1. INTRODUCTION
Bioconversion of lignocellulosic waste (LCW) biomass represents a vast arena of violent exploration in research, development, and commercialization with distinct avant-garde activities at streaming ages [1]. Lignocellulosic biomass is one of the most generous substances in nature. It includes variant compositions of cellulose (40–50%), hemicellulose (25–30%), and lignin (15–20%). All components have been depicted as a higher amount of bioenergy resources towards sustainability. Therefore, the availability of renewable bioenergy assets has been drafted through these lignocellulosic biomass efficiencies [2-4]. LCWs are being comprehended with multiform categories of residues such as industrial waste, agricultural waste, domestic waste, agroindustrial waste, and municipal waste. Huge amounts of fermentable carbohydrates have been rescued from agricultural and food industrial LCW after the pre- and post-cultivation methods [5]. At the same time from these LCWs, immense amounts of value-added products have been generated. For example: Lipids, biodegradable plastic components, carbohydrates, protein, amino acid, resins, medicine, cosmetics, methane, ethanol, organic acids, biopesticides, surfactants, biofertilizers, feed additives, etc. [6].
In 2007, The Energy Independence and Security Act has declared that in 2008 from 9 billion gallons to 36 billion gallons in 2022, sustainable fuel volume has been raised by cellulosic biomass. That is why researchers are mainly focusing to attain a higher yield of bioenergy genesis throughout the cellulose conversion procedure [7] by the hydroxylation and ring cleavage mechanism the phenolic compounds through the usage of algal species [5]. Lignin and hemicellulose breaks down to release cellulose. However, it fails to release the cellulose from LCW following physical, chemical, biological, physicochemical, mechanical, irradiation, extrusion, and ozonolysis pre-treatment procedures time to time. With this enzyme-substrate interaction processes ennoble the hydrolysis possibilities for upward biodiesel formation [8]. After the transformation of lignocellulosic biomass to low fermentable sugars, also another microbial catalyst has been used, for example, Zymomonas mobilis, Saccharomyces cerevisiae, Clostridium thermocellum, Clostridium phytofermentans, Trichoderma reesei, etc. Clostridium acetobutylicum was used for butanol generation from hemicellulosic materials. Basfia succiniproducens and Actinobacillus succinogenes are used for fermentation during succinic acid generation [9,10]. Lignin contents in Phanerochaete chrysosporium produce high value-added products during the catabolic pathways metabolism [11].
Besides during the pretreatment methods for bioenergy consumption, high electric power supplementation, expensive equipment, and some high cost supplementary solvents reclamation are played as a barrier to LCW remediation [12]. Furthermore, there are many drawbacks for using these bacteria, fungi like microorganisms because they are costly, not economically feasible, as well as during the analysis period they should be highly contaminated and discharge some allergens materials toward impel the chances of health hazards. Dealing with these physical, chemical, physico-chemical issues, biologically active, and exclusive catalyst alga is being adopted for LCW conversion and bioenergy genesis. In addition, in this research work, a broad section has been briefly accelerated and examined the main reasons behind choosing algal regimes and how the algal isolates can break the lignocellulosic biomass toward mild green environmental energy generation. Some well-known algal species have demonstrated higher lipid origination yield with the lignocellulosic enzymatic assimilation [13,14]. Chlamydomonas moewusii, Scenedesmus vacuolatus, Dunaliella salina, Dunaliella parva, Tetracystis aeria, Chlorella vulgaris, Chlamydomonas reinhardtii, Hydrodictyon reticulum, Arthrospira platensis, Lyngbya spp., Cladophora spp., Tetracystis isobilateralis, Tetracystis aggregate, Tetracystis illinoisensis, Tetracystis dissociate, Chlorococcum echinozygotum, Chlorococcum elkhartiense, Chlamydomonas noctigama, Tetracystis aplanospora, Desmodesmus communis, Scenedesmus ovaltemus, Acutodesmus obliquus, Neospongiococcum gelatinosum, Chlorococcum oleofaciens, Lobochlamyssegnis spp., Chloromonas chlorococcoides, Spirulina platensis, Wollea saccata, Anabaena oryzae, Nostoc spp., Oscillatoria spp. are the most effective algal species for their enzymatic activities besides bioremediation. Some naive algal species can produce hemicellulase, cellulose, xylanase, glucosidase, mannose, lignin peroxidase, and laccase like various enzymes. Those are highly efficient for biofuel generation using LCW components [15]. The usages of strong biocatalyst like algal regimes for the generation of LCW to biodiesel are the most well accepted third-generation liquid biofuel considering its immense beneficial aspects likely renewability, biodegradability, non-toxicity, high flash point, and sustainability compared to conventional petroleum-based diesel [16]. It could be a most promising future biofuel in the transportation sector as it elucidates common features of petrochemical-derived diesel having lower greenhouse gas emissions during combustion and emission of the lower transferable carcinogenic end products [17,18].
Likewise green and blue-green algae play a very pivotal role as third-generation biodiesel feedstock which shows few distinct benefits, that is, non-dependent on arable or agricultural fields for production purposes, higher photosynthetic efficacies, higher biomass yield having faster growth rate, higher lipid productivity, and higher cetane number. Algal biodiesel deals with around ten oxygen atoms in different fatty acids present and no aromatic constituents. Therefore, algal biodiesel combustion is comparatively higher environmentally benign [19,20]. Besides, algae-based lipids are predominantly neutral lipids or triacylglycerol which can easily be esterified to FAMEs with the most preferable primary C16 and C18 chain lengths for high quality biodiesel production [21]. Algae can tolerate extensive environmental stresses and display flexibility toward mass cultivation. It can grow on marginal land in open or closed water reserves (i.e., waste waters), waste gases (i.e., flue gases), organic wastes (i.e., crude glycerol), etc. [22]. Even diverse metabolic routes of algae have shown greater adaptation, higher biomass yields, and lipid productivities. In general, algae can grow photoautotrophically using carbon dioxide as a carbon source and light as an energy source. Few algal regimes can grow heterotrophically utilizing organic ingredients whereas few can grow mixotrophically considering both organic compounds and carbon dioxide to carry out their growth cycle [23]. However, the first- and second-generation biodiesel production approaches require huge areas of arable agricultural lands. To this end, biomass-derived biodiesel generation is a highly beneficial alternative approach due to extensive unique advantages including carbon neutrality, minimized greenhouse gas emission (i.e., carbon monoxide, carbon dioxide, and sulfur oxides), spontaneous cheaper biomass feedstock availability, and biosafety of biomass production through agriculture [24]. To this end, algae are impressive microbial candidates for biodiesel generation as algae grow without competing with the food chain, inherently faster growth rate, and have higher lipid content under diverse nutrient-limiting environments [25]. Based on this current scenario, the present study focuses on the characterization of biomass and Lipid production as well as emphasized the tremendous possibilities of lignocellulosic biomass degradation ability during the biological treatment procedure utilizing native algal isolates from the mangrove ecosystem for improving biodiesel production. Moreover, these mangrove algal isolates are promising as potential biofuel producers utilizing LCW biomass under variable open climatic circumstances. Hence, the present research work is a preamble attempt to demonstrate that mangrove algal isolates could be a potential microbial cell factory for cellulose degradation and higher lipid production toward potential sustainable biodiesel production using LCWs utilization in near future.
2. MATERIALS AND METHODS
2.1. Cultivation and Microscopic Observation On BS3 and BS4 Algal Isolates
The experimental study had been initiated through water sample collection from the mangrove ecosystem in Sundarban islands. Two distinct types of water samples from the river stream have been collected and stored in a cold box for transportation to the research laboratory. First, these two river water samples were collected and had been filtered through Whatman filter paper (Grade 113, Sigma-Aldrich) to sieve out plant leaves and plant-associated debris. Afterward, 2 ml river water samples (filtered) were inoculated in 8 ml sterile BBM [26] algal growth media (pH 7.2) and incubated at 16 W/m2 light intensity in shaking mode (50 rpm) for 7 days. After 7 days of incubation period, green color appears in the culture tubes. Then, these cultures from two different locations have been undergone for pure culture development on BBM agar plate (pH 7.2). 1 ml sample was transferred to a BBM agar media plate and spread evenly across the surface. Inoculated plates were placed in a temperature-controlled incubator (20–25°C, approximately) where the algae were allowed to grow for about 7–10 days. Grown algal cultures were streaked using a sterile technique onto additional sets of nutrient media plates and placed back in the incubator for isolation. This streaking method was repeated until isolation into an axenic algal culture was achieved. Subsequently, anaxenic algae isolates (BS3 and BS4) had been preserved in 15% glycerol stock at −20°C freezer for further usage. The pure isolates BS3 and BS4 had been regularly sub-cultured to maintain the optimal growth for light microscopic analysis (×400, ×1000 magnification in Magnus MLXi Plus, Magcam DC3) and optical density-biomass analysis (using UV-Vis spectrophotometer from SHIMADZU, Model No: UV-1800).
2.2. Determination of Growth and Lipid Contents in BS3 and BS4 Algal Isolates
The growth profile of novel algae isolates (i.e., BS3 and BS4) had been determined following a spectrophotometric approach by preparing a standard curve between optical density (O.D 700 nm) and biomass (g/ml). Biomass quantification had been done using heating and drying in a hot air oven on filter paper [27]. Biomass content estimation was essential to determine cellular lipid content in regular time intervals during culture sample collection. In vivo qualitative estimation of lipid droplets or lipid accumulation had been determined by the classical Sudan black staining method [28]. It had been observed that cellular algal lipid droplets take blue-black or light blue color whereas non-lipid cellular materials look light pink under the light microscope. Lipid quantification had been carried out using standard lipid extraction and quantification approaches [29,30]. Cellular lipids constituents from algal isolates (BS3 and BS4) had been extracted using dried algae cells (0.5 g) using chloroform methanol solvent (15 ml) extraction method in 2:1 ratio (v/v) following sonication (30 Hz for 20 min on ice pack). Cellular lipid quantification had been carried out using the following molecular standard formula [31] where WL (g) is the weight of lipid and WB (g) the algal biomass dry weight.
C1 (g/g) = WL/WB (1)
2.3. Qualitative Assessment on Cellulose Degradation Potentiality of BS3 and BS4 Algal Isolates
Cellulases randomly act concerning three different ways. The glycosyl bonds were hydrolyzed by exoglucanases, cellodextrinases (1, 4-β-D-glucanhydrolases) and cellobiohydrolases (1, 4-β-D-glucan cellobiohydrolase), and end of the cellulose polymer chain 1, 4-β-glucan-4-glucanhydrolases split the chain through the endoglucanases inwards. However, cellulose degradation was mainly convenient to the catalytic domains of the universal cellulase complex, which was a fourth and non-catalytic element. The β-glucoside glycohydrolases and 1, 4-β-D glucan cellobiohydrolases help to cleave the cellobiose glycosyl bond. Predominantly, in the cellulase complex, a carbohydrate binding module must be present. The attachment and position of the enzymes onto the cellulose and expedite of single molecules discharge helped enzymatic lysis [32-34]. Confirmation of cellulose degradability had been initiated through cellulose Congo red assay where cell-free supernatants had been used for the in vitro degradation assay including carboxy-methyl cellulose (CMC) as sole carbon substrate with a concentration of 1% (w/v). BS3 and BS4 algal isolates from an axenic glycerol mother stock had been picked and inoculated to BBM agar plate (to attain active subculture during experimentation) using an inoculating needle and inoculated onto basal broth medium (modified from Mandels medium) containing CMC sodium salt incubation period at 25°C (±2) for 48 h. Afterward, these exponential BS3 and BS4 algal axenic cultures had undergone for cell free supernatant generation under an aseptic environment. Then, cell-free supernatant of BS3 and BS4 algal isolates had been applied on CMC selective plates bearing groves and incubated for 48 h at 25°C (±2). Finally, these incubated selective plates had been stained with 1% (w/v) Congo-red solution for 15 min and 1 M NaCl (15 min) have been used for discoloration [35-39]. The Congo-red assay had been used for the quantitative determination of the dye decolorization potentiality of the isolate BS4. The dye was degraded by cellulase enzyme produced from the aforementioned algal isolates in the selective BBM medium. Moreover, the Congo red breakdown rate was observed for 0, 3, 6, and 9 days at 417 nm wavelength as a standard outlet [40] including equation 2.
Decolourization (%) = (Initial absorbance – Final absorbance)/Initial absorbance × 100 (2)
Moreover, cellulose degradation potentiality had also been quantified using the di-nitro salicylic acid (DNS) assay to measure the release of reducing sugars after the breakdown of cellulose in the BBM media due to the potential activity of cellulase in BS4 algal isolates. In standard Bradford assay had been followed for estimation the protein concentration of BS4 isolates [41,42]. Afterward, we had measured the cellulase activity following standard protocol [43] including equation 3.
Cellulose enzyme activity as U/ml =W*1000/V. t. M (3)
Where, W = Amount of release glucose, M = Molecular weight of the glucose, V = Volume of the sample, T = Reaction time.
2.4. Studies on Phylogenetic Analysis of Algal Cellulase and Microscopic Observation of BS3 and BS4 Algal Isolates
The endo-b-1, 4-glucanases of gut termites (i.e. UniProtKB_Q64I76) and earthworms (UniProtKB_O77045; UniProtKB_B9A7E3) were the most potent and effective cellulase enzymes toward the degradation of lignocellulosic biomass [44]. These potential enzymes were belonging to the GH protein family 9 (GHF9). Protein sequence homology search among the GHF9 family proteins had helped to identify a few algae-based cellulase and/or endoglucanase group of enzymes, that is, Cel9Bs, Cel9Cs, respectively; however, exact function of these proteins was not yet clear or properly characterized. Based on this context and homology search, algal-based few cellulases and/or endoglucanase sequences, that is, Cel9Bs and Cel9Cs had been collected from online NCBI repositories and depicted in Supplementary Data file S1. Cel9Bs and Cel9Cs sequence alignment for molecular phylogenetic analysis had been generated using Clustal W and Clustal Omega [45] and optimized using MEGA5 [46]. Multiple sequence alignment dataset had been shown in the Supplementary Data file S2. Neighbor-joining (NJ), maximum parsimony, and maximum likelihood (ML) statistical models had been used to infer the phylogenetic relationship among strains of closely associated green algae based on rbcL gene sequences. A substitution model for phylogenetics of each nucleotide sequence data set had been selected using the jModel test [47] with model selection in MEGA5. Representative phylogenetic trees had been drawn using the NJ and ML methods. However, the main objective of doing this phylogenetic analysis was to classify the diverse ranges of algal cellulase and/or endoglucanase enzymes. Recently, Cel9B, Cel9C, and Cel9D-based cellulolytic activity in Chlamydomonas reinharditii has been experimentally proven. However, the exact role of Cel9D on cellulolytic activity had still clearly needed to be understood [48]. Therefore, Cel9B and Cel9C sequences of C. reinharditii were considered in this phylogenetic analysis as a positive set of control. Moreover, the phylogenetic analysis may also be conjugated with the comparative microscopic observation of BS3 and BS4 algal isolates using pre-morphologically characterized algal strains from literature due to non-availability of 16s rRNA sequencing dataset in this current wet research work. This conjugated approach may help to indirectly narrate that BS3 and BS4 belong to which class of algal regimes.
2.5. Statistical Analysis
Statistical analysis was executed using IBM SPSS software (version 26.0). Correlation coefficient test (Pearson Correlation) had been accomplished for the confirmation of significance correlation at 0.05 significance levels (2-tailed) among the day, biomass concentration, and lipid concentration of pure axenic algal cultures inoculated in sterile BBM medium for small-scale experimentations [49].
3. RESULTS AND DISCUSSION
3.1. Studies on Morphological Characterization of BS3 and BS4 Algal Isolates through Microscopic Observation and Phylogenetic Analysis Using Putative Algal Cellulase Enzymes
Microscopic observation had played a very important role to identify the morphological characterization of algal communities through physical appearance with or without staining applications. Two mangrove algal isolates, that is, BS3 and BS4 had been successfully visualized under bright field microscopy without staining as algae contain photosynthetic pigments in its cellular biomass. Microscopic examination had demonstrated the morphological and physiological diversity of two different isolates at ×400 [Figure 1] and ×1000 magnification [Figure 1b and e] using algae culture of the 9th day. Microscopic observation had shown that BS4 has a very close morphological resemblance with the microscopic view [50] of the algal regime Gonium pectorale [Supplementary Figure S1, Figure 2c and d]. However, BS3 did not show any similarity considering the microscopic characteristics of chosen algal communities. Interesting phylogenetic analysis had also shown that G. pectorale algae [50] were closely associated clade to C. reinharditii as phylogenetic analysis considering Cel9B and Cel9C cellulase and/or endoglucanase protein sequences in this present study [Figure 2a and 2b]. However, it would be impossible to place this alga taxonomically based on microscopic observation and in-silico molecular phylogenetic analysis. Therefore, more work on the molecular and physiological levels would be required to firmly establish the taxonomy of this strain. For the purposes of this article, this strain will be referred to as BS4 algal isolate from the mangrove ecosystem.
 | Figure 1: Microscopic observation of mangrove ecosystem derived algal isolates (a) BS3 isolate’s microscopic view at ×400 magnification; (b) BS3 isolate’s microscopic view after Sudan black staining under ×400 magnification; (c) BS3 isolate’s microscopic view after SB stain in ×1000; (d) BS4 isolate’s microscopic view under ×400; (e) BS4 isolate’s microscopic view after staining; (f) BS4 isolate’s microscopic view after SB stain in ×1000). [Click here to view] |
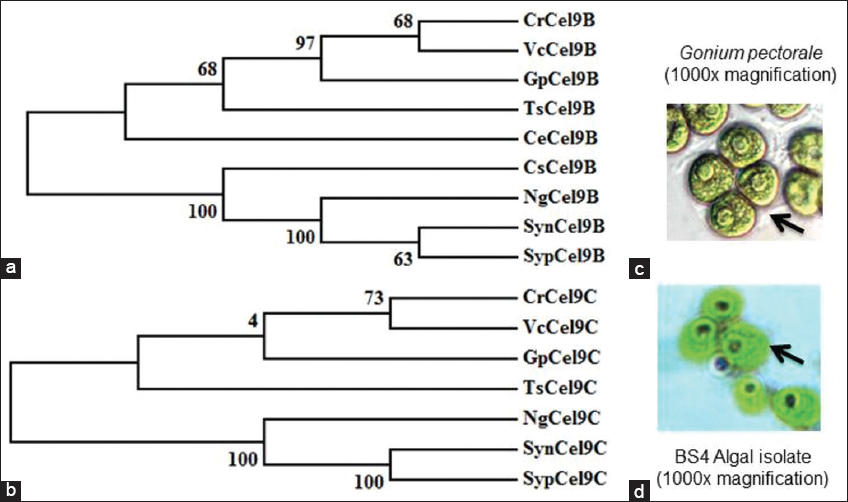 | Figure 2: Comparative overview on characterization of BS3 and BS4 algal isolates through microscopic observation and phylogenetic analysis (Phylogenetic tree has generated based on putative algal Cel9Bs and Cel9Cs protein sequences. The final phylogenetic tree presented here has been developed using MEGA5 with 500 times bootstrap replication and a substitution model p-distance (substitution type amino acids, complete deletion of gaps/missing data treatments). The evolutionary history was inferred using the Neighbor-Joining method [52]. The optimal tree with the sum of branch length is ~2.53. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches [53]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree; algae based Cel9Bs and Cel9Cs abbreviation are mentioned with all possible information in supplementary data file S1 microscopic observation depicts the morphological resemblance of BS4 algal isolate with G. pectorale strain under ×1000 magnification. Microscopic views are not in uniform scale. Here is the abbreviation of protein sequences and strains consider for phylogenetic tree development CrCel9B: Chlamydomonas reinhardtii; VcCel9B: Volvox carteri; GpCel9B: G. pectorale; TsCel9B: Tetrabaena socialis; CeCel9B: Chlamydomonas eustigma; CsCel9B: Chlorella sorokiniana; NgCel9B: Nannochloropsis gaditana; SynCel9B: Synechococcus sp.; SypCel9B: Synechocystis sp.; CrCel9C: Chlamydomonas reinhardtii; VcCel9C: Volvox carteri; GpCel9C: G. pectorale; TsCel9C: Tetrabaena socialis; NgCel9C: Nannochloropsis gaditana; SynCel9C: Synechococcus sp.; SypCel9C: Synechocystis sp. (strain PCC 6803/Kazusa). Moreover, microscopic views of Gonium pectorale (reference point) have been adopted from mentioned references in supplementary Figure S1 [50] as standard reference to make comparative overview in comparison to BS4 isolate in the present study. [Click here to view] |
3.2. Qualitative Analysis on Cellulolytic Activities of BS3 and BS4 Algal Isolates
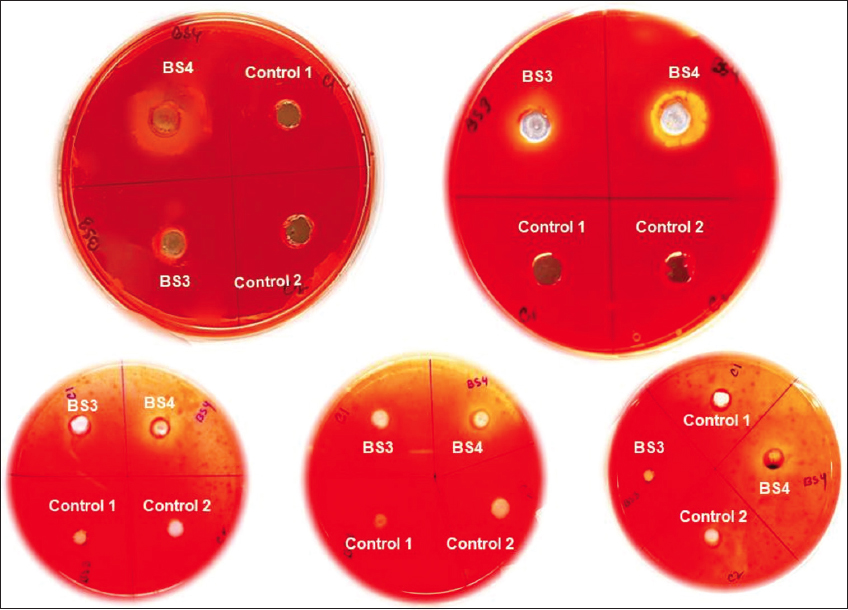
A qualitative analysis of cellulolytic enzymatic activities had been investigated for BS3 and BS4 algal isolates of the mangrove ecosystem considering Congo red plate assay approach. A clear hollow round zone had been observed on selective plates for BS4 algal isolate after 48 h of photo incubation at 25°C (±2) following Congo red plate assay technique. A clear visible yellow-color degradation zone had been shown around the grove where cell-free supernatant of BS4 algal isolate had been applied. Thus, it had clearly and qualitatively confirmed that BS4 algal isolate and its cell free supernatant did have enormous potential to degrade or hydrolyze cellulose as potential cheaper feedstock. However, no zone had been observed for BS3 isolate on selective plates compared to the control sets (i.e., negative control 1 refers to sterilized distilled Milli Q water whereas negative control 2 refers to the sterile BBM medium). Whereas BS3 algal isolate did not show any cellulose-degrading ability while performing a similar experimental design. Extracellular secretion of active endoglucanases and/or cellulases degrading water soluble cellulose moieties had been indicated the presence of an extracellular secretory enzymatic platform toward extracellular cellulose decomposition [Figure 3, Supplementary Figures S2 and S3] in BS4 algal isolates. The dye decolorizing percentage (%) of BS4 algal isolate was attained as 96.04% on 9th day in this present study [Figures 4a-d and 5]. Simultaneously, it had also been shown as the continuation of this study where it had been shown that degradation products (i.e., precursor and/or intermediate hexose or pentose monomers) of these celluloses can be utilized toward value-added biomolecule generations likely biodiesel in this present study. Released reducing sugar had been estimated by routine DNS assay and BS4 released the highest amount of reducing sugar on 9th day (i.e., 276.72 μg.ml-1) [Figure 6 and Supplementary Figure S4]. The protein estimation had been done by Bradford assay, to measure the total protein concentration of algal enzyme hydrolysate the concentration from BS4 90.77mg.ml-1 [Supplementary Figure S5] [43]. However, the present result had specifically shown that higher cellulolytic activities were observed in the case of algal isolate BS4 0.284 (±0.001) U.mg-1 [Table 1]. In comparison, BS3 was not able to accelerate any enzymatic activities for the breakdown of the substrates for cellulose degradation. For BS3 isolate, the low and average molecular weight fragments formation process had been decreased by the split of glycosidic bonds because these fragments were soluble in water. Furthermore, during the photosynthesis growth condition, BS4 isolate helped to produce organic carbon acquisition toward degrading the cellulase. Hydrolysis of in vitro study had been focusing on CMC utilization for algal cellular higher growth rate and revealed that the secretion of cellulase and/or endoglucanase [51].
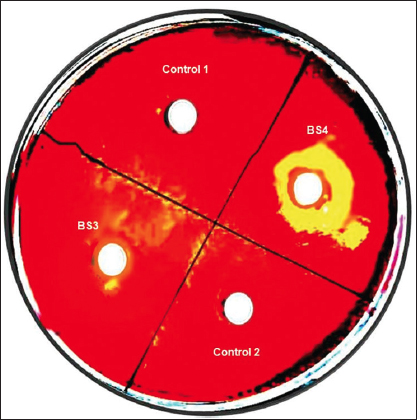 | Figure 3: Profile on secretion of cellulolytic enzymes of BS4 and BS3 algae isolates using CMC supplementation and Congo red plate assay (Experimental results show that BS4 is the potential algal isolate which has extracellular cellulase and/or endoglucanase activity while BS3 does not bear this phenotypic trait through natural selection. Moreover, negative control 1 refers to sterilized distilled MiliQ water which reflects metal constituents do not show any impact on extracellular enzyme secretion and/or on its enzymatic activity; whereas negative control 2 refers to the sterile BBM medium which confirm not contaminants do exist in BBM medium, respectively). [Click here to view] |
 | Figure 4: Dye decolorization analysis using algal isolate BS4 (0–9 days). These pictures signify the congo red decolorizing activity (96.04%) of BS4 in BBM algal growth medium within 9 days. Dye decolorization assay before the treatment (0 days) without Congo red (a), with Congo red (b) and dye decolorization assay after the treatment (9 days) Without Congo red (c), With Congo red (d). [Click here to view] |
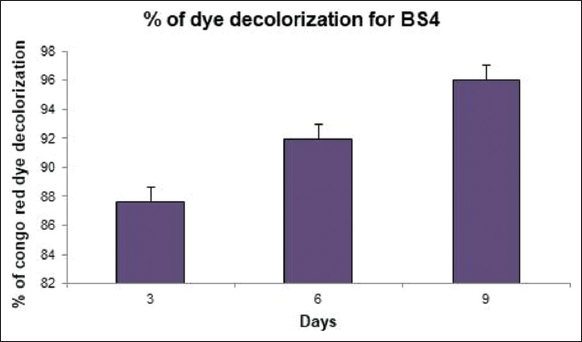 | Figure 5: Determination of dye decolorization analysis using algal isolate BS4 (Decolorizing activity was highest (96.04%) of BS4 in BBM algal growth medium within 9 days; n = 3 where n is the number of best replicates). [Click here to view] |
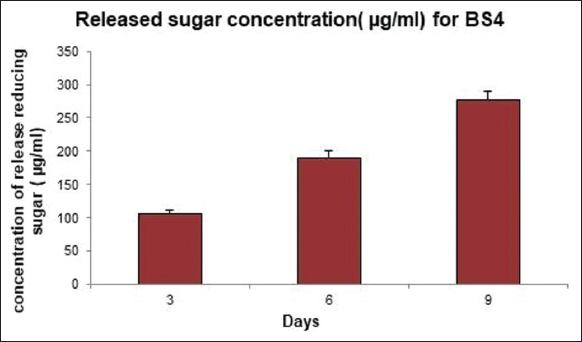 | Figure 6: Measurement on reducing sugar release after cellulose degradation using algal isolate BS4 (Released reducing sugar has been estimated by DNS assay and BS4 released the highest amount of reducing sugar within 9 days, that is, 276.72 mg.ml-1; n = 3 where n is the number of best replicates). [Click here to view] |
Table 1: Comparative overview on productions of biomass and lipid with cellulase enzyme activities in diverse algal assemblages.
| Algal assemblage | Biomass production rate (g.L-1.day-1)* | Lipid production rate (g.L-1.day-1)* | Cellulase Enzymatic activity in U.mg-1* (ND refers undetermined) | References |
|---|---|---|---|---|
| Chlorella sorokiniana | 0.23–1.47 | 0.044 | ≥240,000 (Cellulase) | [54,55] |
| Chlamydomonas reinhardtii | 0.672 | 0.141 | 0.036 (crude enzyme) | [48,56,57] |
| Nannochloropsis gaditana | 0.045 | 0.014 | ND | [58,59] |
| Algal isolate BS4 | 2.77 (±0.04) | 1.93 (±0.03) | 0.284 (±0.001) (crude enzyme) | Current study |
* we have unified the values based on the dataset available in original references.
3.3. Growth and Lipid Profiles of BS3 and BS4 Algal Isolates with Statistical Constraints
Growth profile studies had shown that BS4 gives comparatively higher biomass production compared to BS3 on day 9th of cultivation [Figures 7a,b and 8a,b]. Extensive studies had also depicted that the biomass productivities for BS4 and BS3 isolates are 2.77 (±0.04) g.L-1.day-1 and BS3 1.88 (±0.02) g.L-1.day-1, respectively. BS4 was comparatively generated with a higher amount of lipid content compared to BS3 [Figures 7c and 8c, Figures 7d and 8d] on day 9th of cultivation. Moreover, BS4 and BS3 had shown lipid productivities, that is, 0.0775 (±0.001) g.g-1.day-1 [i.e., 1.93 (±0.03) g.L-1.day-1] and 0.0588 (±0.002) g.g-1.day-1, respectively. However, experimental results had clearly shown that lipid production is highly proportional to biomass production in mangrove algal isolates BS3 and BS4. Preliminary screening had shown lipid droplet accumulation in the cellular fraction of two novel isolates, that is, BS3 and BS4. After Sudan black staining approach shows BS4 accumulates comparatively higher amounts of lipid droplet accumulation inside the algal cells than do the BS3. Algal isolates showed diverse intensity in stain (blue or blue- black) uptake considering the presence of different lipid content in BS3 [Figure 1b and c] and BS4 [Figure 1e and f], respectively. In addition, statistical analysis especially the correlation coefficient test had been done for the confirmation of significant correlation among the day, biomass concentration and lipid concentration by IBM SPSS 26 statistical software at 0.05 significance level and the result showed that there is significant relations present among all the three parameters in both BS3 and BS4 algal assemblages [Supplementary Tables 1 and 2] [45].
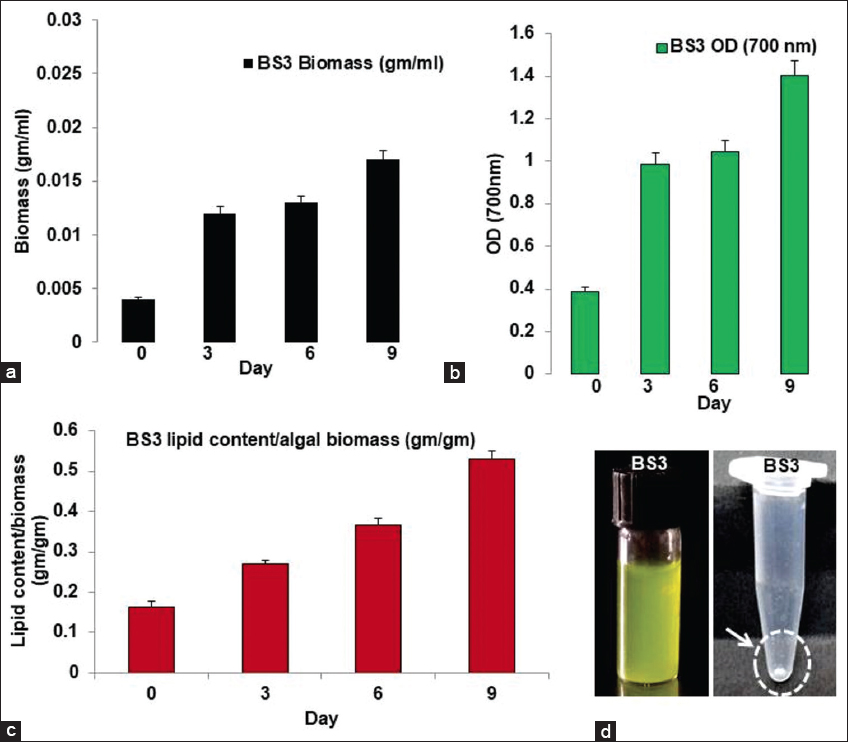 | Figure 7: Biomass and lipid production profile using BS3 algal isolate from mangrove ecosystem (a) BS3 isolate’s growth curve where day-wise biomass increases; (b) BS3 isolate’s growth curve where day-wise OD increases; (c) BS3 growth curve while day-wise lipid content/algal biomass increases; (d) BS3 algal pure culture and the extracted lipid as dried white pellet). [Click here to view] |
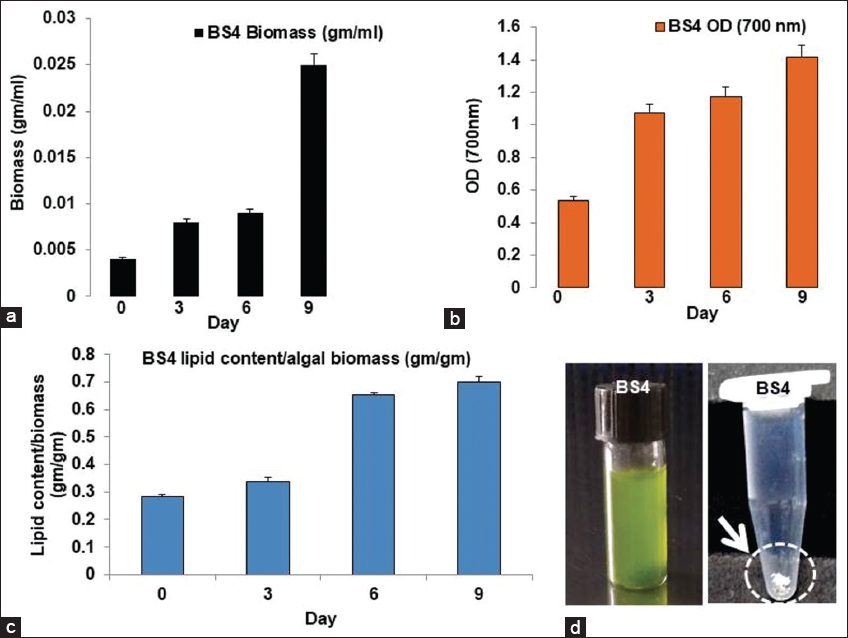 | Figure 8: Biomass and lipid production profile using BS4 algal isolate from mangrove ecosystem (a) BS4 isolate’s growth curve where day-wise biomass increases; (b) BS4 isolate’s growth curve while day-wise O.D. increases; (c) BS4 growth curve where day-wise lipid content/algal biomass increases; (d) BS4 pure culture and the extracted lipid as dried white pellet). [Click here to view] |
4. CONCLUSION
The present study has revealed several intriguing features of mangrove algal isolates, namely, BS4 and BS3. The BS4 algae isolate exhibits two distinct characteristics: Likely cellulose (the predominant constituent of LCW) degradation and higher lipid generation productivities as a single pot microbial cellular factory. Furthermore, at mesophilic and neutrophilic conditions, BS4 algal isolate has produced higher lipid contents, higher biomass productivities, and shown higher cellulose degradation. In contrast, BS3 does not show any cellulolytic efficacy though it produces a good amount of lipid and biomass during batch cultivation operations. Therefore, it seems BS4 algae isolate could be one of the most promising aspirants toward sustainable biodiesel production utilizing lignocellulosic biomass. However, microscopic observation along with in silico comparative phylogenetic analysis tentatively and indirectly infers that BS4 is the closest clade of G. pectorale. However, it would be impossible to place this alga taxonomically based on microscopic observation alone, so molecular phylogenetic analysis needs to be carried out. However, BS4 exhibits the most similar phenotypic traits, such as cellulolytic efficacies harboring extracellular endoglucanase and/or cellulase activities (i.e., Cel9B and Cel9C), as seen in Chlamydomonas reinharditti. Moreover, the function of another cellulose degrading module, that is, Cel9D, needs to be explored not only to understand the catalytic efficiency but also to comprehend the regulatory mechanism. Therefore, this study needs to be researched further to improve biomass and lipid productivities using BS4 and BS3 mangrove algal isolates.
4.1. Supplementary Information
Supplementary Dataset S1; Supplementary Dataset S2; Supplementary Figure S1; Supplementary Figure S2; Supplementary Figure S3; Supplementary Figure S4; Supplementary Figure S5; Supplementary Table 1; Supplementary Table 2.
5. ACKNOWLEDGMENTS
The authors would like to thank JIS University and JIS Group of Educational Initiatives. The author would also like to show their special gratitude to Mr. Somnath Das for his views on statistical analysis.
6. AUTHORS’ CONTRIBUTIONS
Ms. Shrestha Debnath: All wet laboratory experimentation, data collection, data analysis, data set tabulation, data validation, investigation, wet laboratory experimental dataset visualization, original drafting, writing, and formatting. Dr. Dipankar Ghosh: Conceptualization, original drafting, writing, editing, reviewing, computation experimentation and computation dataset analysis, computational results visualization, and supervision.
7. FUNDING
There is no funding to report.
8. CONFICTS OF INTEREST
The authors declare no competing interests.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data supporting this study are available on request.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Vélez-Mercado MI, Talavera-Caro AG, Escobedo-Uribe KM, Sánchez-Muñoz S, Luévanos-Escareño MP, Hernández-Terán F,
2. Tayyab M, Noman A, Islam W, Waheed S, Arafat Y, Ali F,
3. Verdini F, CalcioGaudino E, Grillo G, Tabasso S, Cravotto G. Cellulose recovery from agri-food residues by effective cavitational treatments. Appl Sci 2021;11:4693. [CrossRef]
4. Sankaran R, Cruz RA, Pakalapati H, Show PL, Ling TC, Chen WH,
5. Mtui GY. Recent advances in pretreatment of lignocellulosic wastes and production of value added products. Afr J Biotech 2009;8:1398-415.
6. Ani PC, Nzereogu PU, Agbogu AC, Ezema FI, Nwanya AC. Cellulose from waste materials for electrochemical energy storage applications:A review. Appl Surf Sci 2022;11:100298. [CrossRef]
7. Prasad RK, Chatterjee S, Mazumder PB, Gupta SK, Sharma S, Vairale MG,
8. Ghosh D, Das S. Genetic and metabolic engineering approaches for improving accessibilities of lignocellulosic biomass toward biofuels generations. In:Genetic and Metabolic Engineering for Improved Biofuel Production from Lignocellulosic Biomass. Netherlands:Elsevier;2020. 13-35. [CrossRef]
9. Bilal M, Iqbal HM. Recent advancements in the life cycle analysis of lignocellulosic biomass. Curr Sust Renew Energ Rep 2020;7:100-7. [CrossRef]
10. Arevalo-Gallegos A, Ahmad Z, Asgher M, Parra-Saldivar R, Iqbal HM. Lignocellulose:A sustainable material to produce value-added products with a zero waste approach-a review. Int J Biol Macromol 2017;99:308-18. [CrossRef]
11. Howard RL, Abotsi ELJR, Van Rensburg EJ, Howard S. Lignocellulose biotechnology:Issues of bioconversion and enzyme production. Afr J Biotechnol 2003;2:602-19. [CrossRef]
12. Mishra S, Singh PK, Dash S, Pattnaik R. Microbial pretreatment of lignocellulosic biomass for enhanced biomethanation and waste management. 3 Biotech 2018;8:458. [CrossRef]
13. Otto B, Beuchel C, Liers C, Reisser W, Harms H, Schlosser D. Laccase-like enzyme activities from chlorophycean green algae with potential for bioconversion of phenolic pollutants. FEMS Microbiol Lett 2015;362:fnv072. [CrossRef]
14. Abd Ellatif S, El-Sheekh MM, Senousy HH. Role of microalgal ligninolytic enzymes in industrial dye decolorization. Int J Phytoremediation 2021;23:41-52. [CrossRef]
15. Afreen S, Shamsi TN, Baig MA, Ahmad N, Fatima S, Qureshi MI, Hassan MI,
16. Shi W, Li J, He B, Yan F, Cui Z, Wu K,
17. Baskar G, Aiswarya R. Trends in catalytic production of biodiesel from various feedstocks. Renew Sust Energy Rev 2016;57:496-504. [CrossRef]
18. Tang Y, Gu X, Chen G. 99% yield biodiesel production from rapeseed oil using benzyl bromide-CaO catalyst. Environ Chem Lett 2013;11:203-8. [CrossRef]
19. Hariskos I, Posten C. Biorefinery of microalgae-opportunities and constraints for different production scenarios. Biotechnol J 2014;9:739-52. [CrossRef]
20. Tredici MR. Photobiology of microalgae mass cultures:Understanding the tools for the next green revolution. Biofuels 2010;1:143-62. [CrossRef]
21. Ge S, Champagne P, Plaxton WC, Leite GB, Marazzi F. Microalgal cultivation with waste streams and metabolic constraints to triacylglycerides accumulation for biofuel production. Biofuel Bioprod Biorefin 2017;11:325-43. [CrossRef]
22. Ge S, Champagne P. Nutrient removal, microalgal biomass growth, harvesting and lipid yield in response to centrate wastewater loadings. Water Res 2016;88:604-12. [CrossRef]
23. Sforza E, Cipriani R., Morosinotto T, Bertucco A, Giacometti GM. Excess CO2 supply inhibits mixotrophic growth of
24. De K, Debnath S, Ghosh D. Elevating algal biomass generation toward sustainable utilization for high value added biomolecules generations. J Appl Biol Biotech 2022;10:1-12. [CrossRef]
25. Buši?A, Kundas S, Morzak G, Belskaya H, Mar?etko N, Šantek MI,
26. Bold HC. The morphology of
27. Abdelaziz AE, Leite GB, Belhaj MA, Hallenbeck PC. Screening microalgae native to Quebec for wastewater treatment and biodiesel production. Bioresour Technol 2014;157:140-8. [CrossRef]
28. Osman ME, Abdel-Razik AB, Zaki KI, Mamdouh N, El-Sayed H. Isolation, molecular identification of lipid-producing
29. Malekzadeh M, Najafabadi HA, Hakim M, Feilizadeh M, Vossoughi M, Rashtchian D. Experimental study and thermodynamic modeling for determining the effect of non-polar solvent (hexane)/polar solvent (methanol) ratio and moisture content on the lipid extraction efficiency from
30. Alam MM, Mumtaz AS, Hallenbeck PC, Shah SA, Abidin SZ, Inam F. Morphological characterization, growth appraisal, and probing biofuels potential of newly isolated
31. Abdelaziz AE, Ghosh D, Hallenbeck PC. Characterization of growth and lipid production by
32. Voget S, Steele HL, Streit WR. Characterization of a metagenome-derived halotolerant cellulase. J biotechnol 2006;126:26-36. [CrossRef]
33. Sharma S, Kumawat KC, Kaur S. Potential of indigenous ligno-cellulolytic microbial consortium to accelerate degradation of heterogenous crop residues. Environ Sci Pollut Res 2022;1-16. [CrossRef]
34. Begemann MB, Mormile MR, Paul VG, Vidt DJ. Potential enhancement of biofuel production through enzymatic biomass degradation activity and biodiesel production by halophilic microorganisms. In:Halophiles and Hypersaline Environments. Berlin, Heidelberg:Springer;2011. 341-57. [CrossRef]
35. Cortes-Tolalpa L, Jiménez DJ, de Lima Brossi MJ, Salles JF, van Elsas JD. Different inocula produce distinctive microbial consortia with similar lignocellulose degradation capacity. Appl Microbiol Biotechnol 2016;100:7713-25. [CrossRef]
36. Liang YL, Zhang Z, Wu M, Wu Y, Feng JX. Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by
37. Menasria T, Monteoliva-Sánchez M, Benhadj M, Benammar L, Boukoucha M, Aguilera M. Unraveling the enzymatic and antibacterial potential of rare halophilic actinomycetes from Algerian hypersaline wetland ecosystems. J Basic Microbiol 2022;62:1202-15. [CrossRef]
38. Gerken HG, Donohoe B, Knoshaug EP. Enzymatic cell wall degradation of
39. McDonald JE, Rooks DJ, McCarthy AJ. Methods for the isolation of cellulose-degrading microorganisms. Methods Enzymol 2012;510:349-74. [CrossRef]
40. Mahalakshmi S, Lakshmi D, Menaga U. Biodegradation of different concentration of dye (Congo red dye) by using green and blue green algae. Int J Environ Res 2015;9:735-44.
41. Gupta P, Samant K, Sahu A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int J Microbiol 2012;2012:578925. [CrossRef]
42. Gusakov AV, Kondratyeva EG, and Sinitsyn AP. Comparison of two methods for assaying reducing sugars in the determination of carbohydrase activities. Int J Anal Chem 2011;2011:283658. [CrossRef]
43. Shrestha S, Rahman M, Qin W. New insights in pectinase production development and industrial applications. Appl Microbiol Biotech 2021;105:9069-87. [CrossRef]
44. Nozaki M, Miura C, Tozawa Y, Miura T. The contribution of endogenous cellulase to the cellulose digestion in the gut of earthworm (
45. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W:Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994;22:4673-80. [CrossRef]
46. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5:Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731-9. [CrossRef]
47. Posada D. jModelTest:Phylogenetic model averaging. Mol Biol Evol 2008;25:1253-56. [CrossRef]
48. Blifernez-Klassen O, Klassen V, Doebbe A, Kersting K, Grimm P, Wobbe L,
49. Berthold DE, Shetty KG, Jayachandran K, Laughinghouse HD 4th, Gantar M. Enhancing algal biomass and lipid production through bacterial co-culture. Biomass Bioenergy 2019;122:280-9. [CrossRef]
50. Müller OF. Terrestrial and river worms, or animals of infusoria, helminthics and crustaceans, not marine,a brief history. Havniae Lisiae 1773;1:74.
51. Wahlström N, Edlund U, Pavia H, Toth G, Jaworski A, Pell AJ,
52. Saitou N, Nei M. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-25.
53. Felsenstein J. Confidence limits on phylogenies:An approach using the bootstrap. Evolution 1985;39:783-91. [CrossRef]
54. Zuorro A, Malavasi V, Cao G, Lavecchia R. Use of cell wall degrading enzymes to improve the recovery of lipids from
55. Xiao M, Shin HJ, Dong Q. Advances in cultivation and processing techniques for microalgal biodiesel:A review. Korean J Chem Eng 2013;30:2119-26. [CrossRef]
56. Zhu Z, Cao H, Li X, Rong J, Cao X, Tian J. A Carbon fixation enhanced
57. Xu L, Cheng X, Wang Q. Enhanced lipid production in
58. Mitra M, Patidar SK, George B, Shah F, Mishra S. A euryhaline
59. Scholz MJ, Weiss TL, Jinkerson RE, Jing J, Roth R, Goodenough U,
60. Hirooka S, Hirose Y, Kanesaki Y, Higuchi S, Fujiwara T, Onuma R,
61. Patras D, Moraru CV, Socaciu C. Bioactive ingredients from microalgae:Food and feed applications. BUASVMCN-FST 2019;76:1-9. [CrossRef]
62. Russell JJ, Theriot JA, Sood P, Marshall WF, Landweber LF, Fritz-Laylin L,
63. Arakaki Y, Kawai-Toyooka H, Hamamura Y, Higashiyama T, Noga A, Hirono M,
64. Zanella L, Vianello F. Microalgae of the genus
65. Touloupakis E, Rontogiannis G, Benavides AM, Cicchi B, Ghanotakis DF, Torzillo G. Hydrogen production by immobilized
66. Chader S, Mahmah B, Chetehouna K, Mignolet E. Biodiesel production using
SUPPLEMENTARY FIGURES AND TABLES
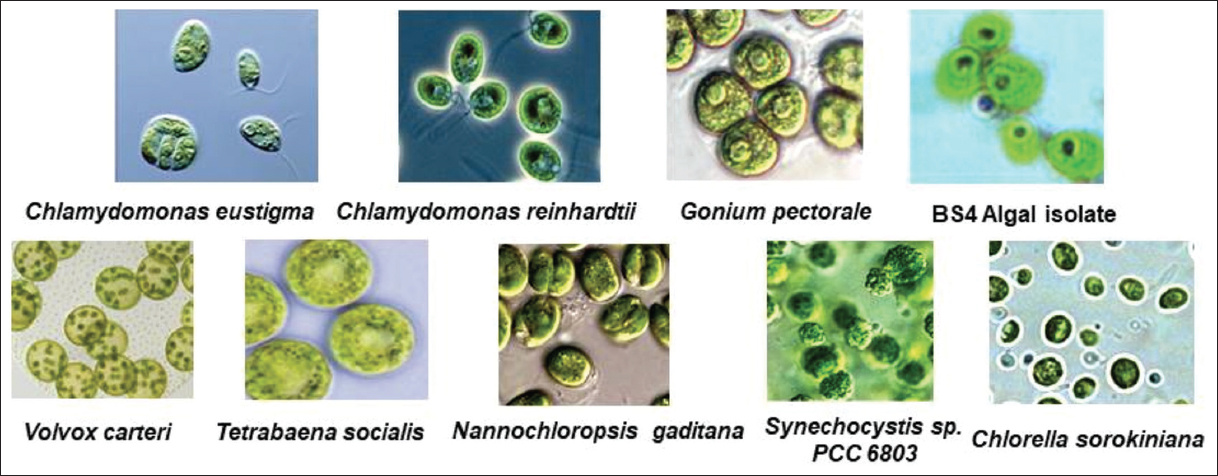 | Supplementary Figure S1: Comparative microscopic overview on diverse algal regimes bearing Cel9B and Cel9C proteins as phenotypic trait to degrade cellulosic materials. Microscopic views are not in uniform scale but photographs have been taken at ×1000 magnification under oil immersion toward comparison with BS4 algal isolate used in this present study and reference microscopic views have been adopted from mentioned references as standard reference to make comparative overview in comparison to BS4 isolate in the present study [50,60-66]. [Click here to view] |
 | Supplementary Figure S2: Cellulase plate assay (qualitative) for Congo red degradation. Breakdown the Congo red mixed BBM agar media enriched with carboxy methyl cellulose shows the orange hollow zone around the groove which signifies the cellulolytic activity of BS4. [Click here to view] |
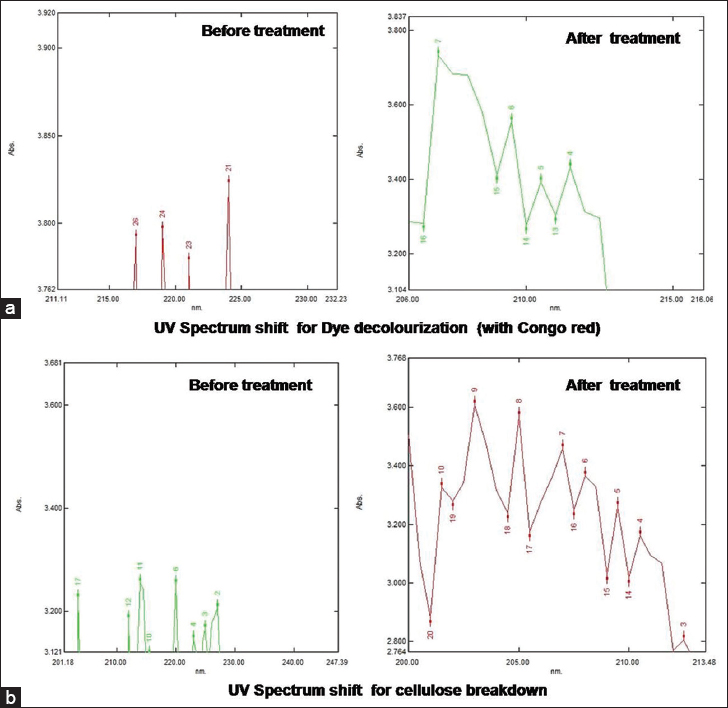 | Supplementary Figure S3: (a) These UV spectrum shifts for dye decolorization (with Congo red) signify the breakdown of Congo red within 9-day time span for BS4 isolate. Before the treatment, the highest UV absorbance spectra of Congo red were 224 nm. That was shifted to 207nm that clearly indicates the breakdown of Congo red. (b) UV Spectrum shift for cellulose breakdown signifies the breakdown of cellulose within 9-day time span BS4 isolate. Before the treatment, the highest UV absorbance spectra 214 nm. That was shifted to 203 nm that clearly indicates the breakdown of cellulose. [Click here to view] |
 | Supplementary Figure S4: DNS assay for sugar estimation. After the treatment, BS4 shows that the amount of released reducing sugar high (deep brown color). [Click here to view] |
 | Supplementary Figure S5: Bradford assay for protein estimation. This picture shows that after reaction of bradford reagent with BS4 extract, it change its color to blue which signifies the presence of high amount of protein. [Click here to view] |
Supplementary Table 1: Correlation among day, biomass concentration, and lipid concentration of BS3.
| Correlation | |||
|---|---|---|---|
| BS3 | Day | Biomass concentration | Lipid concentration |
| Day | |||
| Pearson Correlation | 1 | 0.930 | 0.398 |
| Sig. (2-tailed) | 0.002 | 0.003 | |
| Biomass Concentration | |||
| Pearson Correlation | 0.930 | 1 | 0.411 |
| Sig. (2-tailed) | 0.002 | 0.001 | |
| Lipid Concentration | |||
| Pearson Correlation | 0.398 | 0.411 | 1 |
| Sig. (2-tailed) | 0.003 | 0.001 | |
Correlation is significant at the 0.05 level (2-tailed). Statistical Analysis by IBM SPSS 26.
Supplementary Table 2: Correlation among day, biomass concentration, and Lipid concentration of BS4.
| Correlation | |||
|---|---|---|---|
| BS4 | Day | Biomass concentration | Lipid concentration |
| Day | |||
| Pearson Correlation | 1 | 1.00 | 0.889 |
| Sig. (2-tailed) | 0.003 | 0.002 | |
| Biomass concentration | |||
| Pearson correlation | 1.00 | 1 | 0.890 |
| Sig. (2-tailed) | 0.003 | 0.001 | |
| Lipid concentration | |||
| Pearson correlation | 0.889 | 0.890 | 1 |
| Sig. (2-tailed) | 0.002 | 0.001 | |
Correlation is significant at the 0.05 level (2-tailed). Statistical Analysis by IBM SPSS 26.