1. INTRODUCTION
Enzymes are biological catalysts that accelerate hundreds of metabolic events in living cells [1]. Enzymes are widely studied for their physiological, analytical, and commercial applications, particularly in microbes due to their diverse biochemistry, simplicity of mass culture, and genetic modification [2]. Microbial enzymes are usually preferred due to their high yields, economic feasibility, consistency, rapid growth of microbes on inexpensive media, ease of product modification and optimization, stability, and higher catalytic activity.
Amylases are a type of hydrolytic enzyme that hydrolyze the glycosidic bonds in starch and similar polysaccharide molecules [3,4]. The primary building blocks of starch are glucose molecules connected by α-1,4- or α-1,6-glycosidic linkages. Based on the various mechanisms of the glycosidic linkage hydrolysis, they are classified as exoenzymes and endoenzymes [5]. Exoenzymes that act on the non-reducing α-1,4-glycosidic bond of starch to create glucose or maltose include glucoamylase (EC 3.2.1.3), β-amylase (EC 3.2.1.2), and α-glucosidase (EC 3.2.1.20). While glycosidic linkages in a variety of substrates, including starch and glycogen, are hydrolyzed by endoenzymes such as α-amylase (EC 3.2.1.1) [6,7]. These enzymes are widely used in industrial processes, including the manufacturing of glucose syrup, detergents, alcohol (from starch), baking, preparation of starch coatings for paints, and in the food industry for liquefaction and gelatinization of starch [8-10].
Despite the fact that amylases can be produced biologically by plants, animals, and microorganisms, enzymes obtained from microorganisms are currently used in the majority of firms [11]. A number of factors, including cost-effectiveness, thermostability, and an easy optimization procedure, make bacterial amylases the most popular and frequently chosen source in industrial applications when compared to other options [6,12,13]. At present, amylase production makes up as much as 30% of the worldwide enzyme industry and is still growing [14]. Microorganisms such as Pseudomonas spp., Streptomyces spp., Escherichia coli, and Bacillus spp., including Bacillus subtilis, Bacillus cereus, Bacillus amyloliquefaciens, and Bacillus megaterium, as well as fungi such as Aspergillus niger, Aspergillus ficuum, Talaromyces emersonii, Penicillium, Rhizopus, and Neurospora spp., were selected as a source to produce amylase due to their accessibility, quick growth rates that result in brief fermentation cycles, ease of use, high yield, and overall handling safety [15-17]. Amylase-producing bacteria are highly studied from a variety of sources, including feces of different animals, soil, decaying wood, and hot spring water, but still the fecal sample of deer (Axis axis) is not studied for the amylase enzyme [18]. Proper microorganism selection is crucial for producing high-quality enzymes. Developing novel microbial strains for commercial enzyme production is endless work. There are two main kinds of amylases found in microorganisms: Glucoamylase and alpha-amylase [19].
The spotted deer (A. axis), which is also locally known as chital, is a unique herbivore found in the Indian subcontinent. Since they mostly consume grasses, young shoots, herbs, leaves, and fruits, as well as having a strong and quick capacity to break down fiber, we speculated that a population of bacteria with a variety of enzymes may have grown in their rumen [20,21]. The present paper comprises the first comprehensive research on the isolation, screening, and characterization of amylolytic bacterial strains from spotted deer (A. axis) feces collected in their natural habitat at the Sayaji Baug Zoo in Vadodara, Gujarat, India. The potential amylolytic bacteria were screened out on the basis of the zone of hydrolysis produced by bacteria on the starch agar plate and the enzyme activity of the bacterial culture supernatant. Enterobacter cloacae AD2 and Aeromonas veronii AD4 bacterial strains were identified on the basis of biochemical characterization and 16S rRNA analysis.
2. MATERIALS AND METHODS
2.1. Substrate and Chemicals
Starch agar, starch soluble, agar powder, citrate agar, glucose phosphate broth (GPB), oxidase discs, triple sugar iron (TSI) agar, Bradford’s reagent, and 3,5-dinitrosalicylic acid (DNS) were purchased from Hi-Media Pvt. Ltd, India. Amylum, beechwood xylan, carboxymethylcellulose (CMC), and 4-nitrophenyl beta-D-glucoside (pNPG) were purchased from Sigma-Aldrich, USA. Thin-layer chromatography plate (TLC Silica gel 60 F254) was purchased from Merck India Ltd. All chemicals and reagents were of analytical grade.
2.2. Sample Collection
The fecal sample of spotted deer (A. axis) was collected from the Sayaji Baug Zoo, Vadodara, Gujarat, India. Presterilized spatula and forceps, along with gloves and plastic zip bags, were used for sample collection.
2.3. Isolation and Screening of Amylolytic Bacteria from Spotted Deer (A. axis)
A fecal sample (1 g) of spotted deer was suspended with 50 mL of (0.85% w/v) sterile NaCl solution in a 250 mL conical flask and kept at 180 rpm and 37°C for 1 h. The fecal sample solution was further subjected to serial dilution up to 10−7 using sterile saline solution. Aliquots of 0.1 mL from each dilution were spread onto starch agar plates and incubated at 37°C for 24 h. Bacterial colonies showing different morphology were further purified by streaking onto starch agar medium to obtain pure colonies [22,23]. A single colony from each starch agar plate was picked up and inoculated into 5 mL of Luria-Bertani broth (LB broth) and incubated at 180 rpm and 37°C for 14 h. Each freshly grown bacterial culture was analyzed qualitatively and quantitatively.
2.4. Qualitative and Quantitative Analysis of Bacterial Culture
For qualitative analysis, a loopful of each culture was streaked on starch agar plates and further incubated at 37°C for 24 h. Then, each culture plate was flooded with Lugol’s iodine solution (0.5%) and checked for the appearance of a clear hydrolytic zone around the bacterial colonies on the starch agar plates [24]. For quantitative analysis, 50 μL of each culture was inoculated into different 5 mL enzyme production media containing (g/L) soluble starch (5), peptone (1), CaCl2.2H2O (0.25), Na2HPO4·2H2O (0.62), MgSO4.7H2O (0.25), and KH2PO4 (0.5). Each inoculated culture was incubated at 180 rpm and 37°C for 48 h. Each grown culture was centrifuged at 10,000 g for 2 min, and cell-free supernatant was collected. The amylase activity of each isolate was determined by the 3,5-DNS method given by Bernfeld [25]. A reaction mixture was prepared by adding 300 μL of cell-free supernatant in 600 μL of soluble starch (1% in 50 mM phosphate buffer, pH 7.0) and incubated at 37°C for 30 min in a water bath. After incubation, 900 μL of DNS solution was added, and immediately all tubes were transferred to the boiling water bath for 15–20 min. 900 μL of distilled water was added, and optical density (OD) was determined at 540 nm using a spectrophotometer. All experiments were performed in triplicate. Bacterial isolates, which produce the best clear hydrolytic zone or high enzymatic activity, were selected for further studies and maintained on slants at 4°C and sub-cultured every 15 days.
2.5. Identification of Amylase-Producing Bacteria
The selected bacteria were subjected to different morphological and biochemical tests and compared with Bergey’s Manual of Systematic Bacteriology [26] to identify their respective species.
2.5.1. Cultural characteristics
The bacterial isolates were characterized with respect to color, size, shape, and pigmentations. Isolates were tested by Gram staining technique and observed under a microscope in oil immersion (×100) magnification. This technique was performed to differentiate between Gram-positive and Gram-negative bacteria [27].
2.5.2. Biochemical characteristics
Biochemical tests of the isolates, such as the catalase test, Methyl Red-Voges Proskaur (MR-VP) test, urease test, indole production, and oxidase test, were identified and compared, as described in Bergey’s Manual of Systematic Bacteriology [26].
2.5.2.1. Indole production
A loopful of the test culture was added to the tryptone broth and incubated for 24 h at 37°C. 1 mL of Kovac’s reagent was added to the culture broth after incubation and observed for the formation of a pink ring [28].
2.5.2.2. MR-VP test
GPB was prepared for the MR-VP reaction containing g/L (peptone 7.0, glucose 5.0, and phosphate buffer 5.0). A loopful of the test culture was inoculated into GPB and incubated at 37°C for 24–48 h. After incubation, the grown culture was examined for the MR and VP tests. For the MR test, five drops of methyl red indicator were added into the grown culture and observed for the development of red color. For the VP test, 0.6 mL of α-naphthol and 0.4 mL of KOH solution were added into the grown culture, and the red color development was observed after 15–60 min [29].
2.5.2.3. Oxidase test
The cytochrome oxidase test is helpful in determining the types of bacteria that produce the cytochrome c oxidase enzyme. A filter paper disc (6 mm diameter) was treated with 1% tetra-methyl-p-phenylenediamine dihydrochloride (TMPPD), a synthetic electron donor, and dried it at room temperature. The bacterial colony from each culture grown on a starch agar plate was picked up using the edge of a sterile wooden toothpick and transferred onto TMPPD filter paper disc. Within 10 seconds, a blue-purple color shift can be observed [30,31].
2.5.2.4. Catalase test
A single bacterial colony from a starch agar plate was picked up with the help of a sterile nichrome wire loop and transferred to the surface of a sterilized dry glass slide. A few drops of a 3% hydrogen peroxide (H2O2) solution were added onto a bacterial glass slide and observed for the formation of gas bubbles or effervescence [26].
2.5.2.5. TSI agar test
A loopful of test culture was streaked on TSI agar slant medium containing g/L (peptone 20.0, hydrolysate 6.0, sodium chloride 5.0, glucose 1.0, lactose 10.0, sucrose 10.0, ferrous citrate 0.30, sodium thiosulfate 0.30, phenol red 0.025, and agar 12.0) and stabbed the same nichrome wire loop on the butt of the slant. The inoculated slant medium was incubated at 37°C for 24 h and was observed for the acid/gas/H2S production in the butt as well as on the slant [32].
2.6. Molecular Identification of Bacterial Strain
Two bacterial isolates with the highest amylase activity were sent to Yaazh Xenomics, Coimbatore, Tamil Nadu, India, for 16S rRNA gene sequencing. These bacteria were identified using polymerase chain reaction amplification of 16S rRNA using the universal primers 27F (5′AGAGTTTGATCCTGGCTCAG3′) and 1492R (5′TACGGTTACCTTGTTACGACTT3′) followed by sequencing with the help of the ABI 3730xl sequencer (Applied Biosystems). The 16S rRNA sequence was subjected to Basic Local Alignment Search Tool (BLAST) analysis against the nr/nt database to find homologous nucleotide sequences. This homologous DNA sequence was downloaded from the GenBank database. The isolated strain’s identification was validated using phylogenetic analysis of the 16S rRNA gene with molecular evolutionary genetics analysis (MEGA) 11.0 [33]. The 16S rRNA nucleotide sequences were subjected to multiple sequence alignment using the CLUSTAL W tool [34]. The phylogenetic tree was constructed by the maximum likelihood method.
2.7. Enzyme Production Medium
The isolates with the best zone of hydrolysis were grown in LB broth medium and incubated overnight at 37°C in a shaking incubator at 180 rpm. The amylase enzyme was produced by inoculating 500 μL of grown culture into 50 mL of enzyme production media containing (g/L) soluble starch (5.0), peptone (1.0), CaCl2.2H2O (0.25), MgSO4.7H2O (0.25), Na2HPO4·2H2O (0.62), and KH2PO4 (0.5) and incubated at 37°C for 48 h at 180 rpm. Grown cultures were centrifuged at 10,000 g for 2 min to collect supernatant for further estimation of enzyme activity [35].
2.8. Effect of Different Parameters on Amylase Production
A one-variable-at-a-time approach was used to optimize the effect of different variables on amylase production. Different variables such as incubation periods (12, 24, 36, 48, 60, 72, and 84 h), temperatures (35, 40, 45, 50, and 55°C), and pH ranges (5, 6, 7, 8, and 9) were selected to enhance enzyme production and enzyme activity. Amylase activity was determined using the 3,5-DNS method [1,36].
2.9. Enzyme Assay
The amylase activity of each isolate was determined by the DNS method given by Bernfeld [25]. A 300 μL of cell-free supernatant was added to 600 μL of soluble starch (1% in 50 mM phosphate buffer) and incubated for 30 min in a water bath. The reaction was stopped by the addition of 900 μL of DNS solution, and immediately, all tubes were transferred to the boiling water bath for 15 min. 900 μL of distilled water was added, and OD was determined at 540 nm using a spectrophotometer. One unit of enzymatic activity (U) is defined as 1 mL of enzyme solution that catalyzes starch hydrolysis to produce 1 μg of maltose per minute under the given assay conditions [37,38].
2.10. Effect of Temperature, pH, and Reaction Time on Amylase Activity
The effect of temperature on amylase activity was investigated by making a 900 μL reaction mixture containing 300 μL cell-free supernatant with 600 μL soluble starch (1% in 50 mM phosphate buffer, pH 7.0) and incubated at various temperatures (35°C, 40°C, 45°C, 50°C, and 55°C) for 20 min. The amount of reducing sugar was determined using the DNS method [39].
The influence of pH on the amylase activity was checked by adding 300 μL of cell-free supernatant into 600 μL of soluble starch (1% in 50 mM phosphate buffer of pH 5.0–8.0 and pH 8.0–9.0 in 50 mM Tris-HCl buffer). The reaction mixture was incubated for 20 min at 40°C, and the amount of sugar reduced was determined using the DNS method [39].
Incubation time had been optimized for the amylase activity by incubating the reaction mixture at different incubation times (10–60 min), and enzyme activity was determined by the DNS method [36].
2.11. Substrate Specificity Determination
Various substrates such as CMC, p-nitrophenyl- β-D-glucopyranoside (pNPG), beechwood xylan, amylum, and apple pectin were used to determine the substrate specificity of the crude extract of bacterial isolates AD2 and AD4 [40]. The 900 μL reaction mixture containing 600 μL of 1% (w/v) substrate dissolved in 50 mM phosphate buffer, pH 7.0, and 300 μL of crude enzyme was initially incubated at 37°C for 20 min and further incubated for 10 min in a boiling water bath after the addition of DNS solution. Finally, absorbance was measured at 540 nm using a spectrophotometer [41].
2.12. Analysis of Hydrolyzed Products by TLC
A reaction mixture containing 300 μL of substrate (1% [w/v] soluble starch) in 50 mM citrate phosphate buffer under their respective optimum conditions and 150 μL of enzyme was incubated for 60 min. The reaction was terminated by incubating the mixture in a boiling water bath for 10 min. 1350 μL of ethanol was added to the reaction mixture and centrifuged at 10,000 g for 10 min [42]. The reaction mixture was concentrated to 500 μL by evaporating in a hot air oven. 0.8 μL of reaction mixture and the standard glucose and maltose (1 mg/mL) were loaded on a TLC plate (TLC Silica gel 60 F254, Merck India Ltd.) [43]. The chromatogram was developed with a mobile phase of n-butanol/ethanol/water in the ratio of 5:3:2. The end product could be seen by immersing the TLC plate in a solution containing 0.2% orcinol in sulfuric acid: methanol (1:9) (v/v), followed by drying the TLC plate in a hot air oven at 95°C for 10 min to reveal the spots [44].
2.13. Data Analysis
The data were analyzed using Microsoft Excel and GraphPad Prism version 8, and all graphs were plotted using GraphPad Prism version 8. The phylogenetic tree was generated using MEGA 11 software.
3. RESULTS AND DISCUSSION
3.1. Isolation and Screening of Amylase-Producing Bacteria
A fecal sample of deer (A. axis) was collected from its natural habitat at the Sayaji Baug Zoo, Vadodara, and spreading of the deer fecal sample on a starch agar plate resulted in four distinct colonies based on differences in colony morphology. Among the four bacterial isolates, only two isolates designated as AD2 and AD4 exhibited clear zones of hydrolysis around their colonies on the starch agar plates after performing Lugol’s iodine staining. Clear zones around the bacterial colonies after staining with Lugol’s iodine on the starch agar plates are shown in Supplementary Figure S1. This finding implies that the isolates have the ability to produce amylase enzymes, which break down starch into simpler sugars. For quantitative analysis, isolates with a significant zone of hydrolysis were grown in enzyme production medium. Out of these four isolates, AD2 and AD4 showed good initial amylase activity after 24 h incubation at 37°C [Table 1].
Table 1: Initial amylolytic activity of amylase-producing isolates after incubation of 24 h at 37°C
| Isolate | Amylase activity (U/mL) |
|---|---|
| AD1 | 1.94 |
| AD2 | 6.64 |
| AD3 | 2.75 |
| AD4 | 9.89 |
The identification of amylase-producing bacteria, namely, isolates AD2 and AD4, in the fecal sample emphasizes the existence of an enzymatic machinery within their gut microbiota that facilitates the breakdown of complex carbohydrates. Amylase activity of bacterial isolates AD2 and AD4 are somewhat similar to the activity of some known amylolytic isolates, for example, Bacillus licheniformis (15.6 U/mL) and Bacillus coagulans (14.5 U/mL) [45], Anoxybacillus mongoliensis (4.4 U/mL) and Anoxybacillus flavithermus (4.3 U/mL) [36], B. subtilis (8.96 U/mL) and A. veronii (7.58 U/mL) [46], Lactococcus chungangensis CAU 28T (8.86 U/mL), and Lactococcus lactis ssp. lactis KCTC 3769T (9.03 U/mL) [47].
3.2. Identification of Potential Amylase-Producing Bacteria
Bacterial isolates were initially identified using colony morphology and the Gram-staining method. The morphology of the isolates was identified using the Gram-staining test [Supplementary Figure 2]. Isolate AD2 was found to be Gram-negative short rods, arranged individually or in pairs, as observed under a microscope in an oil immersion lens. Isolate AD4 was found to be rod-shaped, non-spore-forming, Gram-negative bacteria under the microscope. Isolate AD2 showed a positive test for indole production, catalase test, and citrate utilization, whereas it was negative for the oxidase test. On the other hand, AD4 showed positive results for methyl red, the Voges-Proskauer test, and indole production while being found to be negative for oxidase and H2S production. The bacterial isolates AD2 and AD4 were identified as Enterobacter spp. and Aeromonas spp., respectively, based on both biochemical and morphological traits when compared to the Bergey’s Manual of Systematic Bacteriology [26]. Cultural characteristics and biochemical characteristics of amylolytic isolates are shown in Table 2.
Table 2: Physiological and biochemical characteristics of isolates from fecal sample of spotted deer (Axis axis).
| Parameters | AD2 | AD4 |
|---|---|---|
| Gram’s reaction | Negative | Negative |
| Endospore staining | - | - |
| Motility | Motile | Motile |
| Cell size | Small | Small |
| Cell arrangement | Single, in pairs | Single |
| Pigmentation | Off-white colour | Creamy white |
| Indole production | + | + |
| Methyl red | - | + |
| Voges-Proskaur | - | + |
| Citrate utilization | + | + |
| Oxidase | - | - |
| Gelatin hydrolysis | - | + |
| Catalase | + | - |
| H2S from TSI | + | - |
| TSI slant color | - | - |
| TSI butt color | + | + |
| TSI gas production | + | + |
+: Positive; -: Negative, TSI: Triple sugar iron
3.3. Molecular Identification of Amylolytic Bacteria
Bacterial isolates (AD2 and AD4) were also subjected to 16S rRNA gene sequencing for molecular-based identification of bacteria. Bacterial isolate AD2 showed 99.17%, while AD4 showed 98.48% similarity in NCBI databases when compared using the BLAST. These results confirmed that AD2 and AD4 bacterial isolates belong to the genera Enterobacter and Aeromonas, respectively. The 16S rRNA gene nucleotide sequences from AD2 and AD4 were submitted in the NCBI’s GenBank database with the accession numbers OR361741 and OR226595, respectively [Table 3]. The phylogenetic tree of isolates AD2 and AD4 was generated using gene sequences in MEGA 11.0 by the maximum likelihood method, as shown in Figure 1.
Table 3: Percentages of identity of 16S rRNA gene sequences of amylolytic bacteria with closely related bacteria and their GenBank accession numbers.
| Bacterial isolate | Closely related bacteria | Query cover (%) | Identity (%) | E value | Deposited GenBank accession no. |
|---|---|---|---|---|---|
| AD2* | Enterobacter cloacae strain S7 | 100 | 99.17 | 0.0 | OR361741 |
| AD4* | Aeromonas veronii strain HTN02 | 100 | 98.48 | 0.0 | OR226595 |
*Strains submitted to Gene Bank-National Canter for Biotechnology Information
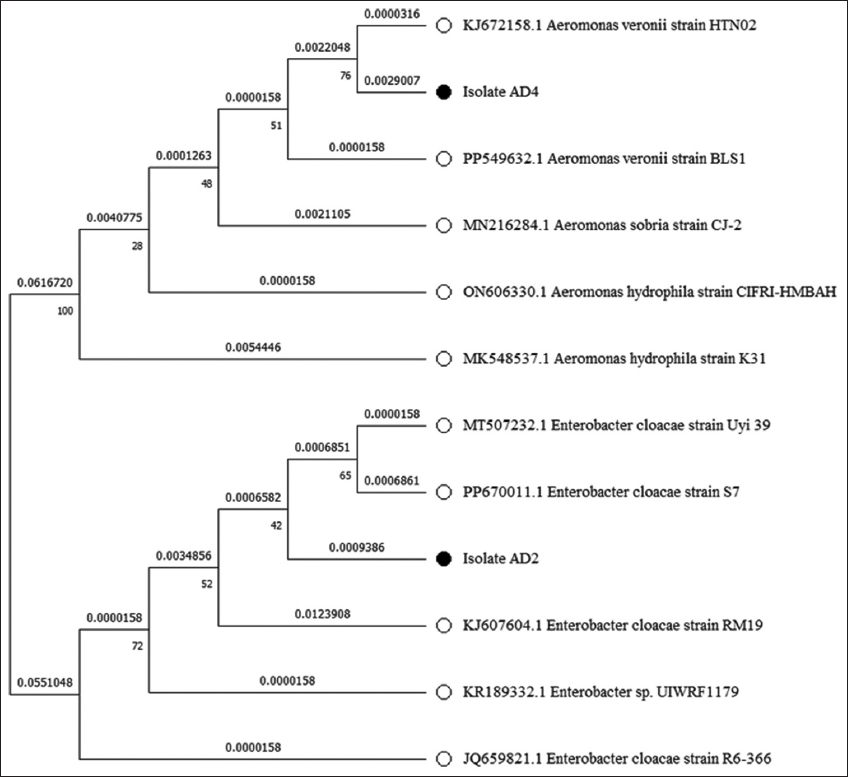 | Figure 1: Phylogenetic tree constructed from a comparative analysis of 16s rRNA gene sequences. 16S rRNA gene sequences showing the relationships of strains AD2 and AD4 with representatives of related genera of the families Enterobacteriaceae and Aeromonadaceae. The tree was made using the maximum-likelihood method. [Click here to view] |
3.4. Optimization of Culture Conditions for Amylase Production
3.4.1. Incubation time
Optimizing the incubation time is crucial for achieving optimum amylase production. E. cloacae produced maximum enzyme at 48 h, while A. veronii produced it at 60 h [Figure 2a]. After optimum incubation time, enzyme production declined substantially, likely due to microorganism cell death, nutrient fatigue, and byproduct accumulation in the cultivated medium. In addition, the cells exhibited decreased amylase enzyme biosynthesis during the decline phase of growth [48]. These varying incubation periods reveal differences in each isolate’s growth and enzymatic activity patterns, highlighting the need to optimize incubation times to achieve desired results. A similar kind of observation was also reported by Yassin et al. [1]; maximum amylase production from Bacillus spp. strain M8 was observed at 48 h of incubation, and Bacillus spp. strain M2 was observed at 60 h of incubation. In another study, maximum amylase production by B. subtilis MTCC 121 was achieved after 48 h of incubation [49].
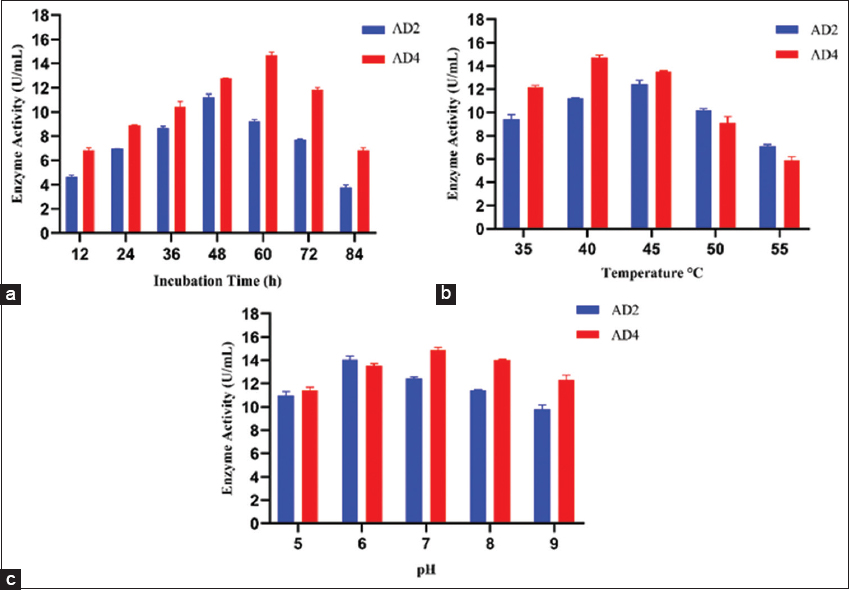 | Figure 2: Effect of different parameters on amylase production (a) effect of incubation time (12–84 h), (b) effect of incubation temperature (35–55°C), and (c) effect of different pH (5.0–9.0). [Click here to view] |
3.4.2. Incubation temperature
Temperature affects amylase synthesis in bacterial strains AD2 and AD4, with maximal enzyme production at 45°C and 40°C, respectively [Figure 2b]. This led to an increase in amylase production as the temperature rose and a modest drop after it exceeded the ideal temperature. Previous reports suggested that E. cloacae IIT-BT 08 exhibited maximum production at 45°C and E. cloacae strain D1 at 40°C [50,51]. Production of amylase by B. amyloliquefaciens peaked between 35°C and 40°C, while B. licheniformis showed maximum production at 40°C [52-54]. Bacillus atrophaeus NRC1 isolated from the honey sample produced maximum amylase at 45°C [1]. This optimal temperature for amylase production can be attributed to their environmental conditions and ecological niches. Many amylolytic bacterial strains isolated from fecal samples and agricultural soil samples showed optimum temperatures between 40°C and 45°C. In addition, bacteria that live in the gut are used to conditions that are close to the host’s body temperature. Spotted deer (A. axis) frequently attain a body temperature of 38.5°C [55]. At these temperatures, the metabolic machinery is adjusted to operate at its best, including the control of gene expression for the manufacture of enzymes. In addition, genetic factors influencing enzyme stability, thermal tolerance, and the bacteria’s ability to adapt to temperature variations likely contribute to these differences in optimal temperature for amylase production [56,57]. Since both isolates in this study were obtained from similar environments, they exhibited comparable optimal temperature ranges. From the industrial standpoint, the ideal temperature range for amylase in the detergent industry is 40–60°C for the best detergent performance. While the amylase enzyme works best at temperatures of 30–60°C for de-sizing processes in textile industries. Therefore, the enzyme discovered in this study might be suitable for the detergent and textile industries [58,59].
3.4.3. Incubation pH
To ascertain the ideal pH for amylase production, the pH is varied between 5.0 and 9.0. It resulted in the maximum amylase production at pH 6.0 for AD2 and pH 7.0 for AD4 [Figure 2c]. The majority of the starch-degrading bacterial strains had a pH range of 6.0–7.0 for proper growth and enzyme production [60]. Adjusting the pH of the medium can improve the efficiency of amylase production and subsequent biochemical processes involving these isolates. This outcome was comparable to E. cloacae IIT-BT 08’s optimal pH of 6.0 for amylase production [50]. A. mongoliensis and A. veronii NS07 were also reported to have optimal enzyme production at pH 7.0 [36,61]. The pH of fresh fecal samples of herbivores was in a range from 5.82 to 8.32, and bacterial diversity has been demonstrated to peak at neutral pH and progressively decline below and above neutral pH [62,63]. Multiple lines of evidence imply that the activity and structure of numerous membrane proteins are influenced by ambient pH, which has direct consequences on bacterial metabolic rates [64,65]. The strain and culture medium also have a major role in determining the ideal pH for growth and amylase synthesis. From a biotechnological perspective, industrial processes are better suited for amylolytic organisms that function ideally at pH 5.0–8.0. Numerous industrial applications, including the textile industry, the production of fuel, the chocolate industry, and liquefaction, are compatible with this pH range. Furthermore, the amylase enzyme works best at a pH range of 5.5–7.0 for de-sizing processes in textile industries. These enzymes have the benefit of being starch-specific, meaning they can eliminate starch without causing harm to the support fabric [58].
After the optimization through OVAT approach, E. cloacae FZAD generated (14.13 U/mL) of enzyme at pH 6.0 and 45°C after 48 h of incubation, which is 2.12-fold from 6.64 U/mL of unoptimized medium. On the other hand, OVAT enhanced the amylase productivity of A. veronii FZD by 1.50-fold, from 9.89 U/mL of unoptimized medium to 14.92 U/mL at pH 7.0, 40°C, and 60 h of incubation. In recent times, many newly isolated amylolytic strains were optimized to enhance amylase production and activity. Saad et al. optimized the production parameter for B. licheniformis WF67 to increase productivity by 1.90-fold from 3.48 U/mL to 6.63 U/mL [66]. In another study with Bacillus aryabhattai KIIT BE-1, 1.39-fold increased enzyme activity (4.16 U/mL) was observed after optimization [67]. Some other examples include Citrobacter portucalensis A-4 (4.96 U/mL), Streptomyces griseorubens NBR14 (7.72 U/mL), and B. cereus spH1 (8.5 U/mL); all observed increased productivity after optimization [68-70].
3.5. Enzyme Characterization
The cell-free extracts of AD2 and AD4 were analyzed for reaction time. As the reaction time increased, the activity of the amylase enzyme steadily decreased. For AD2 and AD4, optimal amylase activity was observed in 20 and 30 min of reaction time, respectively [Figure 3a]. The amylase activities of AD2 and AD4 cell-free extracts were analyzed for optimum temperature. Cell-free extracts of both AD2 and AD4 isolates showed maximum activity at 40°C [Figure 3b]. Due to differences in their native environments and metabolic adaptations, different species of bacteria have different preferences for temperatures. Previous reports on the effect of temperature are in line with our finding; according to reports, maximum amylase activities by Aeromonas vernoii and Aeromonas jandaei peaked between 35 and 40°C [46]. On the other hand, optimum amylase activities by E. cloacae strain D1 and E. cloacae IIT-BT 08 were observed at 37°C [50]. According to a different investigation carried out in the early 21st century by Coronado et al., Halomonas meridiana showed the best amylase activity at 40°C [71].
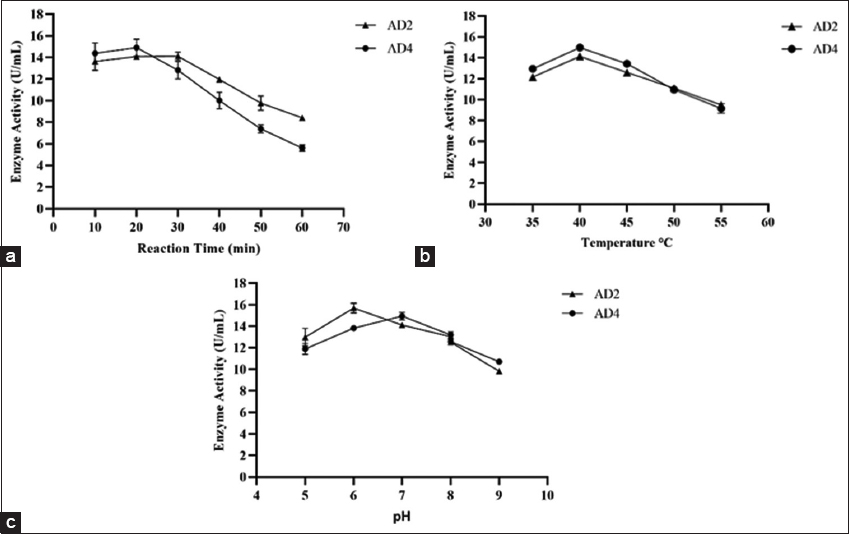 | Figure 3: Effect of different parameters on amylolytic activity (a) effect of reaction time on enzymatic activity, (b) effect of temperature on amylase activity, and (c) effect of pH on amylase activity. [Click here to view] |
The cell-free extracts of AD2 and AD4 were analyzed for optimum pH. AD2 exhibited maximal amylase activity at pH 6.0, while the optimum pH for AD4 was pH 7.0 [Figure 3c]. These findings are in line with previous reports by Suman et al. [46]. According to the report, A. vernoii, A. jandaei, and Aeromonas hydrophila had the optimal enzyme activity at pH 7.0. Another finding suggested that while Bacillus spp. WA21 and B. atrophaeus NRC1 exhibit maximum amylase activity at pH 6.0, B. amyloliquefaciens had the optimal pH of the amylase at 7.0 [72,73]. Pseudomonas fluorescens was also reported to have optimal enzyme activity at pH 7.0 [74]. Under neutral pH, amylolytic bacterial strains A. veronii and E. cloacae were both stable and gave maximum activity. This was comparable to the amylolytic activity of other amylase-producing bacterial species, which remained active and stable at neutral pH conditions. Consequently, the amylases found in this study were more suited for applications requiring moderate conditions, including those seen in the industrial sectors such as food and baking industries [39,72].
3.6. Substrate Specificity
The ability of the enzyme to breakdown several substrates compared with starch as 100% hydrolysis was examined [Table 4]. The substrate specificity investigation of the amylase-producing isolates AD2 and AD4 yielded intriguing results. Both isolates exhibited a great tendency for amylum as a substrate, as evidenced by their high specificity toward amylum. This is consistent with the role of amylase enzymes in breaking down starch molecules into simpler sugars. However, AD2 and AD4 showed very little activity when tested against apple pectin. Pectin, a complex polysaccharide present in plant cell walls, had poor activity with these isolates, suggesting a restricted capacity to breakdown this substrate. Neither isolate was significantly selective against other substrates such as pNPG, beechwood xylan, or CMC. Cellulase and xylanase enzymes frequently employ CMC and beechwood xylan as model substrates, respectively. The absence of noteworthy activity against these substrates implies that AD2 and AD4 have a more specific role in starch hydrolysis and are effective in breaking down cellulose or xylan. This information is useful for understanding the enzymatic activities of both isolates, and it can help guide their possible uses in sectors that need starch hydrolysis.
Table 4: Substrate specificity of amylase enzyme.
| Substrate | Relative activity (%) AD2 | Relative activity (%) AD4 |
|---|---|---|
| Starch | 100 | 100 |
| Carboxymethylcellulose | 1.82 | 1.99 |
| p-nitrophenyl-b-d-glucopyranoside | 0.64 | 0.27 |
| Apple pectin | 8.26 | 7.35 |
| Beechwood xylan | 1.12 | 1.22 |
3.7. TLC Analysis of Enzyme
The utilization pattern of starch by E. cloacae FZAD and A. veronii FZD was analyzed by running the supernatant on TLC [Figure 4]. TLC analysis revealed the presence of maltose as the only hydrolysis end product from starch. These findings suggest that it can be either β-amylase (4-α-D-glucan maltohydrolase) as it catalyzes the breakdown of alternate α-1,4-glycosidic linkages in starch at the non-reducing end or maltogenic amylase (glucan 1,4-α-maltohydrolase) acts on α-1,4 glycosidic bonds in starch and produces maltose as the main end product. Similar results were reported, where starch was hydrolyzed by β-amylase from Bacillus spp. KYJ, Lactobacillus fermentum, and only maltose were detected as major a end product [74-76].
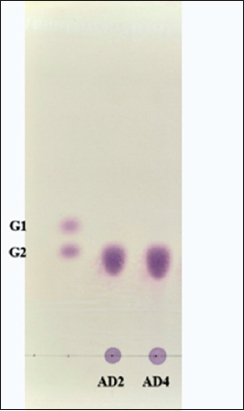 | Figure 4: Analysis by thin layer chromatography of the hydrolysis products by the amylase produced by Enterobacter cloacae FZAD and Aeromonas veronii FZD. G1- Glucose; G2- Maltose; the mobile phase of n-butanol/ethanol/water in the ratio of 5:3:2 was used. [Click here to view] |
4. CONCLUSION
Fecal sample analysis resulted in the screening of two amylolytic bacterial strains, AD2 and AD4. Isolates AD2 and AD4 were identified as E. cloacae and A. veronii after the morphological, biochemical, and molecular characterization. This research identified the precise circumstances in which these amylolytic bacteria exhibited their highest level of amylase production and activity. The optimal enzyme production was observed at 45°C, a pH of 6.0, and 48 h of incubation for AD2 and 40°C, a pH of 7.0, and an incubation of 60 h for AD4. The crude extract of E. cloacae FZAD exhibited maximum amylase activity of 14.13 U/mL, while crude extract from A. veronii FZD displayed maximum activity of 14.92 U/mL. Both bacterial isolates possessed several distinct benefits over other amylolytic bacteria, including activity over a wide range of pH and temperature, which makes them suitable for a variety of industrial processes, including detergent industries, brewing, sugar production, and de-sizing processes in textile industries. Furthermore, further research is needed to ramp up the production of amylase for the commercial use of these bacterial strains, including purification and kinetic investigations on free enzymes.
5. ACKNOWLEDGMENTS
We would like to thank Mr. Manav Mehta (Education Officer) and Dr. Pratyush Patankar (Curator) of Sri Sayaji Baug Zoo, Vadodara Municipal Corporation, Gujarat, for co-operating with us and supporting us during the collection of the fecal samples.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
8. CONFLICTS OF INTEREST
The author reports no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This article does not contain any studies involving animals performed by any of the authors.
10. DATA AVAILABILITY
Data of 16S rRNA gene sequences supporting the results of this research are available in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers OR361741 and OR226595. This published article contains all other data produced during the course of this study.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Yassin SN, Jiru TM, Indracanti M. Screening and characterization of thermostable amylase-producing bacteria isolated from soil samples of Afdera, Afar region, and molecular detection of amylase-coding gene. Int J Microbiol. 2021;1-14. [CrossRef]
2. Eichler J. Biotechnological uses of archaeal extremozymes. Biotechnol Adv. 2001;19(4):261-78. [CrossRef]
3. Nguyen T, Loan T, Van Thuoc D. High amylase production by a novel strain of Bacillus amyloliquefaciens M37 isolated from can Gio mangrove forest, Vietnam. Biointerface Res Appl Chem. 2021;12(4):4675-85. [CrossRef]
4. Silaban S, Marika DB, Simorangkir M. Isolation and characterization of amylase-producing amylolytic bacteria from rice soil samples. J Phys Conf Ser. 2020;1485(1):012006. [CrossRef]
5. Van Der Maarel MJ, Van Der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L. Properties and applications of starch-converting enzymes of the a-amylase family. J Biotechnol. 2002;94(2):137-55. [CrossRef]
6. Joshi N, Andhare P, Marchawala F, Bhattacharya I, Upadhyay D. A study on amylase:Review. Int J Biol Pharm Allied Sci. 2021;10(4):333-40. [CrossRef]
7. Benjamin S, Smitha RB, Jisha VN, Pradeep S, Sajith S, Sreedevi S, et al. A monograph on amylases from Bacillus spp. Adv Biosci Biotechnol. 2013;4(2):227-41. [CrossRef]
8. Alves KJ, Da Silva MC, Cotta SR, Ottoni JR, Van Elsas JD, De Oliveira VM, et al. Mangrove soil as a source for novel xylanase and amylase as determined by cultivation-dependent and cultivation-independent methods. Braz J Microbiol. 2019;51(1):217-28. [CrossRef]
9. Kafilzadeh F, Dehdari F. Amylase activity of aquatic actinomycetes isolated from the sediments of mangrove forests in south of Iran. J Aquat Res. 2015;41(2):197-201. [CrossRef]
10. Paul JS, Gupta N, Beliya E, Tiwari S, Jadhav SK. Aspects and recent trends in microbial a-amylase:A review. Appl Biochem Biotechnol. 2021;193:2649-98. [CrossRef]
11. Islam T. Isolation of Amylase Producing Bacteria from Soil and Identification by 16S rRNA Gene Sequencing and Characterization of Amylase. Dhaka, Bangladesh:BRAC University;2016.
12. Ashwini K, Gaurav K, Karthik L, Bhaskara Rao KV. Optimization, production and partial purification of extracellular a-amylase from Bacillus sp. Marini. Arch Appl Sci Res. 2021;3:33-42.
13. Alves PD, de Faria Siqueira F, Facchin S, Horta CR, Victória MN, Kalapothakis E. Survey of microbial enzymes in soil, water, and plant microenvironments. Open Microbiol J. 2014;8:25-31. [CrossRef]
14. Abdullah R, Shaheen, N, Iqtedar M, Naz S, Iftikhar T. Optimization of cultural conditions for the production of a-amylase by Aspergillus niger (BTM-26) in solid state fermentation. Pak J Bot. 2014;46:1071-8.
15. Pandey A. Solid-state fermentation. Biochem Eng J. 2003;13:81-4. [CrossRef]
16. Saxena R, Singh R. Amylase production by solid-state fermentation of agro-industrial wastes using Bacillus sp. Braz J Microbiol. 2011;42(4):1334-42. [CrossRef]
17. Asad W, Asif M, Rasool S. Extracellular enzyme production by indigenous thermophilic bacteria:Partial purification and characterization of a-amylase by Bacillus sp. WA21. Pak J Bot. 2011;43:1045-52.
18. Abo-Kamer AM, Abd-El-Salam IS, Mostafa FA, Mustafa AE, Al-Madboly LA. A promising microbial a-amylase production, and purification from Bacillus cereus and its assessment as antibiofilm agent against Pseudomonas aeruginosa pathogen. Microb Cell Fact. 2023;22(1):141. [CrossRef]
19. Anto H, Trivedi U, Patel K. a-Amylase production by Bacillus cereus MTCC 1305 using solid-state fermentation. Food Technol Biotechnol. 2006;44(2):241-5.
20. Grubb P. Artiodactyla. In:Species of the World. A Taxonomic and Geographic Reference. 3rd ed. Baltimore, USA:Johns Hopkins University Press;2005. 637-722.
21. Raman TS. Chital Axis axis. In:Mammals of South Asia. Vol. 2. Hyderabad:Universities Press, 2015. 192-222.
22. Kanimozhi M, Johny M, Gayathri N, Subashkumar R. Optimization and production of a -amylase from Halophilic Bacillus species isolated from mangrove soil sources. J Appl Environ Microbiol. 2014;2(3):70-3.
23. Ogbonna CN, Okpokwu NM, Okafor CU, Onyia CE. Isolation and screening of amylase producing fungi obtained from garri processing site. Int J Biotechnol Food Sci. 2014;2(5):88-93.
24. Aynadis TH, Tilahun BG, Gulelat DH. Thermostable alpha-amylase from geothermal sites of Ethiopia (Afar Region):Isolation, purification and characterization. Greener J Biol Sci. 2013;3:61-73. [CrossRef]
25. Bernfeld P. Amylases, a and b. Methods Enzymol 1955;1:149-58. [CrossRef]
26. Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, et al. Bergey's Manual of Systematic Bacteriology. New York:Springer-Verlag;2011.
27. Bartholomew JW, Mittwer T. The Gram stain. Bacteriol Rev. 1952;16(1):1-29. [CrossRef]
28. Biswas S, Saber MA, Tripty IA, Karim MA, Islam MA, Hasan MS, et al. Molecular characterization of cellulolytic (endo- and exoglucanase) bacteria from the largest mangrove forest (Sundarbans), Bangladesh. Ann Microbiol. 2020;70(1):68.[CrossRef]
29. Tille PM. Bailey and Scott's Diagnostic Microbiology. Missouri:Mosby;2021.
30. Roy B, Das T, Bhattacharyya S. Overview on old and new biochemical test for bacterial identification. J Surg Case Rep. 2014;6:23-8. [CrossRef]
31. Winn WC, Allen SC, Janda WM, Koneman EW, Procop GW, Schreckenberger P, et al. Color Atlas and Textbook of Diagnostic Microbiology. Philadelphia, PA:Lippincott Williams and Wilkins;2006.
32. Swenson JM, Patel JB, Jorgensen JH. Special phenotypic methods for detecting Antibacterial resistance. In:Manual of Clinical Microbiology. Washington, DC:ASM Press;2011. [CrossRef]
33. Tamura K, Dudley J, Nei M, Kumar S. MEGA4:Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596-9. [CrossRef]
34. Thompson J, Higgins DG, Gibson TJ. CLUSTAL W:Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673-80. [CrossRef]
35. Bozic N, Ruiz J, Lopez-Sant?n J, Vujcic Z. Production and properties of the highly efficient raw starch digesting a- amylase from a Bacillus licheniformis ATCC 9945. Biochem Eng J. 2011;53(2):203-209. [CrossRef]
36. Sharif S, Shah AH, Fariq A, Jannat S, Rasheed S, Yasmin A. Optimization of amylase production using response surface methodology from newly isolated thermophilic bacteria. Heliyon. 2023;9(1):e12901. [CrossRef]
37. Wang J, Bao F, Wei H, Zhang Y. Screening of cellulose-degrading bacteria and optimization of cellulase production from Bacillus cereus A49 through response surface methodology. Sci Rep. 2024;14(1):7755. [CrossRef]
38. Singh S, Moholkar VS, Goyal A. Isolation, identification, and characterization of a cellulolytic Bacillus amyloliquefaciens strain SS35 from rhinoceros dung. ISRN Microbiol. 2013;3:1-7. [CrossRef]
39. Klinfoong R, Thummakasorn C, Ungwiwatkul S, Boontanom P, Chantarasiri A. Diversity and activity of amylase-producing bacteria isolated from mangrove soil in Thailand. Biodivers J. 2022;23(10):5519-5531. [CrossRef]
40. Yin LJ, Huang PS, Lin HH. Isolation of cellulase-producing bacteria and characterization of the cellulase from the isolated bacterium Cellulomonas Sp. YJ5. J Agric Food Chem. 2010;58(17):9833-7. [CrossRef]
41. Miller GL. Use of dinitrosalisylic acid reagent for determination of reducing sugars. Anal Chem. 1959;31(3):426-428. [CrossRef]
42. Kumar K, Rajulapati V, Goyal A. In vitro prebiotic potential, digestibility and biocompatibility properties of laminari-oligosaccharides produced from curdlan by b-1,3-endoglucanase from Clostridium thermocellum. 3 Biotech. 2020;10(6):241. [CrossRef]
43. Chakraborty S, Khopade A, Kokare C, Mahadik K, Chopade B. Isolation and characterization of novel a-amylase from marine Streptomyces sp. D1. J Mol Catal B Enzym. 2008;58(1-4):17-23. [CrossRef]
44. Silva TM, De Oliveira M, Somera AF, Jorge JA, Terenzi HF, De Lourdes TM, et al. Thermostable saccharogenic amylase produced under submerged fermentation by filamentous fungus Penicillium purpurogenum. Braz J Microbiol. 2011;42(3):1136-40. [CrossRef]
45. Medda S, Chandra AK. New strains of Bacillus licheniformis and Bacillus coagulans producing Thermostable α-amylase active at alkaline pH. J Appl Bacteriol. 1980;48(1):47-58. [CrossRef]
46. Suman S, Rani L, Hussain MZ, Kashyap S, Verma R. Biochemical analysis and optimization of Aeromonas and Bacillus species for a-amylase production isolated from soil samples of Ranchi, Jharkhand. Eur Chem Bull. 2022;11:1447-1453. [CrossRef]
47. Konkit M, Kim W. Activities of amylase, proteinase, and lipase enzymes from Lactococcus chungangensis and its application in dairy products. J Dairy Sci. 2016;99(7):4999-5007. [CrossRef]
48. Aiyer PV. Amylases and their applications. Afr J Biotechnol. 2005;4(13):1525-9.
49. Raul D, Biswas T, Mukhopadhyay S, Das SK, Gupta S. Production and partial purification of alpha amylase from Bacillus subtilis (MTCC 121) using solid state fermentation. Biochem Res Int. 2014;2014:568141. [CrossRef]
50. Kumar N, Das D. Production and purification of a-amylase from hydrogen producing Enterobacter cloacae IIT-BT 08. Bioprocess Eng. 2000;23(2):205-8. [CrossRef]
51. Ezebuiro V, Obichi EA, Minimah SO, Ezebuiro NC, Akinido E. Optimization of a-amylase production by Enterobacter cloacae strain D1 isolated from cassava effluent-impacted soil using response surface methodology. Asian J Biotechnol. 2022;8:68-80. [CrossRef]
52. Uygut MA, Tanyildizi M?. Optimization of alpha-amylase production by Bacillus amyloliquefaciens grown on orange peels. J Sci Technol Trans A Sci. 2016;42(2):443-9. [CrossRef]
53. Nusrat A, Rahman SR. Comparative studies on the production of extracellular and alpha Amylase by three mesophilic Bacillus isolates. Bangladesh J Microbiol. 1970;24(2):129-32. [CrossRef]
54. Ikram-Ul-Haq N, Ashraf H, Iqbal J, Qadeer MA. Production of alpha amylase by Bacillus licheniformis using an economical medium. Bioresour Technol. 2002;87(1):57-61. [CrossRef]
55. Arnemo J, Kreeger T. Handbook of Wildlife Chemical Immobilization. 5th ed. United Kingdom:Sunquest Publishing;2007.
56. Singh S, Bajaj BK. Medium optimization for enhanced production of protease with industrially desirable attributes from Bacillus subtilis K-1. Chem Eng Commun. 2014;202(8):1051-60. [CrossRef]
57. Fatokun E, Nwodo U, Okoh A. Classical optimization of cellulase and xylanase production by a marine Streptomyces species. Appl Sci. 2016;6(10):286. [CrossRef]
58. Mojsov K. Application of enzymes in the textile industry:A review. In:II International Congress Engineering, Ecology and Materials in the Processing Industry. 2011. 230-9.
59. Dahiya P, Rathi B. Characterization and application of alkaline alpha-amylase from Bacillus licheniformis MTCC1483 as detergent additive. Int Food Res J. 2015;22(3):1293-7.
60. Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B. Microbial a-amylases:A biotechnological perspective. Process Biochem. 2003;38(11):1599-616. [CrossRef]
61. Samie N, Noghabi KA, Gharegozloo Z, Zahiri HS, Ahmadian G, Sharafi H, et al. Psychrophilic a-amylase from Aeromonas veronii NS07 isolated from farm soils. Process Biochem. 2012;47(9):1381-7. [CrossRef]
62. Ndlela LL, Schmidt S. Evaluation of wild herbivore faeces from South Africa as a potential source of hydrolytically active microorganisms. SpringerPlus. 2016;5(1):118. [CrossRef]
63. Luan L, Jiang Y, Dini-Andreote F, Crowther TW, Li P, Bahram M, et al. Integrating pH into the metabolic theory of ecology to predict bacterial diversity in soil. Proc Nat Acad Sci. 2023;120(3):e2207832120. [CrossRef]
64. Padhi S, Reddy LK, Priyakumar UD. pH-mediated gating and formate transport mechanism in the Escherichia coli formate channel. Mol Simulat. 2017;43(13-16):1300-6. [CrossRef]
65. LüW, Du J, Wacker T, Gerbig-Smentek E, Andrade SL, Einsle O. PH-Dependent gating in a FOCA formate channel. Science. 2011;332(6027):352-4. [CrossRef]
66. Saad WF, Othman AM, Abdel-Fattah M, Ahmad MS. Response surface methodology as an approach for optimization of a-amylase production by the new isolated thermotolerant Bacillus licheniformis WF67 strain in submerged fermentation. Biocatal Agric Biotechnol. 2021;32:101944. [CrossRef]
67. Ojha SK, Singh PK, Mishra S, Pattnaik R, Dixit S, Verma SK. Response surface methodology-based optimization and scale-up production of amylase from a novel bacterial strain, Bacillus aryabhattai KIIT BE-1. Biotechnol Rep. 2020;27:e00506. [CrossRef]
68. Liaqat UE, Naz S, Hussain HI. Optimization and production of highly stable amylase isolated from Citrobacter portucalensis using low-cost agro-industrial wastes as substrates:Prospective therapeutics. Kuwait J Sci. 2023;51(1):100156. [CrossRef]
69. El-Sayed MH, Gomaa AE, Atta OM, Hassane AM. Characteristics and kinetics of thermophilic actinomycetes?amylase production on agro-wastes and its application for ethanol fermentation. World J Microbiol Biotechnol. 2024;40(8):205. [CrossRef]
70. Hmida BB, Mabrouk SB, Fendri A, Hmida-Sayari A, Sayari A. Optimization of newly isolated Bacillus cereus a-amylase production using orange peels and crab shells and application in wastewater treatment. 3 Biotech. 2024;14(4):119. [CrossRef]
71. Coronado MJ, Vargas C, Hofemeister J, Ventosa A, Nieto JJ. Production and biochemical characterization of an α-amylase from the moderate halophile Halomonas meridiana. FEMS Microbiol Lett. 2000;183(1):67-71. [CrossRef]
72. Luang-In V, Yotchaisarn M, Saengha W, Udomwong P, Deeseenthum S, Maneewan K. Isolation and identification of amylase-producing bacteria from soil in Nasinuan community forest, Maha Sarakham, Thailand. Biomed Pharmacol J. 2019;12(3):1061-8. [CrossRef]
73. Abd-Elaziz AM, Karam EA, Ghanem MM, Moharam ME, Kansoh AL. Production of a novel a-amylase by Bacillus atrophaeus NRC1 isolated from honey:Purification and characterization. Int J Biol Macromol. 2020;148:292-301. [CrossRef]
74. Bakare M, Adewale I, Ajayi A, Shonukan O. Purification and characterization of cellulase from the wild-type and two improved mutants of Pseudomonas fluorescens. Afr J Biotechnol. 2005;4(9):898-904.
75. Kim YJ. Thin layer chromatogram by an extracellular b-amylase of Bacillus sp. KYJ 963 and its amino acid composition. J Life Sci. 2001;11(2):92-3.
76. Kocabay S, Çetinkaya S, Akkaya B, Yenidünya AF. Characterization of thermostable b-amylase isozymes from Lactobacillus fermentum. Int J Biol Macromol. 2016;93:195-202. [CrossRef]