1. INTRODUCTION
Soybean is the leading oilseed crop with a total production of 339.0 million metric tons in 2019–20 [1]. Crude oil extracted from soybean seeds is the source of most concentrated natural phospholipids, as it contains 1.5%–3.0% phospholipids, which is higher than any other oil. Phospholipids from crude soybean oil are removed during the degumming process and commercially known as lecithin, which is a mixture of phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidic acid (PA), and a fraction of crude vegetable oil [2]. Among different phospholipids present in lecithin, PC is the major component and has widespread uses in food, cosmetics, pharmaceuticals, plastic, and rubber industries [3,4]. PC also provides several health benefits as it helps in fetal brain development, reduces risk of cardiovascular disease and symptoms of ulcerative colitis, and improves liver health and lipolysis [5]. Therefore, soybean genotypes with high concentration of PC are desired.
After extraction of oil from soybean grains, soymeal left is high in protein. Among all the protein sources used as animal feed, soymeal accounts for approximately 69%, followed by rapeseed meal (13%), cottonseed meal (6%), sunflower meal (5%), and peanut meal (2%) [6]. In general, soybean seeds contain 38%–40% protein content on dry weight basis. Soymeal from Argentina, Brazil, China, and USA contains 46.7%, 49.3%, 45.1%, and 47.3% protein, respectively, while average protein content of Indian soymeal is 49.5% [7]. Increment in protein content in soymeal requires high protein soybean genotypes as protein content in soymeal depends upon its level in the seeds.
In recent years, due to high protein content and nutraceutical properties, soybean is highly recommended for human consumption. One of the major obstacles in utilization of soybean for human consumption is off-flavor generated due to the presence of lipoxygenase isozymes. Among three lipoxygenase isozymes, lipoxygenase-2 isozyme is a major contributor to the off-flavor. Although it is heat labile, heat inactivation adversely affects solubility and functionality of proteins. So, breeding programs aiming to develop soybean genotypes free from off-flavor generating lipoxygenase-2 isozymes are desired. Protein and PC are quantitative traits, while lipoxygenase-2 activity is controlled by a single dominant gene. Mapping of quantitative trait loci (QTL) associated with PC and protein content is important to develop soybean genotypes with high PC and high protein content. For this purpose, parental polymorphism survey (PPS) is the first prerequisite. PPS is also desired for the introgression of null allele of lipoxygense-2 in desirable genetic background through marker-assisted backcross breeding as it helps in the identification of recovery of recurrent parent genome. At ICAR-Indian Institute of Soybean Research, Indore, Madhya Pradesh, India, high protein genotype NRC151 was developed. Similarly, high PC and low PC soybean genotypes were identified. AVKS215, a lipoxygenase-2 free advance breeding line, was also developed at same institute. In the present study, five different parental combinations, namely NRC151 × IC275, PS1476 × IC275, NRC151 × IC574373, JS20-34 × AVKS215, and JS20-98 × AVKS215, were selected for PPS, which comprise seven soybean genotypes with difference in PC content, protein content, and presence and absence of off-flavor-generating lipoxygensase-2 isozymes.
2. MATERIALS AND METHODS
Seven soybean genotypes, namely NRC151, PS1476, IC275, IC574373, AVKS215, JS20-34, and JS20-98 with different characteristics were selected for the PPS. NRC151 is a high protein (48%) and high PC (8.8 mg/g soy flour) soybean genotype developed at ICAR-Indian Institute of Soybean Research, Indore, Madhya Pradesh, India. PS1476, IC275, and IC574373 are soybean germplasm accessions. PS1476 is high PC (8.4 mg/g soy flour) soybean genotypes, while IC275 is low protein (35%) and low PC (2.04 mg/g soy flour) soybean genotype. IC574373 is also low protein (37%) soybean genotypes. AVKS215 is lipoxygenase-2 free (Lox-2 −ve) soybean genotype, while JS20-34 and JS20-98 are high-yielding soybean varieties in which lipoxygenase-2 isozyme is present (Lox-2 +ve). From these seven soybean genotypes, five parental combinations, namely NRC151 × IC275 (parental combination 1), PS1476 × IC275 (parental combination 2), NRC151 × IC574373 (parental combination 3), JS20-34 × AVKS215 (parental combination 4), and JS20-98 × AVKS215 (parental combination 5) were made for PPS.
Genomic DNA from tender leaves of each soybean genotype was extracted following cetyl trimethyl ammonium bromide method [8]. Concentration of purified DNA was quantified using spectrophotometer (Perkin Elmer, Model Lambda 35, MA, USA) and adjusted to approximately 25 ng/ml. Subsequently, DNA was subjected to polymerase chain reaction (PCR) amplification in 10 μl reaction mixture composed of 2 μl DNA, 1 μl 10× PCR buffer, 1.1 μl MgCl2 (25 mM), 0.1 μl deoxynucleotide triphosphates (dNTPs) (25 mM), 0.5 μl of forward and reverse SSR primers (30 ng/ml), 0.068 μl Taq DNA polymerase (5 U/μl), and 4.732 μl PCR grade double-distilled water. For the amplification of genomic DNA, a thermocycler (HiMedia, LA948 Prima duo) was used. Initially, DNA was denatured at 94°C for 5 minutes, followed by 35 cycles each comprising denaturation at 94°C for 55 seconds, primer annealing at 55°C for 55 seconds and elongation at 72°C for 55 seconds. After completion of 35 cycles, final elongation step was carried out at 72°C for 7 minutes. Amplified PCR products so obtained were resolved on 3% DNA grade agarose (HiMedia, Catalogue no. MB080) and gel was stained with nucleic acid stain (Gbioscience, LabSafe™ Nucleic Acid Stain, catalogue no. 786-409). Gel was analyzed in gel documentation system (Syngene, Cambridge, UK). For PPS, a total of 695 SSR marker pairs were selected across all the 20 linkage groups (LGp) [9] at an average density of 5 cm.
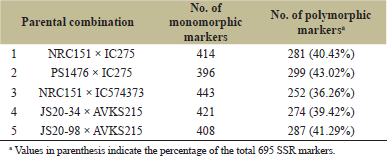 | Table 1: PPS of five parental combinations using 695 SSR markers. [Click here to view] |
3. RESULTS AND DISCUSSION
For PPS of all the five parental combinations, 695 SSR marker pairs were assessed across the 20 LGp. Out of these, the number of polymorphic markers observed in parental combinations 1, 2, 3, 4, and 5 were 281, 299, 252, 274, and 287, respectively. Percentage polymorphism observed for parental combination 1, 2, 3, 4, and 5 was 40.43%, 43.02%, 36.26%, 39.42%, and 41.29%, respectively (Table 1). Similar results were reported by Kumar et al. [10] in five parental combinations, namely NRC7 × PI542044, JS97-52 × PI542044, MACS450 × PI542044, JS93-05 × PI542044, and DS97-12 × PI542044 exhibiting 43.38%, 45.04%, 48.14%, 44.00%, and 46.48% polymorphisms, respectively. The authors developed these parental combinations for introgression of null allele of Kunitz trypsin inhibitor from donor parent PI542044 into NRC7, JS97-52, MACS450, JA93-05, and DS97-12 through marker-assisted backcross breeding. Rawal et al. [11] also observed 49.24% parental polymorphism in parental combination JS97-52 × PI596540, which was developed to introgress null allele of lipoxygenase-2 gene in recipient parent JS97-52 from donor parent PI596540.
Thirty-five SSR markers from different LGp were polymorphic across all the five parental combinations (Table 2). The highest number of seven SSR markers was detected polymorphic on LGp A2, followed by five on LGp A1, while only one polymorphic marker (Fig. 1) was observed on LGp B1, LGp B2, LGp E, LGp F, LGp G, LGp I, and LGp M across all the five parental combinations. Non-uniform distribution of polymorphic SSR markers has also been reported in earlier studies [10,11].
The number of polymorphic markers observed in each of the five cross combinations is presented in Table 3. Varied numbers of polymorphic markers were found on different LGps in different parental combinations. For parental combinations 1 and 2, the maximum number of polymorphic markers was observed on LGp A2 (26) and F (27), respectively. For parental combination 3, the maximum number of polymorphic markers was observed on LGp A2 (22), LGp D1b (22), and LGp D2 (22), while for the parental combination 4 and 5, the maximum number of polymorphic markers 24 and 29 was detected on a single LGp G, respectively.
 | Table 2: Details of common polymorphic SSR markers for all five parental combinations across different LGps. [Click here to view] |
In parental combination 1 (NRC151 × IC275) and parental combination 3 (NRC151 × IC574373), NRC151 is a high protein (48%) soybean genotype, while IC275 (35%) and IC574373 (37%) are low protein soybean genotypes. Therefore, crossing between these two parental combinations would generate mapping population (F2/RILs) for identification of QTLs associated with high protein content. Besides, NRC151 has high PC content (8.8 mg/g of flour), while IC275 has very low PC content (2.04 mg/g of flour). Therefore, mapping population generated from NRC151 × IC275 can also be utilized for tagging QTLs associated with PC content. Polymorphic SSR markers (252) observed for parental combination 3 can be used for genotyping the mapping population (F2/RILs) generated for identification of QTLs associated with PC content in soybean, while polymorphic SSR markers (281) identified for parental combination 1 can be used for genotyping the mapping population (F2/RILs) generated for identification of QTLs associated with both PC content and protein content in soybean.
In parental combination 2 (PS1476 × IC275), PS1476 possesses high PC content (8.40 mg/g soy flour), while IC275 has low PC content (2.04 mg/g soy flour). Therefore, polymorphic SSR markers (299) observed for parental combination 2, besides parental combination 1, and can also be utilized for genotyping of mapping population (F2/RILs) generated for the identification of QTLs associated PC content in soybean. In parental combination 4 and 5, AVKS215 (derived from JS93-05 × PI596540), a lipoxygenase-2 free soybean genotype, could be used as donor parent for null allele of lipoxygenase-2 gene, while JS20-34 and JS20-98 are high-yielding released varieties having lipoxygenase-2 isozymes (responsible for production of beany off-flavor in soy food). Therefore, parental combination 4 (JS20-34 × AVKS215) and parental combination 5 (JS20-98 × AVKS215) can be used for introgression of null allele of lipoxygenase-2 from AVKS215 into JS20-34 and JS20-98 through marker-assisted backcross breeding. Polymorphic SSR markers observed in parental combinations 4 and 5 can be used in background selection for the recovery of recurrent parents’ genome content (i.e., of JS20-34 and JS20-98).
 | Figure 1: PPS using SSR markers. Lane 1–7 represents amplified products of SSR markers of NRC151, IC275, PS1476, IC574373, AVKS215, JS20-34, and JS20-38, respectively. L indicates 50 bp DNA ladder. [Click here to view] |
 | Table 3: Number of polymorphic SSR markers for each parental combination across different LGps. [Click here to view] |
Polymorphism of SSR markers depends upon the type of motifs present in the SSR marker and number of repeats present in the motif [12]. In the present study, it has been found that SSR markers with tri-nucleotide repeats were more polymorphic than SSR markers with di-nucleotide repeats. Out of 35 common polymorphic SSR markers among all the 5 parental combinations, 21 markers contained tri-nucleotide repeats, and 14 contained di-nucleotide repeats. Among the di-nucleotide repeats, SSR markers with 12–33 repeats, i.e., (AT12–33), were found to be more polymorphic than others, while in tri-nucleotide repeats SSR markers with 9–23 repeats, i.e., (ATT9–23), appears to be more polymorphic than others. These results are in consonance with previous studies in soybean [10,11].
4. CONCLUSION
Soybean grains with high protein content and PC content are much sought after raw material as feed and nutraceutical to improve nutritive value. On the other hand, removal of the off-flavor generating lipoxygenase-2 from soybean truly enhances the utilization of this economical source of protein for human consumption. In the present study, a total of 695 SSR markers across 20 LGps were surveyed for parental polymorphism in all the 5 parental combinations varying for protein, PC and lipoxygenase-2 exhibiting 40.43%, 43.02%, 36.26%, 39.42%, and 41.29% polymorphisms. Polymorphic markers identified for parental combinations 1, 2, and 3 can be utilized for genotyping of F2/RILs population generated for development and identification of genomic regions associated with high protein content and PC content. Polymorphic markers observed in parental combinations 4 and 5 would be used for the recovery of recurrent parent genome content of respective recurrent parent utilized for development of lipoxygenase-2 free high-yielding soybean genotypes.
5. ACKNOWLEDGMENT
The authors are grateful to ICAR-Indian Institute of Soybean Research, Indore, Madhya Pradesh, India, for providing facilities to carry out the study.
6. CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest in relation to this article.
7. FUNDING
None.
8. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design; acquisition, analysis and interpretation of data; taking part in drafting the article and revising it critically for important intellectual content; agreed to submit to the current journal and gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
REFERENCES
1. USDA. Oilseeds: world markets and trade. 2021. Available via https://apps.fas.usda.gov/psdonline/circulars/oilseeds.pdf (Accessed 23 April 2021).
2. Shurtleff W, Aoyagi A. History of lecithin and phospholipids (1850-2016). California, USA: Soyinfo Center; 2016.
3. van Hoogevest P, Wendel A. The use of natural and synthetic phospholipids as pharmaceutical excipients. Eur J Lipid Sci Technol 2014;116(9):1088–107. CrossRef
4. List GR. Soybean lecithin: food, industrial uses, and other applications. In: Ahmad MU, Xu X (ed.). Polar Lipids, Urbana, Illinois: Elsevier; 2015, pp 1–33. CrossRef
5. Küllenberg D, Taylor LA, Schneider M, Massing U. Health effects of dietary phospholipids. Lipids Health Dis 2012;11(1):3. CrossRef
6. Cromwell D. Soybean meal–an exceptional protein source. 2017. Available via https://www.soymeal.org/wp-content/uploads/2018/04/soybean_meal_an_exceptional_protein_source.pdf (Accessed 23 April 2021).
7. Lagos LV, Stein HH. Chemical composition and amino acid digestibility of soybean meal produced in the United States, China, Argentina, Brazil, or India. J Anim Sci 2017;95(4):1626–36. CrossRef
8. Doyle JJ, Doyle JL. Isolation of plant DNA from the fresh tissue. Focus 1990;12:14–5.
9. Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, et al. A new integrated genetic linkage map of the soybean. Theore Appl Genet 2004;109(1):122–8. CrossRef
10. Kumar V, Rani A, Mourya V, Rawal R, Verma K, Marana S, et al. Marker-assisted selection for development of kunitz trypsin inhibitorfree soybean varieties: I. parental polymorphism survey using SSR markers. Indian J Genet Plant Breed 2011;71(4):372–6.
11. Rawal R, Kumar V, Rani A, Gokhale SM, Husain SM. Marker assisted backcross selection for genetic removal of lipoxygenase-2 from popular soybean (Glycine max) variety JS 97-52: parental polymorphism survey and hybridity validation. Indian J Agric Sci 2014;84(3):414–7.
12. Xu J, Liu L, Xu Y, Chen C, Rong T, Ali F, et al. Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Res 2013;20(5):497–509. CrossRef