ARTICLE HIGHLIGHTS
This work was carried out in different captive wild animal species during winter, summer, and monsoon seasons.
Of 29 samples were positive for acid–fast bacilli out of the total 126 fecal samples screened.
Real time-polymerase chain reaction (PCR) targeting IS900 gene detected 14 samples (5 carnivores, 7 herbivores, and 2 omnivores) positive.
The overall prevalence of Mycobacterium avium subspecies paratuberculosis (MAP) infection was 11.11% (11.11% carnivores, 29.16% herbivores, 66.66% omnivores, 0% birds, and 0% reptiles).
Molecular typing (IS1311 PCR_REA) revealed all positive samples belonged to “Indian Bison type,” biotype.
1. INTRODUCTION
Johne’s disease (JD) was first time reported in ruminants and assumed that it was caused by as an etiological agent [1]. Mycobacterium avium subspecies paratuberculosis (MAP) has emerged as a major pathogen having substantial zoonotic importance due to its association with Crohn’s disease (CD) [2,3]. The disease has been declared of major global importance by OIE, in view of its socioeconomic and public health significance. MAP is worldwide in distribution and is considered a major threat to livestock trade, dairy business, and beef meat industry [3,4].
JD is an incurable, chronic debilitating granulomatous enteropathy of domestic animals, ruminant and non-ruminant wild species, and wild carnivores [5-9]. In ruminants, JD is insidious and does not show clinical signs at early stage of disease acting as a silent killer [2,10,11]. Although disease is more than 100 years old, research studies have been conducted from the past 50 years generating information that has been pivotal regarding basic understanding of the disease only and its control or elimination has not been possible thereby remaining a threat to animal and human health [10,12,13].
Diseases in captive animals have a number of negative effects on health, longevity, and if not properly diagnosed or timely intervened can prove fatal. Chronic diseases act as pre-disposing factor to secondary deficiency diseases resulting in inefficient reproduction (hampering fast multiplication of animals with good germplasm an important aspect for specialized breeding programs meant for increasing number of captive and endangered species), negative energy balance, and protein loosing enteropathy. The present study was undertaken to have a better understanding about the epidemiology of the MAP affecting different species of captive wild animals that will be helpful for effective management of this chronic life-threatening disease in captive wild animal health and minimizing public health concerns.
2. MATERIALS AND METHODS
2.1. Sampling Strategy and Captive Wild Animals Involved in the Study
Epidemiological study of MAP from captive wild animals and birds of the Zoo was carried out based on individual/pooled fecal samples (n = 42) from different captive wild animal species during winter, summer, and monsoon seasons for a period of 10 months from January to October 2021. The study was undertaken with prior approval from office of the Chief Conservator of Forests, The Zoo Authority of Maharashtra, Nagpur, vide no. CCOF/Technical/P. No. 3B/31/2021-22 dated: 05 May 2021.
2.2. Collection of Fecal Samples
Based on color and moisture content, interior portion (surface portion removed) of fresh feces (0–12 h) was collected in sterile whirl pack polythene bags using sterile spatulas and stored at 4°C until further processing.
2.3. Direct Microscopy (Ziehl–Neelsen Staining)
Smears of processed samples were prepared on clean dry glass slides and then heat fixed. Smear was flooded with carbol fuchsin solution (1.0%) and heated to steam for 7–10 min keeping smear wet by adding sufficient stain intermittently. Slides were cooled and washed with distilled water and decolorized with 3% acid alcohol or 25% H2SO4 for 10–30 s or until pink color stopped coming. Slide washed with distilled water, counter stained for 1–2 min with methylene blue (0.1%), and again washed with distilled water and blotted. Cells appearing pink colored, short rods in bunch or clumps dispersed singly under oil-immersion objective were MAP [14].
2.4. Isolation of DNA from Fecal Samples
Feces (2 g) were homogenized in sterilized 1 × PBS (4 ml) using sterilized pestle mortar, and contents were transferred to 15 ml centrifuge tube containing 8 ml sterilized 1 × PBS for centrifugation at 3500 rpm (45 min) to concentrate bacilli. The tube was kept undisturbed at room temperature for 1–2 h, supernatant was discarded, and semisolid middle layer was collected with sterilized swab in Eppendorf tube containing 300 μl of 1 × PBS that was heated at 95°C for 10 min. 40 μl of lysozyme (20 mg/ml) were added and contents were incubated at 37°C for 2 h with continuous shaking. The mixture was added 20 μl proteinase K (10 mg/ml) and 50 μl 10% SDS and incubated at 56°C for 2 h with continuous shaking followed by the addition of CTAB-NACL (64 μl) and incubation for 30 min at 65°C. The mixture was added equal volume of chloroform-isoamyl alcohol (24:1), contents centrifuged at 10000 rpm for 20 min at 4°C, and the aqueous layer (upper layer) was remounted into the fresh 1.5 ml Eppendrof tube, to which 0.6 volume of isopropanol was added with gentle mixing and allowed for precipitation of DNA at −20°C overnight. The contents again centrifuged at 10000 rpm for 20 min at 4°C to get the DNA pellet that was washed with 1 ml of 70% ethanol followed by centrifugation at 10000 rpm for 10 min at 4°C, and finally, the DNA pellet was resuspended in 30 μl I× TE buffer and stored at 4°C overnight for dissolving the DNA followed by storage at −20°C for further use [15].
2.5. Agarose Gel Electrophoresis
Two gram agarose was weighed and dissolved in 100 ml 1× TAE and heated in microwave until a clear transparent solution appeared. Agarose was allowed to cool (55–60°C) and added with ethidium bromide (0.5 μg/ml in 100 ml 1× TAE) slowly to avoid bubble formation and then poured into the arranged casting tray. The Gel was allowed to solidify and later placed in horizontal gel electrophoresis unit filled with 1× TAE. The DNA mixed with DNA loading dye was loaded into the wells and electrophoresis was carried out for 45min at 80 Volts and gel was then visualized under UV light in the Jel documentation system (Biorad) [16].
2.6. 1 IS900 Polymerase Chain Reaction (PCR) Protocol for MAP
DNA was amplified by PCR targeting IS900 sequence (IS900 15C/921) as per the protocol and primers of Vary et al with slight modifications (IS900/15C: 5’-CCG CTA ATT GAG AGA TGC GAT TGG-3’; IS900/921: 5’-AAT CAA CTC CAG CAG CGC GGC CTC G-3’) [17].
2.7. IS1311 Polymerase Chain Reaction Restriction Endonuclease Analysis
IS1311 PCR was carried out using M56 and M119 primers as per with some modification using the primers (M56: 5’-GCG TGA GGC TCT GTG GTG AA-3’; M119: 5’- ATG ACG ACC GCT TGG GAG AC-3’). For PCR-REA, a 30.0 μl reaction mixture was prepared consisting of various ingredients comprising of 10× Buffer (3.0 μl), HinfI (2.0 units; Fermentas), MseI (2.0 units; Fermentas), PCR product (20 μl) and HPLC grade water. Also control reactions, one consisting of PCR products without enzyme and another containing enzyme mixture with PCR products were carried out. The digestion of reaction mixture for PCR-REA was carried out at 37°C for 1.5 h in water bath followed by mixing of digested PCR products with loading dye (2.0 μl) for electrophoreses in 4.0% agarose gel at 3–5 V/cm for 1.5 h, and finally, the gel was visualized under gel documentation system (Biorad, USA). Interpretation of band patterns was done as described by researcher who has divided MAP isolates into 3 groups based on the generation of 3 kinds of band patterns for different MAP strains: Sheep type/Type I/III (2 bands of 285bp and 323 bp), Cattle type/Type II (4 bands of 67 bp, 218 bp, 285 bp, and 323 bp), and Bison type/B type (3 bands of 67 bp, 218 bp, and 323 bp) [18].
3. RESULTS AND DISCUSSION
A total of 126 fecal samples consisting of Carnivores (45), Herbivores (24), Omnivores (3), Birds (21), and Reptiles (33) were examined by direct microscopy revealing 29 as acid–fast bacilli [Table 1]. Samples positive under microscopy were further confirmed by IS900 PCR [Figure 1] that revealed 14 samples positive for MAP, comprising of 5 carnivores, 7 herbivores, and 2 omnivores fecal samples revealing an overall prevalence of MAP as 11.11% (11.11% carnivores, 29.16 % herbivores, 66.66% omnivores, 0 % birds, and 0 % reptiles). Lower prevalence (2.41%) of MAP in wildlife was reported by Carta et al. [8] whereas comparatively higher prevalence of MAP in wild mammals (38%) and birds (18%) from three farms in the South Island of New Zealand was reported [19,20]. Kruze et al. reported that 13.3% of captive ungulates were positive on culture examination [21] while Pradenas et al. observed that two samples were culture positive from farmed deer suspected for MAP [22]. Deutz and Spergser have investigated different species of MAP-positive wild animals and concluded that transmission could have been from contaminated pastures having grazed by cattle [23]. Mir has reported three positive samples (two from Hyenas and one from Jungle cat) with a prevalence of 1.9% and reported the MAP infection first time in captive Hyenas and Jungle cats from India while studying the prevalence of MAP in free captive wild animals [24].
Table 1: Breakdown of MAP detection in various captive animal species of Siddharth zoo.
| S. No | Animal Group | Species | Microscopy | Molecular | |||||
|---|---|---|---|---|---|---|---|---|---|
| Neg. | 1+ | 2+ | 3+ | 4+ | PCR | Genotyping | |||
| 1 | Carnivores | Tiger (n=30) | 18 | 9 | 3 | 0 | 0 | 2 | Indian Bison type |
| Leopard (n=6) | 3 | 2 | 1 | 0 | 0 | 1 | Indian Bison type | ||
| Jackal (n=4) | 2 | 2 | 0 | 0 | 0 | 2 | Indian Bison type | ||
| Indian Civetcat (n=3) | 3 | 0 | 0 | 0 | 0 | 0 | - | ||
| 2 | Herbivores | Sambar Deer (n=9) | 3 | 4 | 2 | 0 | 0 | 3 | Indian Bison type |
| Blackbuck (n=3) | 2 | 1 | 0 | 0 | 0 | 1 | Indian Bison type | ||
| Spotted Deer (n=3) | 1 | 0 | 2 | 0 | 0 | 2 | Indian Bison type | ||
| Indian Porcupine (n=6) | 6 | 0 | 0 | 0 | 0 | 0 | - | ||
| Nilgai (n=3) | 0 | 1 | 2 | 0 | 0 | 1 | Indian Bison type | ||
| 3 | Omnivores | Monkey (n=3) | 3 | 0 | 0 | 0 | 0 | 2 | Indian Bison type |
| 4 | Birds | Emu (n=3) | 3 | 0 | 0 | 0 | 0 | 0 | - |
| Indian Peafowl (n=3) | 3 | 0 | 0 | 0 | 0 | 0 | - | ||
| Painted Storck (n=3) | 3 | 0 | 0 | 0 | 0 | 0 | - | ||
| Little Egret (n=3) | 3 | 0 | 0 | 0 | 0 | 0 | - | ||
| Spoon Bill (n=3) | 3 | 0 | 0 | 0 | 0 | 0 | - | ||
| White Necked Storck (n=3) | 3 | 0 | 0 | 0 | 0 | 0 | - | ||
| Grey Heron (n=3) | 3 | 0 | 0 | 0 | 0 | 0 | - | ||
| 5 | Reptiles | Snakes (n=27) | 27 | 0 | 0 | 0 | 0 | 0 | - |
| Crocodile (n=6) | 6 | 0 | 0 | 0 | 0 | 0 | - | ||
| Total (n=126) | 97 | 19 | 10 | 0 | 0 | 14 | |||
*n: Number of samples, *Neg: Negative samples, PCR: Polymerase chain reaction
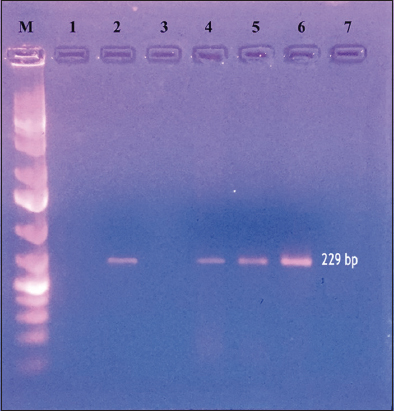 | Figure 1: Traditional IS900 polymerase chain reaction for the diagnosis of Mycobacterium avium subspecies paratuberculosis infection: Lane M: 100bp Marker, Lane 1: Blank, Lane 2: Positive Control, Lane 3: Negative control, Lane 4–6: Positive samples. [Click here to view] |
The present study is reporting MAP infection first time in tigers from India, although earlier MAP has been reported throughout the world by different researchers from various wild animal species such as in fox and stoat [25], in Addax [26], in fallow deer [27], in brown bears [5], in captive Bengal tiger at Republic of Korea [28], in brushtail possums, hares, and hedgehogs, cats, rabbits and among birds in paradise shelducks [19], in wild mammals [8,9], in Alpaca [29], in wild boars [30], in gazelle and Red deer [19,21], and in farmed deer [22]. All the animals were positive on PCR in winter season when again screened in summer and monsoon seasons, some of them showed negative PCR results although they were positive on microscopy (2+) an observation also reported by Mir [24].
Molecular characterization and genetic diversity of 14 positive MAP isolates using IS1311 PCR restriction endonuclease analysis revealed all were Indian Bison type [Table 1 and Figure 2]. Similar findings were also reported by researchers who reported Indian Bison type strain of MAP in two animals from Abohar Wildlife Sanctuary Fazilka, Punjab [24]. Researchers observed bison and cattle-type MAP genotypes from the Wild Boars in Korea which are the predominant MAP genotypes infecting domestic species in Korea [30], O’Brien et al. reported bovine strain genotype of MAP from intestinal lymphatic tissues of 74 JD infected farmed red deer from New Zealand [22] and also first time established the experimental infection of MAP in red deer and observed strong infectivity of MAP bovine-strain for cervines [31]. Woodbury et al. reported the cattle strain of MAP in asymptomatic farmed white-tailed deer [32]. Fritsch et al. observed that cattle type genotype was shared between cattle and wild red deer in Germany [33]. The present study revealed Indian Bison type strain among carnivores in tiger, leopard, and jackal; among herbivores in black buck, sambar deer, spotted deer, and nilgai; and among omnivores in monkeys. However, MAP could not be detected from different species of birds and reptiles; a finding has been correlated to the preference for pastoral habitats in certain species of birds that increase the chances of getting infection and also possibly due to good managemental practices being followed at zoo premises that prevent spillover of infection [19]. The study highlights the potential of cross infection of Indian Bison type strain of MAP between herbivores, carnivores, and omnivores, and therefore, proper managemental strategies need to be adopted at zoo premises to prevent the occurrence and spread of infection. MAP could be a risk to the health and welfare of wild animals kept under captivity with respect to negative effects on health, longevity and if not properly diagnosed or timely intervened can prove fatal or compromises the conservation programs meant for endangered species.
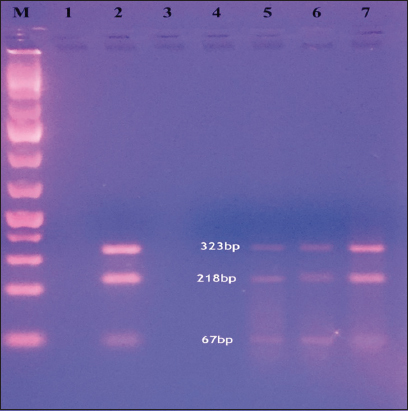 | Figure 2: IS1311 polymerase chain reaction-REA using restriction enzyme HinfI and MseI: Lane M: 100 bp Marker, Lane 1: Blank, Lane 2: Positive control, Lane 3: Negative control, Lane 5–7: Positive samples. [Click here to view] |
4. CONCLUSIONS
The study is first time reporting MAP infection in tigers from India and it is highlights, the silent interspecies transmission of MAP “Indian Bison type” biotype from herbivores to omnivores and carnivores. These findings may help in the implementation of effective management strategies for the control of MAP in captive wild animals and minimize spread and reduce burden of this chronic life-threatening disease in captive animal species.
5. ACKNOWLEDGMENTS
Authors are grateful to Principal Chief Conservator of Forests, Maharashtra, for giving the permission to undertake this investigation.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
Certificate vide no. CCOF/Technical/P. No. 3B/31/2021-22 dated: 05- 05-2021 from The Chief Conservator of Forests, The Zoo Authority of Maharashtra, Nagpur.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Johne H, Frothingham L. A particular case of tuberculosis in a cow. Dtsch Z Tiermed Pathol 1895;21:438-54. [CrossRef]
2. Lombard JE. Epidemiology and economics of paratuberculosis. Vet Clin North Am Food Anim Pract 2011;27:525-35, v. [CrossRef]
3. Eslami M, Shafiei M, Ghasemian A, Valizadeh S, Al-Marzoqi AH, Shokouhi Mostafavi SK, et al. Mycobacterium avium paratuberculosis and Mycobacterium avium complex and related subspecies as causative agents of zoonotic and occupational diseases. J Cell Physiol 2019;234:12415-21. [CrossRef]
4. Richardson E, Mee J, Sánchez-Miguel C, Crilly J, More S. Demographics of cattle positive for Mycobacterium avium subspecies paratuberculosis by faecal culture, from submissions to the Cork Regional Veterinary Laboratory. Ir Vet J 2009;62:398-405. [CrossRef]
5. Kopecna M, Ondrus S, Literak I, Klimes J, Horvathova A, Moravkova M, et al. Detection of Mycobacterium avium subsp. paratuberculosis in two brown bears in the central European Carpathians. J Wildl Dis 2006;42:691-5. [CrossRef]
6. Witte CL, Hungerford LL, Rideout BA. Association between Mycobacterium avium subsp. paratuberculosis infection among offspring and their dams in nondomestic ruminant species housed in a zoo. J Vet Diagn Invest 2009;21:40-7. [CrossRef]
7. Singh PK, Singh SV, Kumar H, Sohal JS, Singh AV. Diagnostic application of IS900 PCR using blood as a source sample for the detection of Mycobacterium avium subspecies paratuberculosis in early and subclinical cases of caprine paratuberculosis. Vet Med Int 2010;74?. [CrossRef]
8. Carta T, Martin-Hernando MP, Boadella M, Fernández-de-Mera IG, Balseiro A, Sevilla IA, et al. No evidence that wild red deer (Cervus elaphus) on the Iberian Peninsula are a reservoir of Mycobacterium avium subspecies paratuberculosis infection. Vet J 2012;192:544-6. [CrossRef]
9. Hernández-Reyes AL, Chávez-Gris G, Maldonado-Castro E, Alcaraz-Sosa LE, Díaz-Negrete MT. First identification of Mycobacterium avium subsp. paratuberculosis in wild ruminants in a zoo in Mexico. Vet World 2022;15:655-61. [CrossRef]
10. Momotani E, Ozaki H, Hori M, Yamamoto S, Kuribayashi T, Eda S, et al. Mycobacterium avium subsp. paratuberculosis lipophilic antigen causes Crohn's disease-type necrotizing colitis in Mice. Springerplus 2012;1:47. [CrossRef]
11. Garvey M. Mycobacterium Avium paratuberculosis:A disease burden on the dairy industry. Animals (Basel) 2020;10:1773. [CrossRef]
12. Kreeger JM. Ruminant paratuberculosis--a century of progress and frustration. J Vet Diagn Invest 1991;3:373-82. [CrossRef]
13. Kaczmarkowska A, Didkowska A, Kwiecie?E, Stefa?ska I, Rzewuska M, Anusz K. The Mycobacterium avium complex-an underestimated threat to humans and animals. Ann Agric Environ Med 2022;29:22-7. [CrossRef]
14. Singh SV, Singh PK, Gupta S, Chaubey KK, Singh B, Kumar A, et al. Comparison of microscopy and blood-PCR for the diagnosis of clinical Johne's disease in domestic ruminants. Iranian J Vet Res 2013;14:345-9.
15. Hemati Z, Meletis E, Derakhshandeh A, Haghkhah M, Kostoulas P, Singh SV, et al. Application of Bayesian modeling for diagnostic assays of Mycobacterium avium subsp. paratuberculosis in sheep and goats flocks. BMC Vet Res 2022;18:47. [CrossRef]
16. Lee PY, Costumbrado J, Hsu CY, Kim YH. Agarose gel electrophoresis for the separation of DNA fragments. J Vis Exp 2012;62:3923. [CrossRef]
17. Vary PH, Andersen PR, Green E, Hermon-Taylor J, McFadden JJ. Use of highly specific DNA probes and the polymerase chain reaction to detect Mycobacterium paratuberculosis in Johne's disease. J Clin Microbiol 1990;28:933-7. [CrossRef]
18. Whittington RJ, Sergeant ES. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp paratuberculosis in animal populations. Aust Vet J 2001;79:267-78. [CrossRef]
19. Nugent G, Whitford EJ, Hunnam JC, Wilson PR, Cross Ml, de Lisle GW. Mycobacterium avium subsp. paratuberculosis infection in wildlife on three deer farms with a history of Johne's disease. N Z Vet J 2011;59:293-8. [CrossRef]
20. Fisher CA, Bhattarai EK, Osterstock JB, Dowd SE, Seabury PM, Vikram M, et al. Evolution of the bovine TLR gene family and member associations with Mycobacterium avium subspecies paratuberculosis infection. PLoS One 2011;6:e27744. [CrossRef]
21. Kruze J, Bermudez N, Hidalgo E, Mella A, Salgado M. Detection of MAP in Captive Wild Ungulates of Metropolitan Region, Chile. In:Proceedings of the 12th International Colloquium on Paratuberculosis, Parma, Italy;2014. p. 77-9.
22. Pradenas M, Navarrete-Talloni MJ, Salgado M, Zamorano P, Paredes E. Paratuberculosis or avian tuberculosis in red deer with chronic diarrhea?Arch Med Vet 2014;46:45-52. [CrossRef]
23. Deutz A, Spergser J. Paratuberculosis in wild animals prevalence, clinical and postmortem findings. Circ Meat Hygiene Food Control 2009;61:12-5.
24. Mir AQ. Molecular Epidemiology and Diagnosis of Bacterial and Parasitic Pathogens from Faeces of Wild Ungulates and Carnivores [PhD Dissertation]. Ludhiana:Guru Angad Dev Veterinary and Animal Sciences University;2016.
25. Beard M, Henderson D, Daniels MJ, Pirie A, Buxton D, Greig A, et al. Evidence of paratuberculosis in fox (Vulpes vulpes) and stoat (Mustela erminea). Vet Rec 1999;145:612-3. [CrossRef]
26. Burton MS, Olsen JH, Ball RL, Dumonceaux GA. Mycobacterium avium subsp. paratuberculosis infection in an addax (Addax nasomaculatus). J Zoo Wildl Med 2001;32:242-4.
27. Marco I, Ruiz M, Juste R, Garrido JM, Lavin S. Paratuberculosis in free-ranging fallow deer in Spain. J Wildl Dis 2002;38:629-32. [CrossRef]
28. Cho HS, Kim YH, Park NY. Disseminated mycobacteriosis due to Mycobacterium avium in captive Bengal tiger (Panthera tigris). J Vet Diagn Invest 2006;18:312-4. [CrossRef]
29. Münster P, Völkel I, von Buchholz A, Czerny CP. Detection of Mycobacterium avium subspecies paratuberculosis by IS900-based PCR assays from an alpaca (Vicugna pacos) kept in a German zoological garden. J Zoo Wildl Med 2013;44:176-80. [CrossRef]
30. Kim JM, Ku BK, Lee HN, Hwang IY, Jang YB, Kim J, et al. Mycobacterium avium paratuberculosis in wild boars in Korea. J Wildl Dis 2013;49:413-7. [CrossRef]
31. O'Brien R, Mackintosh CG, Bakker D, Kopecna M, Pavlik I, Griffin JF. Immunological and molecular characterization of susceptibility in relationship to bacterial strain differences in Mycobacterium avium subsp. paratuberculosis infection in the red deer (Cervus elaphus). Infect Immun 2006;74:3530-7. [CrossRef]
32. Woodbury MR, Chirino-Trejo M, Mihajlovic B. Diagnostic detection methods for Mycobacterium avium subsp. paratuberculosis in white-tailed deer. Can Vet J 2008;49:683-8.
33. Fritsch I, Luyven G, Köhler H, Lutz W, Möbius P. Suspicion of Mycobacterium avium subsp. paratuberculosis transmission between cattle and wild-living red deer (Cervus elaphus) by multitarget genotyping. Appl Environ Microbiol 2012;78:1132-9. [CrossRef]