1. INTRODUCTION
During the recent years, water bodies have been largely polluted due to the increasing industrialization. The textile industry is one among the leading contributors to such pollutions. Even a minute quantity of organic dyes can form intense color in the water bodies and make the water aesthetically unpleasant which can eventually cause possible harmful effects in the whole ecosystem [1]. Some of the dyes used in the textile industries in coloration process are toxic, mutagenic, and even carcinogenic which can badly affect the life of human, animals, and plants. All these conditions recommend the necessity of dye contaminated water to be treated before entering into any ecosystem.
During the past decades, many physicochemical decolorization techniques have been developed for the remediation of dyes; however, very few methods have been accepted by the textile industries [2,3]. The continuous progress of these accepted techniques slowly revealed their disadvantages as they are expensive, formation of excessive sludge and toxic substances [4]; furthermore, some of these methods are ineffective of removing their organic metabolites completely which are the ultimate cause of the secondary pollution [5].
Nowadays, it is well understood that the water pollution affects the quality of life in many aspects including the health and life span of all living beings; therefore the remediation of toxic pollutants will require much attention in the near future. On this backdrop, there is an urgent need for the cost effective and environmentally friendly methods for the removal of toxic dyes. As an appreciable alternative technique, biological methods which include several taxonomic groups of microbes such as bacteria and fungi have been received increasing attention due to their cheap cost, production of very less sludge, and their environmental friendliness [6]. Bacteria from many trophic groups are reported for their higher degree of dye-degradation and can process a complete degradation of many dyes under standardized conditions [7]. In the recent years, remediation of textile effluent using microbial sources has been considered as the most potential method than any other physiochemical techniques [8].
Reactive Black 5 is a bis(azo) compound with two aryldiazenyl moieties placed at positions 2 and 7 on a multi-substituted naphthalene with its absorption maximum at 595 nm. It has a molecular formula of C16H21N5Na4O19S6 with a molecular weight of 991.8 g/mol. It is one of the most commonly used synthetic dyes in the textile industries of many developing nations, like India; furthermore, huge amounts of this dye are being used and discharged in the water columns every year. Therefore, it requires an urgent step to develop an effective treatment method since small amounts of this dye are clearly visible and detrimental to the water environment [8]. Based on these backdrops, the present investigation aimed to isolate and characterize a potential Reactive Black 5 dye degrading bacterium from textile industry effluents that might be considered for use in the industrial dye based bioremediation process.
2. MATERIALS AND METHODS
2.1. Sample Collection
Three effluent sediment samples were procured with a 100 m distance from a textile industry situated at Salem, Tamil Nadu, India, using sterile spatula where all the possible aseptic techniques were followed while collecting the samples to avoid contamination. These samples were immediately transported to the laboratory in 250 ml pre-sterilized bottle containers which were stored at 4°C until further processing.
2.2. Isolation of Dye Degrading Bacteria
The collected effluent samples were processed immediately in the laboratory in which 1 g of samples was serially diluted using pre-sterilized bacteriological saline water and 0.1 ml of dilutions were spread plated individually on the pre-sterilized nutrient agar Petri plate medium. After 48 hours incubation at 37°C, individual colonies with distinct colony morphology were chosen and further purified by culture on fresh nutrient agar Petri plates medium and axenic cultures were lyophilized for further studies.
2.3. Screening and Identification of Potential Dye Degrading Bacterium
All the axenic bacterial strains were individually tested for the biodegradation of Reactive Black 5 dye. Nutrient broth was used as the culture medium supplemented with 10 μg/ml concentration of Reactive Black 5 dye and the screening process was carried out in 30 ml screw cap tubes. The dye degrading potentials of the isolated strains were studied after 4 days incubation at 37°C using the remaining dye concentration in the cell free supernatant which were read at an optical density (OD) of 595 nm under UV Visible spectrophotometer. Furthermore, the standards were prepared using known values of Reactive Black 5 in distilled water for the study of dye degradation percentage. The potential strain showing the best dye degradation percentage was identified using Bergy’s Manual of Determinative Bacteriology [9].
2.4. Growth Kinetics Profile as a Function of Time on Dye Degradation
The potential bacterium was optimized for its peak time of dye degradation with reference to the cell growth which was studied under regular time interval of 12 hours from lag to decline growth phase. The experiment was performed in a 250 ml conical flask with 100 ml working volume of nutrient broth as the medium supplemented with 10 μg/ml Reactive Black 5 dye concentration along with the basal conditions of pH 7.0, 37°C temperature and 150 rpm agitation. The inoculum was prepared using the log growth phase culture of the potential bacterium in the same nutritive medium conditions, where the (OD 620 nm) of the inoculum was adjusted to 0.1 according to McFarland turbidity 0.5 standards which was equivalent to 1 × 108 cfu/ml bacterial concentration. The evaluations were monitored from a portion of cultured broth followed by the separation of cell pellet and cell free supernatant using centrifugation at 3,000 rpm for 15 minutes. Bacterial growth estimations were done using the dry weight of cell biomass concentration obtained from centrifuged cell pellets which was hot air oven dried at 50°C for 30 minutes and dye degradation efficiency was determined in cell free supernatant as per the screening procedure mentioned above. All the experiments were performed in triplicate and the results were represented as mean ± standard deviation.
2.5. Degradative Enzymes Study
At the above-mentioned regular time intervals on the study of growth kinetic profile, a set of seven degradative enzymes were also evaluated. A portion of cultured broth was sonicated at 4°C under 20 KHz for 45 seconds using Ultra-sonicator (Hielscher, USA) and centrifuged the lysed broth at 3,000 rpm for 15 minutes. The supernatant was studied for the activities of seven different enzymes using the following experimental assay procedures mentioned below. All enzyme assays were performed in triplicate, and the results were represented as mean ± standard deviation.
2.5.1. Laccase assay
Laccase activity was evaluated using the oxidation method of 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid (ABTS) [10]. The nonphenolic dye, ABTS is oxidized to a more stable cation radical state by laccase enzyme. The concentration of the cation radical is directly proportional to the intense blue-green color development which can be read at 420 nm [11]. The reaction mixture has 2.450 ml citrate phosphate buffer (pH 4.0), 500 μl ABTS (1.5 mM), and 50 μl of lysed cell supernatant and incubated for 5 minutes. The estimation of ABTS oxidation was evaluated by the increase in A420 (ε420 = 36mM−1 cm−1). The absorbance was monitored at 420 nm and the enzyme activity was expressed as Unit/ml/min.
2.5.2. Veratryl alcohol oxidase assay
The veratryl alcohol oxidase activity was spectrophotometrically examined by the changes in the absorbance at 310 nm at 30°C [12]. The reaction mixture has 1.0 ml of veratryl alcohol (10 mmol/l), 0.4 ml of lysed broth supernatant and 1.5 ml of sodium tartrate buffer (100 mmol/l, pH 3.0), and then the reaction was initiated by the addition of 0.1 ml of H2O2 (10 mmol/l). Veratryl alcohol oxidase activity was expressed in international units (U), one unit is equal to 1 μmol of veratryl alcohol oxidized per minute. The standards were done using veratraldehyde which is the oxidation product of veratryl alcohol.
2.5.3. Lignin peroxidase assay
The lignin peroxidase activity was studied using the oxidation of azure B dye [13]. The reaction mixture has 1 ml of 125 mM sodium tartarte buffer (pH 3.0), 500 μl of 0.160 mM azure B, 500 μl of the lysed broth supernatant, and 500 μl of 2 mM hydrogen peroxide. The reaction was initiated by the addition of 10 mmol/l hydrogen peroxide and one unit of enzyme is equal to the O.D. decrease of 0.1 units/minute/ml of the lysed broth supernatant.
2.5.4. Manganese peroxidase assay
Manganese peroxidase activity toward 2,6-dimethoxyphenol (DMP) was performed as per the method of Wariishi et al. [14]. Oxidation of DMP by manganese peroxidase results in the coerulignone formation, which can be spectroscopically measured by the absorbance at 469 nm. Reaction mixture has 200 μl of 50 mM sodium tartrate (pH 7.0) containing 5 μg/ml manganese peroxidase extract from lysed broth supernatant, 0.5 mM DMP, 1 mM MnSO4, and 0.1 mM H2O2 and the reaction was performed at 30°C. One unit of manganese peroxidase activity is equal to the amount of enzyme oxidize 1 mmol of 2,6-DMP/minute.
2.5.5. Polyphenol oxidase assay
sThe enzymatic activity was estimated as per the method earlier described by Liu et al. [15]. The reaction mixture has 20 μl lysed broth supernatant and 180 μl substrate solution (0.2 M L-tyrosine in 0.05M phosphate buffer solution at pH 6.8). The reaction was performed at 25°C, the absorbance was monitored at 410 nm and the activity was estimated using a microplate spectrophotometer. One unit of enzyme activity was represented as an increase of 0.001 absorbance units/minute/ml.
2.5.6. Azoreductase assay
The azoredutase activity was examined using a UV–Vis spectrophotometer [16]. Reaction mixture has lysed broth supernatant which was carried out in 1 ml final volume containing 50 mM pH 7.0 sodium phosphate buffer, 150 μM NADH, 50 μM FMN, and 25 μM methyl red at 30°C. The reaction was initiated by adding NADH prepared in 50 mM sodium phosphate buffer at pH 7.0 to a final volume of 1 mM. Azoreductase activity was expressed by the reduction of absorbance at 430 nm over 1 minute duration with the help of UV/Vis spectrophotometer. The extinction coefficient for methyl red was 23,360 M−1 cm−1 at 430 nm. A unit of enzyme activity was expressed as the reduction in azo dye concentration (μM/min).
2.5.7. NADH-DCIP reductase assay
Nicotinamide adenine dinucleotide + hydrogen - 2,6-dichlorophenolindophenol (NADH-DCIP) reductase activity was evaluated by the reduction of DCIP concentration at 590 nm. The reaction mixture has lysed broth supernatant which was performed in the final volume of 3 ml containing 50 M DCIP, 50 mM potassium phosphate buffer solution at pH 7.4 and 50 M NADH [17]. One unit of NADH-DCIP reductase activity is equal to one microgram of DCIP reduced/minute/ml.
Table 1. Identification of the potential bacterium, B. cereus SKB12 based on morphological and biochemical characterization, indications: + and – indicates positive and negative results.
| Identification test | B. cereus SKB12 |
|---|---|
| Gram stain | Gram positive |
| Shape | Rod |
| Indole | − |
| Catalase | + |
| Gelatin digested | + |
| Acid production from | |
| Arabinose | − |
| Glucose | + |
| Glycogen | + |
| Mannitol | − |
| Maltose | − |
| Mannose | − |
| Rhamnose | − |
| Salicin | + |
| Sucrose | + |
| Xylose | − |
| Enzyme activity | |
| β-Galactosidase | − |
| Leucine arylamidase | − |
| Glycine arylamidase | − |
3. RESULTS AND DISCUSSION
3.1. Isolation, Screening and Identification of a Potential Dye Degrading Bacterium
All the collected three sediment effluent samples were individually studied for the bacterial strains. After incubation, the agar plates were visualized for the distinct colony morphologies which were isolated and axenic cultured on the same culture medium conditions. Totally, 22 pure cultured strains were isolated and named as SKB1–22. These strains were screened to identify the most promising Reactive Black 5, dye-degrading bacterium. Among these isolates, SKB12 showed appreciable activity of 73% dye degradation followed by SKB16 revealed 66.5% dye degradation, but the other strains showed poor activities within 10%–30% dye degradation. Based on this potentiality, the strain SKB12 was chosen for further dye degradation and its degradative enzymes study, further, the potential strains were identified as Bacillus cereus based on the morphological and biochemical uniqueness as specified in the Bergy’s Manual of Determinative Bacteriology [9].
The biochemical characterization results of the B. cereus SKB12 was shown in the Table 1. A similar investigation was carried out by Kumar and Saravanan [18] isolated 13 bacterial strains from the textile effluent of Kanchipuram, Tamil Nadu, India in which the strain no. DD4 revealed the potential biodegradation of the malachite green used in their study and it was identified as Staphylococcus species. Likewise, Olukanni et al. [19] isolated 18 bacterial strains from a textile effluent which were belonging to the genera of Bacillus, Acinetobacter, Staphylococcus, Legionella, and Pseudomonas. These strains were investigated for the textile effluent degrading potentiality, in which Bacillus and Legionella were found to be effective in effluent bioremediation treatments.
 | Figure 1: Conical flasks showing the biodegradation of Reactive Black 5 dye using the potential strain, B. cereus SKB12 at the start (0 hour) and end (144 hours) of the dye degradation experiment. [Click here to view] |
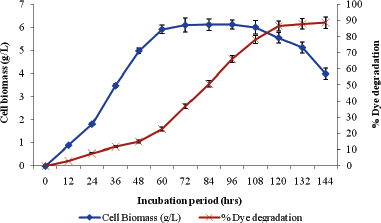 | Figure 2: Growth kinetics profile as a function of time on biodegradation of reactive black 5 dye using B. cereus SKB12. [Click here to view] |
3.2. Growth Kinetic Profile with Reference to Dye Degradation Potential
The growth kinetic profile with reference to Reactive Black 5, dye degradation by the strain B. cereus SKB12 as a function of time revealed that the dye degradation was initiated during the first hours of lag phase and observed till the end period (144 hours) of decline growth phase and the peak time of dye degradation was observed from the middle of the logarithmic growth phase (48 hours) till the end of exponential phase (108 hours) (Figs. 1 and 2). Furthermore, the maximum cell biomass concentration of 6 ± 0.13 g/l was observed during 60 to 108 hours of time incubation which constituted the exponential growth phase of the B. cereus SKB12 and maximum dye degradation of 88.7% was evidenced at the end of decline growth phase.
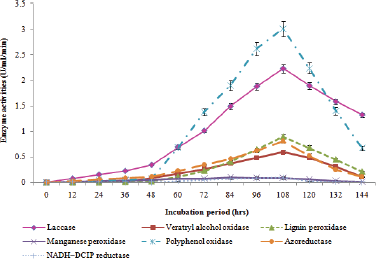 | Figure 3: Quantitative biosynthesis of seven different degradative enzymes during biodegradation of Reactive Black 5 dye. [Click here to view] |
The results revealed that the metabolites involved in the dye degradation process of B. cereus SKB12 were both primary and secondary metabolites. A similar investigation was done by Chen et al. [20] identified Aeromonas hydrophila as the promising dye decolorizing bacteria and the maximum color removal was estimated under anaerobic conditions. More than 90% degradation of Red RBN dye was achieved in 8 days and this strain revealed a similar pattern of growth kinetics with reference to dye degradation.
3.3. Degradative Enzymes Synthesis of the Potential Bacterium
Dye decolorization is mainly associated with interaction of enzyme molecules with the dye substrates resulting into the formation of dye products [21]. There were seven degradative enzymes were studied during the biodegradation of Reactive Black 5 with an intervals of 12 hours from lag to decline growth phase of B. cereus SKB12. Among the studied enzymes, the potential bacterium showed promising production of laccase enzyme with its peak synthesis of 2.23 U/ml at 108 hours; furthermore, the bacterium showed growth dependent production of laccase (Fig. 3).
Likewise, this strain showed the potential production of veratryl alcohol oxidase which was maximum at 108 hours with its 0.59 U/ml/minute, further, the biosynthesis of veratryl alcohol oxidase was observed during the exponential growth phase which representing it as secondary metabolite (Fig. 3). Similarly, potential production of lignin peroxidase was observed in this study which was high during 108 hours with its 0.9 U/ml/minute; furthermore, the production was evidenced that as secondary metabolite (Fig. 3).
Moreover, appreciable biosynthesis of polyphenol oxidase and azoreductase were also observed which were high at 108 hours with their 3.01 U/ml/minute and 0.81 U/ml/minute, respectively. Furthermore, the polyphenol oxidase production acts as secondary metabolite whereas the azoreductase biosynthesis acts as primary metabolite (Fig. 3).
Furthermore, the strain SKB12 showed poor synthesis of manganese peroxidase and NADH-DCIP reductase enzymes (Fig. 3). Interestingly, none of the studied seven degradative enzymes were synthesized in control which was not added with Reactive Black 5 dye in the culture medium. This observation clearly evidenced that these degrading enzymes were produced with the presence of a textile dye in the growth medium.
Likewise, Thanavel et al. [22] revealed a significant increase in the level of dye degradation enzymes such as tyrosinase, laccase, lignin peroxidase, riboflavin reductase, and azoreductase which was observed in A. hydrophila SK16 during the biodegradation of textile dyes, viz., Reactive Red 180, Reactive Black 5 and Remazol Red. Phugare et al. [23] developed a microbial consortium using Providencia sp. SDS and Pseudomonas aeuroginosa BCH which were isolated from a dye contaminated soil. The consortium was found to be effective in decolorizing the Red HE3B when compared to the individual strains. Furthermore, the consortium revealed 100% decolorization of Red HE3B and evidenced a significant increase in the synthesis of veratryl alcohol oxidase, laccase, azoreductase and DCIP reductase when compared to control.
4. CONCLUSION
In the present study, the isolated bacterium, B. cereus SKB12 from a textile effluent showed significant Reactive Black 5 dye degradation capability. This strain revealed dye degradation from the first hours till the end of decline phase of bacterial growth. Furthermore, the strains showed promising activities of five different dye degradation enzymes, viz., laccase, veratryl alcohol oxidase, lignin peroxidase, polyphenol oxidase, and azoreductase enzymes. Taking all together, the isolated bacterium, B. cereus SKB12 evidenced its strong dye degradations efficiency and showed the possibilities of an effective effluent dye bioremediation process in near future.
ACKNOWLEDGMENT
The authors very gratefully acknowledge Geobiotechnology Lab, PG & Research Department of Biotechnology, National College (Autonomous), Tiruchirapalli - 620001 Tamil Nadu, India, for providing lab facilities and supporting our research.
CONFLICT OF INTEREST
Authors declare that they do not have any conflicts of interest.
FINANCIAL SUPPORT
None.
REFERENCES
1. Uddin MT, Islam MA, Mahmud S, Rukanuzzaman M. Adsorptive removal of methylene blue by tea waste. J Hazard Mater 2009;164(1):53–60. CrossRef
2. da Silva CG, Faria JL. Photochemical and photocatalytic degradation of an azo dye in aqueous solution by UV irradiation. J Photoch Photobio A 2003;155(1–3):133–43. CrossRef
3. Okazaki SY, Nagasawa SI, Goto M, Furusaki S, Wariishi H, Tanaka H. Decolorization of azo and anthraquinone dyes in hydrophobic organic media using microperoxidase-11 entrapped in reversed micelles. Biochem Eng J 2002;12(3):237–41. CrossRef
4. Senan RC, Abraham TE. Bioremediation of textile azo dyes by aerobic bacterial consortium aerobic degradation of selected azo dyes by bacterial consortium. Biodegradation 2004;15(4):275–80. CrossRef
5. Saratale RG, Saratale GD, Kalyani DC, Chang JS, Govindwar SP. Enhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium-GR. Bioresource Technol 2009;100(9):2493–500. CrossRef
6. Kalyani DC, Telke AA, Dhanve RS, Jadhav JP. Ecofriendly biodegradation and detoxification of Reactive Red 2 textile dye by newly isolated Pseudomonas sp. SUK1. J Hazard Mater 2009;163(2–3):735–42. CrossRef
7. Asad S, Amoozegar MA, Pourbabaee A, Sarbolouki MN, Dastgheib SM. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresource Technol 2007;98(11):2082–8. CrossRef
8. Shah MP. Microbial degradation of textile dye (Remazol Black B) by Bacillus spp. ETL-2012. J Appl Environ Microbiol 2013;1(1):6–11.
9. Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST. Bergey’s manual of determinative bacteriology. 9th edition, Williams & Wilkins, Baltimore, MD, 1994.
10. Naghdi M, Taheran M, Brar SK, Kermanshahi-pour A, Verma M, Surampalli RY. Immobilized laccase on oxygen functionalized nanobiochars through mineral acids treatment for removal of carbamazepine. Sci Total Environ 2017;584:393–401. CrossRef
11. Majcherczyk A, Johannes C, Hüttermann A. Oxidation of polycyclic aromatic hydrocarbons (PAH) by laccase of Trametes versicolor. Enzyme Microb Tech 1998;22(5):335–41. CrossRef
12. Tien M, Kirk TK. 1988. Lignin peroxidase of Phanerochaete chrysosporium. In: Methods in enzymology, Academic Press, Cambridge, MA, 161, pp 238–49, 2009. CrossRef
13. Archibald FS. A new assay for lignin-type peroxidases employing the dye azure B. Appl Environ Microbiol 1992;58(9):3110–6. CrossRef
14. Wariishi H, Valli K, Gold MH. Manganese (II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J Biol Chem 1992;267(33):23688–95.
15. Liu F, Han Q, Ni Y. Comparison of biochemical properties and thermal inactivation of membrane-bound polyphenol oxidase from three apple cultivars (Malus domestica Borkh). Int J Food Sci Technol 2018;53(4):1005–12. CrossRef
16. Qi J, Schlömann M, Tischler D. Biochemical characterization of an azoreductase from Rhodococcus opacus 1CP possessing methyl red degradation ability. J Mol Catal B: Enzym 2016;130:9–17. CrossRef
17. Bhosale S, Saratale G, Govindwar S. Biotransformation enzymes in Cunninghamella blakesleeana (NCIM-687). J Basic Microb 2006;46(6):444–8. CrossRef
18. Kumar NM, Saravanan D. Isolation of dye degrading bacteria from textile effluent. J Chem Pharmaceut Res 2015;7(3):2214–8.
19. Olukanni OD, Osuntoki AA, Gbenle GO. Textile effluent biodegradation potentials of textile effluent-adapted and non-adapted bacteria. Afr J Biotechnol 2006;5(20):1980–84.
20. Chen KC, Wu JY, Liou DJ, Hwang SC. Decolorization of the textile dyes by newly isolated bacterial strains. J Biotechnol 2003;101(1):57–68. CrossRef
21. Khan R, Bhawana P, Fulekar MH. Microbial decolorization and degradation of synthetic dyes: a review. Rev Env Sci Bio/Technol 2013;12(1):75–97. CrossRef
22. Thanavel M, Kadam SK, Biradar SP, Govindwar SP, Jeon BH, Sadasivam SK. Combined biological and advanced oxidation process for decolorization of textile dyes. SN Appl Sci 2019;1(1):97. CrossRef
23. Phugare SS, Kalyani DC, Patil AV, Jadhav JP. Textile dye degradation by bacterial consortium and subsequent toxicological analysis of dye and dye metabolites using cytotoxicity, genotoxicity and oxidative stress studies. J Hazard Mater 2011;186(1):713–23. CrossRef