (~85%) for Eriochrome Black T (Fig. 6c). Dye decolorization decreased at lower pH (5–6) and very high pH (10–11). It was observed that in general, azo dye show better decolorization from neutral to alkaline conditions. Similar results were observed for decolorization of several azo dyes by Micrococcus sp. [37]. Since alkaline environment is required for binding of most azo dyes to fibers, dye degrading bacteria are better adapted to alkaline environment [38]. However, the dye degradation pattern for all the dyes treated with EBT-2 strain are different from each other, which prove that degradation is dependent on the chemical structure and reactivity of dyes as well.
 | Figure 4: (a) Effect of dye concentration on dye decolorization (EBT: 10–50 mg/l). (b) Effect of dye concentration on dye decolorization (EBT: 60–100 mg/l). (c) Effect of dye concentration on dye decolorization (EBT: 200–500 mg/l). [Click here to view] |
 | Figure 5: (a) Effect of agitation on dye decolorization (MO; 100 mg/l). (b) Effect of agitation on dye decolorization (CR; 100 mg/l). (c) Effect of agitation on dye decolorization (EBT; 100 mg/l). [Click here to view] |
3.5. Effect of Temperature on Dye Decolorization
EBT-2 completely decolorized MO at both 35°C and 45°C in 120 hours (Fig. 7a). It also decolorized EBT completely at both 35°C and 45°C. However, decolorization at 45°C was achieved much faster, i.e., in 72 hours as compared to 120 hours at 35°C (Fig. 7c). It decolorized CR completely only at 35°C in 120 hours (Fig. 7b). For all the three dyes, decolorization efficiency increased with increase in temperature from 25°C to 35–45°C. This is in accordance with results previously given for decolorization of Acid Orange by Staphylococcus hominis and decolorization of azo dyes by Micrococcus sp. [30,37,39]. Increase in temperature can increase bacterial growth hence increasing decolorization efficiency [19]. No decolorization was observed at 55°C for all three dyes which can be caused due to loss of Azoreductase enzymatic activity at high temperature or decrease in cell viability [39].
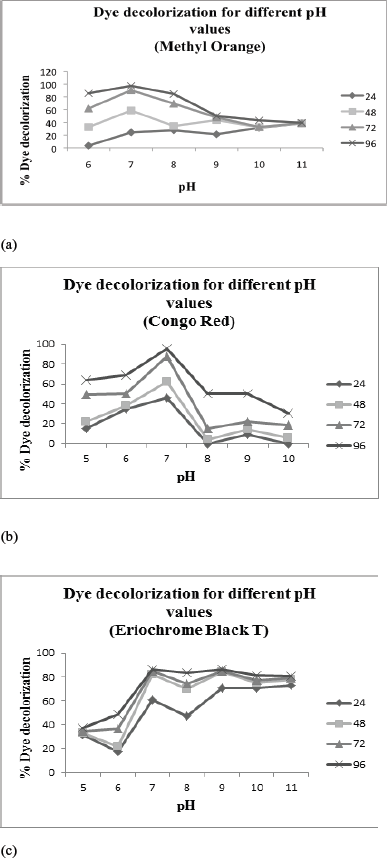 | Figure 6: (a) Effect of pH on dye decolorization (MO; 100 mg/l). (b) Effect of pH on dye decolorization (CR; 100 mg/l) (c) Effect of pH on dye decolorization (EBT; 100 mg/l). [Click here to view] |
 | Figure 7: (a) Effect of temperature on dye decolorization (MO; 100 mg/l) (b) Effect of temperature on dye decolorization (CR; 100 mg/l) (c):Effect of temperature on dye decolorization (EBT; 100 mg/l). [Click here to view] |
4. CONCLUSION
EBT-2 strain was identified as Citrobacter sp. It showed complete decolorization of all the three dyes up to 100 mg/l dye concentration in 96–120 hours. It was able to decolorize MO, CR, and EBT up to 300, 500, and 500 mg/l of dye concentration, respectively. Optimum pH for decolorization was 7, 7, and 9 for MO, CR, and EBT, respectively. Similar results were observed for decolorization of several azo dyes by Micrococcus sp. [37]. Effect of agitation on decolorization was studied under static and shaking (200 rpm) condition. More than 90% decolorization was observed at static condition for each dye but only 10.3%, 18.47% and 35.92% decolorization was observed at shaking condition in MO, CR, and EBT respectively in 96 hours. This agrees with the results previously demonstrated by bacterial degradation of Reactive Red 141 and Amaranth dyes [34,35]. Complete decolorization was obtained for MO and EBT at 35°C and 45°C. CR showed complete decolorization only at 35°C. This is in accordance with results previously given for decolorization of Acid Orange by S. hominis and decolorization of azo dyes by Micrococcus sp. [30,37,39].Temperature increase can stimulate bacterial proliferation and increase decolorization efficiency [19].
The results conclude that Citrobacter sp. can be used for successful dye decolorization of Azo dyes; primarily MO, CR, and EBT under optimum physiochemical conditions. Further work needs to be done on complete mineralization of dyes. Scaling up the process also remains a challenge. Since metal ions like Copper, Lead, and Cadmium are present in high concentration in industrial effluents [23], it is also important to study their effects individually on azo dye bioremediation.
5. AUTHORS’ CONTRIBUTION
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
6. ETHICAL APPROVAL
This study does not involve the use of animals or human subjects.
7. CONFLICT OF INTEREST
The authors report no conflicts of interest in this work.
8. FUNDING
None
REFERENCES
1. Bafana A, Devi SS, Chakrabarti T. Azo dyes: past, present and the future. Environ Rev 2011;19:350–70. CrossRef
2. Tripathi A, Srivastava SK. Biodegradation of orange G by a novel isolated bacterial strain Bacillus megaterium ITBHU01 using response surface. Afr J Biotechnol 2012;1:1768–78. CrossRef
3. Zollinger H. Colour chemistry synthesis properties and application of organic dyes and pigments. VCH, New York, NY, pp 92–102, 1991.
4. Pandey A, Singh P, Iyengar L. Bacterial decolorization and degradation of azo dyes. Int Biodeterior Biodegradat 2007;59:73–8. CrossRef
5. Rajeshwari K, Subashkumar R, Vijayaraman K. Biodegradation of mixed textile dyes by bacterial strain isolated from dye waste effluent. Res J Environ Toxicol 2011;5:97–107. CrossRef
6. de Campos Ventura-Camargo B, Marin-Morales MA. Azo dyes: characterization and toxicity-a review. Text Light Ind Sci Technol 2013;2:85–103.
7. Chang SJ, Lin YC. Decolorization kinetics of recombinant Escherichia coli strain harboring azo dye decolorization determinants for Rhodococcus sp. Biotechnol Lett 2001;23:631–6. CrossRef
8. Boer CG, Obici L, Souza CG, Peralta RM. Decolourization of synthetic dyes by solid state cultures of Lentinula (Lentinus) edodes producing manganese peroxidase as the main lignolyticenzyme. Bioresour Technol 2004;94:107–12. CrossRef
9. Wong PK, Yuen PY. Decolorization and biodegradation of methyl red by Klebsiella pneumoniae RS-13. Water Res, 1996;30:1736-44. CrossRef
10. Bell J, Plumb JJ, Buckley CA, Stuckey DC. Treatment and decolourization of dyes in anaerobic baffled reactor. J Environ Eng 2000;126:1026–32. CrossRef
11. Kalyani DC, Telke AA, Dhanve RS, Jadhav JP. Ecofriendly biodegradation and detoxification of reactive red 2 textile dye by newly isolated Pseudomonas sp. SUK1. J Hazard Mater, 2008;163:735–42. CrossRef
12. Champagne PP, Nesheim ME, Ramsay JA. Effect of a non-ionic surfactant, Merpol, on dye decolorization of Reactive blue 19 by laccase. Enzyme Microb Technol 2010;46:147–52. CrossRef
13. Saratale RG, Saratale GD, Chang JS, Govindwar SP. Decolorization and biodegradation of textile dye Navy blue HER by Trichosporonbeigelii NCIM-3326. J Hazard Mater 2009;166:1421–8. CrossRef
14. Amin KA, Abdel Hameid H, Abd Elsttar AH. Effect of food azo dyes tartrazine and carmoisine on biochemical parameters related to renal, hepatic function and oxidative stress biomarkers in young male rats. Food Chem Toxicol 2010;48:2994–9. CrossRef
15. Robinson T, McMullan G, Marchant R, Nigam P. Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative. Bioresour Technol 2001;77:247–55. CrossRef
16. Dubrow SF, Boardman GD, Michelsen DL. Chemical pretreatmentand aerobic–anaerobic degradation of textile dye wastewater. In: Reife A, Reife A, Freeman HS (ed.). Environmental chemistry ofdyes and pigments. Wiley, New York, NY, pp 75–102, 1996.
17. Celik C, Ozturk A, Abdullah MI. Biodegradation of reactive red 195 azo dye by the bacterium Rhodopseudomonas palustris 51ATA. Afr J Microbiol Res 2012;6:120–6. CrossRef
18. Wang H, Zheng XW, Su JQ, Tian Y, Xiong XJ, Zheng TL. Biological decolorization of the reactive dyes reactive black 5 by a novel isolated bacterial strain Enterobacter sp. EC3. J Hazard Mater 2009;171:654–9. CrossRef
19. Asad S, Amoozegar MA, Pourbabaee AA, Sarbolouki MN, Dastqheib SM. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresour Technol 2007;98:2082–8. CrossRef
20. Solis M, Solis A, Perez HI, Manjarrez N, Flores M. Microbial decoloration of azo dyes: a review. Process Biochem, 2012;47:1723–48. CrossRef
21. Stolz A. Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol 2001;56:69–80. CrossRef
22. Van der Zee Frank P, Villaverde S. Combined anaerobic–aerobic treatment of azo dyes-A short review of bioreactor studies. Water Res 2005;39:1425–40. CrossRef
23. Zhuang M, Sanganyado E, Zhang X, Xu L, Zhu J, Liu W, et al. Azo dye degrading bacteria tolerant to extreme conditions inhabit nearshore ecosystems: optimization and degradation pathways. J Environ Manage 2020;261:1–9. CrossRef
24. Chung KT, Cerniglia CE. Mutagenicity of azo dyes: structure-activity relationships. Mutat Res 1992;277:201–20. CrossRef
25. Mall ID, Srivastava VC, Agarwal NK, Mishra IM. Removal of congo red from aqueous solution by bagasse fly ash and activated carbon: kinetic study and equilibrium isotherm analyses. Chemosphere 2005;61:492–501. CrossRef
26. Langer I, Atassi G, Robberecht P, Resibois A. Eriochrome black T inhibits endothelial cell growth through S-phase blockade. Eur J Pharmacol 2000;399:85–90. CrossRef
27. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007;24:1596–9. CrossRef
28. Bergey DH, Buchanan RE, Gibbons NE. Bergey’s manual of determinative bacteriology. Williams and Wilkins, Baltimore, MD, 1974.
29. Harazono K, Nakamura K. Decolorization of mixtures of different reactive textile dyes by the white-rot basidiomycete Phanerochaetesordida and inhibitory effect of polyvinyl alcohol. Chemosphere 2005; 59:63–8. CrossRef
30. Singh RP, Singh PK, Singh RL. Bacterial decolorization of textile azo dye acid orange by Staphylococcus hominis RMLRT0. Toxicol Int 2014;21:160–6. CrossRef
31. Ramya M, Iyappan S, Manju A, Jiffe John S. Biodegradation and decolorization of acid red by Acinetobacter radioresistens. J Bioremediat Biodegrad 2010;1:1–6. CrossRef
32. Dubin P, Wright KL. Reduction of azo food dyes in cultures of Proteus vulgaris. Xenobiotica 2008;59:563–71. CrossRef
33. Dawkar VV, Jadhav UU, Jadhav SU, Govindwar SP. Biodegradation of disperse textile dye brown 3REL by newly isolated Bacillus sp. VUS. J Appl Microbiol 2008;105:14–24. CrossRef
34. Telke A, Kalyani D, Jadhav J, Govindwar S. Kinetics and mechanism of reactive red 141 degradation by a bacterial isolate Rhizobium radiobacter MTCC 8161. Acta Chim Slov 2008;55:320–9.
35. Ghodake G, Jadhav U, Tamboli D, Kagalkar A, Govindwar S. Decolorization of textile dyes and degradation of mono-azo dye amaranth by Acinetobacter calcoaceticus NCIM 2890. Indian J Microbiol, 2011;51:501–8. CrossRef
36. Chang SJ, Lin YC. Fed-batch bioreactor strategies for microbial decolorization of azo dye using a Pseudomonas luteola strain. Biotechnol Prog, 2000;16:979–85. CrossRef
37. Olukanni OD, Osuntoki AA, Gbenle GO. Decolorization of azo dyes by strain of Micrococcus isolated from a reuse dump soil. J Biotechnol 2009;8:442–8. CrossRef
38. Aksu Z. Reactive dye bioaccumulation by Saccharomyces cerevisiae. Process Biochem 2003;38: 1437–44. CrossRef
39. Saratale RG, Saratale GD, Chang JS, Govindwar SP. Bacterial decolorization and degradation of azo dyes: a review. J Taiwan Inst Chem Eng 2011;42:138–57. CrossRef