ARTICLE HIGHLIGHTS
Isolation of oleaginous bacteria from marine water samples which are potential accumulators of polyunsaturated fatty acids (PUFAs).
De novo synthesis of fatty acids through PUFA synthase/polyketide synthase pathway does not require desaturation and elongation of saturated fatty acids.
Triphenyl tetrazolium chloride method was used for screening of oil producers from seawater.
Bacillus paralicheniformis (OQ202112) was identified as the most potent lipid accumulator.
Lignocellulosic agro waste was used as a source of nutrition for lipid accumulators.
The major components of fatty acid profile were oleic acid and linoleic acid.
Transesterification of fatty acids under optimum conditions produced 86.2% fatty acid methyl ester (biodiesel).
Isolated organism can be used as a single-cell oil factory to produce biodiesel and thus minimize dependence on non-renewable energy resources.
The cetane rating obtained for the biodiesel was in the range that marks good quality of the fuel.
1. INTRODUCTION
High rise in energy requirements and decreasing amount of fossil fuels seems troublesome for the future. Energy crunch and climate crisis are results of over consumption of renewable energy sources. This compelled researchers to look for green energy production. Biodiesel has been proven advantageous, as it is more maintainable and economically efficient [1]. The idea of using mineral-based fuels is gradually falling out of favor due to the overuse, depletion, and associated environmental effects of fossil fuels [2]. Plant and animal fats can be transesterified to produce biodiesel and this product can be used after mixing it with petroleum diesel. Biodiesel is beneficial for society and environment as it is biodegradable, safe, renewable, and less polluting [3].
Production of biodiesel can be done from various organisms based on the multiplication capacity of organisms as well as their ability to accumulate lipids as faster growth results in higher lipid accumulation, eventually, increasing the production yield coefficient [4,5]. Ideal fatty acid accumulation for biodiesel production can be achieved by suitable organism possessing aforesaid properties. Algae and plant seeds possessing higher amounts of lipids can also be used as fatty acid sources for transesterification. However, there may be several problems in using algae and seeds such as varying climatic changes and land requirements [6]. Literature has reviewed many yeast strains possessing higher amount of lipids but bacterial contamination while growing yeast causes difficulty and becomes the reason for lower production as cell growth gets inhibited [7-9]. Above all, oleaginous bacteria show alluring properties such as low space requirements, speedy growth rate, economic feasibility, and less dependence on seasons [10]. Moreover, many bacterial species are reported to be suitable candidates for high oil production like Rhodococcus opacus PD630 which accumulated oil up to 80% cell dry weight (CDW) [11,12]. Considering above-mentioned benefits, bacteria are suitable for the production of future fuel [12]. Microorganisms’ ability to increase fatty acids varies with varying growth conditions even if the organism is the same [13]. Yeasts and bacteria grow at the same rate so it is feasible to study their genes and metabolic engineering at the genetic level [14]. Several genes are studied well and can be altered for more production of fatty acids. These genetic fragments are responsible to code for specific proteins, which play a role in fatty acid synthesis [15,16]. In maritime environment, omega-3 polyunsaturated fatty acids (PUFAs) are contributed by microorganisms, which are considered the main producer of it. As far as sources of long-chain polyunsaturated fatty acids are concerned, bacteria seem more potent than fish as downstream processing in the case of bacteria is inexpensive [17-21]. Gammaproteobacteria such as Colwellia, Moritella, Shewanella, Vibrio, and Psycromonas are known for their PUFAs accumulating ability. Moreover, fatty acid metabolism in the aforementioned microorganisms has also been studied [22]. The accumulation of LC-PUFAs (long-chain PUFAs) in these organisms is believed to be responsible for increasing membrane fluidity to adapt organisms to psychrophilic environments [23-25]. Secondary metabolites are synthesized by a PUFA synthase through the anaerobic polyketide synthase pathway in seawater microbes [26]. Genes, involved in fatty acid synthesis, were firstly decoded in Shewanella sp. (SCRC-2738) and are organized in 38Kbp gene segment containing operon for EPA synthesis [27]. The gene cluster, which was found in Shewanella sp. (SCRC-2738), possesses five genes sequentially and its presence has been discovered in eukaryotes and bacterial strain [26,28,29]. This data show that PUFA producers from seawater have not been fully discovered and there is a need to explore lipid accumulators from marine water to save environment by providing renewable sources of biofuel.
The current question is to find supreme substrate, which is cheaper to produce biodiesel for green environment [30,31]. Researchers are experimenting to find novel economical substrate for fatty acid accumulation in bacteria such as lignocellulosic agro wastes [32-34]. The procedure becomes more efficient and less expensive when lignocellulosic biomass is used to produce lipids. It also has a smaller negative environmental impact [35]. Crude glycerol, dairy waste, etc. can also be used instead of non-renewable resources [36,37]. Agribusiness is the main occupation in India. Hence, abundant amount of agro waste is generated and burnt annually leading to pollution and difficulty for breathing. This warrants the need for biodiesel production using renewable waste like lignocellulose.
In the present research, to reduce harmful effects on environments, a promising effort has been made to produce biodiesel by utilizing marine microorganisms. These microorganisms have the ability to accumulate long-chain PUFAs using agro waste. Unlike previous studies, the substrate was not subjected to any pre-treatment to accumulate PUFA for biodiesel production. There has not been a single study conducted yet on B. paralicheniformis as a potential lipid accumulator.
2. MATERIALS AND METHODS
2.1. Sample Collection and Enrichment for Lipid Accumulator for Screening
Seawater sample was collected from various sites, i.e., (1) seawater from Somnath, (2) seawater from seashore near Diu, and (3) water sample from the point where sea and river water mixes near village Simar. Marine Artificial Seawater (MASW) agar plates containing 25.5 g of instant ocean salt, 5 g peptone, 1 g yeast extract, and 15 g of bacteriological agar were prepared for bacterial isolation. 0.1 ml sea water was taken for inoculation. Plates were incubated at 28°C for 3 days in aerobic condition [38]. On the basis of phenotypic heterogeneity, 23 isolates were screened and streaked over MASW plates for 5 days and incubated at 28°C. Purification and conservation were done for further analysis of this sample. All these isolates were tested for lipid accumulation ability by performing Sudan black B staining and TTC (triphenyl-tetrazolium chloride) screening [39,40].
2.2. Lipid Synthesis
The isolates selected by Sudan Black B staining and TTC screening were further checked for their lipid accumulation ability. Each of the selected strains from every sample was activated by Luria-Bertani broth. Activation was done for different time periods of 24 h, 48 h, and 72 h for every strain to check effective inoculum age as well as they were screened for higher production using selected agro wastes (groundnut husk and rice bran) and glucose as a standard. The production medium containing (g/L) KH2PO4 (0.4), K2HPO4 (1.6), MgSO4.7H20 (0.2), MnSO4.H2O (0.05), CuSO4.5H2O (0.001), ZnSO4.7H2O (0.001), and CaCl2 (0.0005) was adjusted to pH ≈ 4.7 [41]. Substrate concentration and inoculum concentration were set 4% for the initial batch and the process was continued for 72 h at 120 rpm.
2.3. Efficient Lipid Synthesis by Optimization and Extraction of Fatty Acids from Fermented Medium for Advanced Application
Groundnut husk powder and rice bran powder were tested as substrates in anticipation of higher PUFA content production. One potential strain from the selected three isolates was taken for the optimization process in search of maximum lipid yield. Optimization was carried out by varying parameters such as substrate concentration (2, 4, 6, 8, 12%), inoculum size (3, 5, 7, 9%), and inoculum age (24, 48, 72, 96 h). Other parameters such as fermentation incubation time, temperature, pH, and agitation speed were also optimized for finding of suitable environmental condition for the screened strain. Fatty acid extraction was performed by Bligh and Dyer method [42]. Centrifugation of fermented media was performed and supernatant was discarded to record CDW. Single-cell oil content was derived by evaporation of organic solvents from extracted lipids using gravimetric method [43].
2.4. Fine-Tuning Biodiesel Production
Transesterification process was performed for the conversion of extracted oil (2 ml) into biodiesel. Reaction mixture was taken in 250 ml Erlenmeyer flask. A strong base catalyst and methanol were thoroughly mixed by providing slight heat. After the dissolution of base in methanol, the mixture was added in extracted sample. Flask was heated at required temperature for required time after tightening with screw cap. After completion of the process, reaction mixture was cooled and set to pH ≈ 7. The solution was washed using warm water for 3–4 times and poured in separating funnel for checking the presence of separated layers. Optimization of the process was continued by varying process parameters such as catalyst type, catalyst concentration, oil: methanol molar ratio, time, and reaction temperature. The following equation was used to calculate the yield of biodiesel production [44].
Biodiesel yield (%v/v)=Amount of biodiesel (ml)/Amount of lipids (ml)
2.5. Comprehensive Characterization of Biodiesel
The purified product was tested by gas chromatography–mass spectrometry (GC-MS) to check the presence of fatty acids. Fourier transform infrared (FT-IR) spectrum analysis was performed to know the nature of fatty acid methyl ester from Bacillus paralicheniformis within the range 4000-400 cm-1. Fatty acid methyl esters (FAME) were further characterized using nuclear magnetic resonance (NMR) spectroscopy. Biodiesel obtained from Bacillus transesterified lipids was confirmed using NMR spectroscopy. Gelbard used the NMR technique for checking transesterified products for the first time [45].
2.6. Biodiesel Revealed: Unraveling Physicochemical Properties of Biodiesel
Certain parameters such as the properties and quality of biodiesel depend on the structural and molecular configuration of fatty acids [46]. The standard methods such as American standard testing material and American oil Chemist’s society were used to check elemental properties of the produced biodiesel such as viscosity, density, iodine value, free fatty acid (FFA) content(%), specific gravity, flash point, acid value (mg KOH g1), pour point, and cetane number [47,48] [Table 1].
Table 1: List of standard methods for Biodiesel quality.
| Properties | Test Method |
|---|---|
| Kinematic Viscosity @ 40°C | ASTM D 445 2018 |
| Density @ 40°C | ASTM D 792 |
| Specific gravity @ 40°C | ASTM D 792 |
| Pour point | ASTM D 97 2017 |
| Cloud point | ASTM D 2500 |
| Flash point | ASTM D 92 2018 |
| Acid number | AOCS Cd 3d 63 2009(RA2017) |
| Iodine number | AOCS Cd 1d 92 2009(RA2017) |
| Cetane number | ASTM D 613 |
ASTM: American Society for Testing and Materials
3. RESULTS AND DISCUSSION
3.1. Selection of Lipid Accumulator for Efficient Biofuel Production
In our study, 23 organisms with potential to accumulate lipids were isolated from 3 different locations. Water sample was collected from the sea, near a place called Somnath located in the state of Gujarat in India. Through various surveys, it is reported that organisms showing maximum lipid accumulation can be collected from seawater sample [49]. Among 23 isolates, three powerful lipid accumulating strains (RKC-1, RKC-2, RKC-3) showing positive results in TTC screening and Sudan black B staining were screened. Among these three isolates, based on its potentiality to increase PUFA, RKC-2 was selected for our further study. The yield of this strain was 3.3 g/L of lipids produced from the dry weight of 5.5 g/L. This finding indicates that the production of lipid does not depend on cell growth [Figure 1a and b]. The yield of each strain obtained at the end of 24 h, 48 h, and 76 h was compared. The results showed that RKC-2 was a fastidious organism which produced lipids faster than the other reported strains.
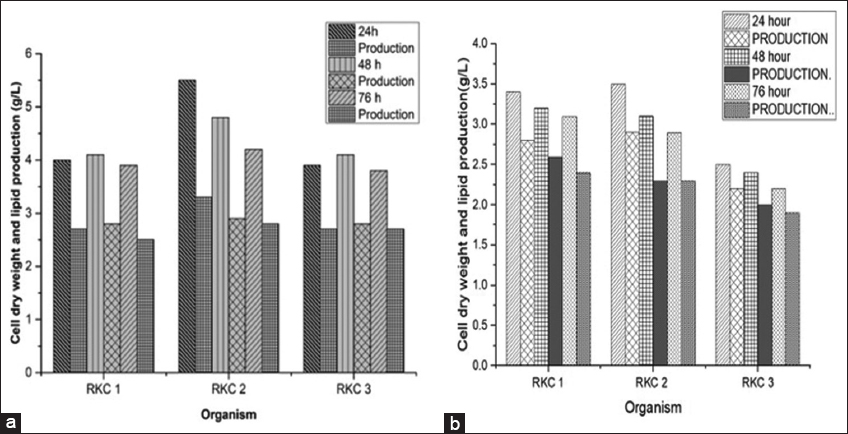 | Figure 1: Effect of various substrates on different selected strain for cell dry weight and oil accumulation. (a) Groundnut husks. (b) Ricebran. [Click here to view] |
The screened strain was checked for its morphological and biochemical characteristics and it was confirmed as B. paralicheniformis after 16s rRNA sequence analysis [Figure 2].
 | Figure 2: Phylogenetic relationships of Bacillus paralicheniformis (OQ202112). [Click here to view] |
The nucleotide sequence was submitted to National Center for Biotechnology Information and assigned accession number for the bacterial strain is OQ202112. Zhang et al. reported B. subtilis HB1310 as an oleaginous microorganism isolated from thin-shelled walnut. The mentioned strain grew well on cotton stalk hydrolysate as a substrate and could enrich fat content 39.8% in 48 h [50]. Moreover, Qadeer showed orange waste as the best possible substrate that can be provided for the growth of Bacillus cereus KM15 for the accumulation of lipids. The process was carried out for 72-h incubation period at neutral pH [51].
3.2. Lipids Unleashed: Optimizing Production and Extraction
Bacterial lipid accumulation and composition of fatty acids are strongly influenced by various culture conditions [52]. PUFA accumulation depends on varying species of microbes [53]. B. paralicheniformis (OQ202112) was able to produce 3.3 g/L lipids using mineral salt medium supplemented with 9% groundnut husks as a substrate, 6% inoculum size, 24 h inoculum age at 120 rpm agitation speed at 28°C. Lipid yield of 2.8 g/L was obtained by using mineral salt medium supplemented with 9% rice bran as a substrate, 7% inoculum size, 24 h inoculum age at 140 rpm agitation speed at 28°C. In case of glucose used as a standard source of carbon, 2.48 g/L lipids were produced. Thus, in our study, groundnut husk showed better results compared to rice bran for fat accumulation by selected strain. Lipid yield in microorganisms can also get affected by various factors such as agitation speed, aeration rate, pH and temperature of medium, nitrogen and carbon source, and C: N ratio. Maximum fat accumulation was achieved at 120 rpm whereas it decreased while decreasing the speed of agitation. This result supports that proper mixing is achieved by high agitation speed and it is required for nutrient and heat transfer [54]. Zhang et al. showed that Bacillus subtilis HB1310 accumulated lipids in its idiophase [50]; whereas, the present study revealed that B. paralicheniformis accumulated lipid in its log phase. According to a study by Udeh et al., biodiesel can be produced using the fungus Aspergillus niger from the lipase-catalyzed groundnut shells [55]. Pocan et al. reported that pomegranate waste peel was used without pre-treatment to accumulate oil by Bacillus spp. [56]. Qadeer et al. reported in the literature that mango waste was used for fat accumulation by different strains of Bacillus (Bacillus cereus KM15) [52]. In our study, agro waste (groundnut husk) is an inexpensive source that gave higher yield of biodiesel without pre-treatment. In the end, oil content was found 5 ml/L after extraction by Bligh and Dyer method and evaporation by Rota evaporator.
3.3. Fine-Tuning Biodiesel Production
Type and utilization of substrate during the cultivation of oleaginous strain decide the fate of production in terms of quantity [57]. The test strain was proven potential as it yielded 86.2% fatty acid methyl ester using groundnut husks as an agro waste. In our study, transesterification which was carried out using 0.3% catalyst concentration with 1:6 fatty acid to alcohol ratio at 68°C temperature in 120 min yielded 86.2% FAME. Biodiesel production can be affected by variation in catalyst and its quantity [58]. In our study, sodium, potassium, and calcium hydroxides were used to increase the rate of reaction and sodium hydroxide was the most effective among all the tested catalysts. Potassium and calcium hydroxide yielded 50.2 and 65.3, respectively, whereas NaOH yielded 86.2% FAME [Figure 3a]. Saponification was found to occur at higher concentration of catalyst [59,60]. It has been reported that more than 1% of catalyst concentration causes the formation of soap from triacylglycerols [61]. Therefore, we used 0.3% concentration of the catalyst to prevent saponification and which was also optimum for transesterification. For low molar ratios, it was shown that the ester production was sensitive to the concentration of NaOH and that it increased as the methanol to vegetable oil ratio increased. [62]. We found that, 1:6 fatty acid methyl ratio was responsible for more product formation [Figure 3b]. Varying temperature ranges from 45°C to 80°C and time from 30 to 180 min was taken for optimization. A number of researchers described the influence of increased temperature on the production of biodiesel in desired manner [63-67]. The ester yields marginally drops above the reaction temperature of 50°C. According to Dorado et al., it might most likely be caused by a negative interaction between temperature and catalyst concentration due to the side reaction of saponification [68]. High process temperatures have the tendency to accelerate the transesterification of the triglycerides by the alkaline catalyst before it is fully completed. However, we found 68°C temperature to be optimum for the maximum yield of ester [Figure 3c]. Some experimenters found that 98% formation of ester completes in 1 h and 80% of it completes in 5 min [63-67]. Similar results were observed in the current study with maximum lipid yield obtained during the first 2 h [Figure 3d].
 | Figure 3: Optimization of fatty acid methyl esters formation. (a) Catalyst. (b) Oil: methanol ratio. (c) Temperature (°C). (d) Time (min). [Click here to view] |
3.4. Comprehensive Characterization of Biodiesel
3.4.1. Analyzing biodiesel by GC-MS
FFAs are of considerable interest due to their suitability for the manufacture of biofuel [11,69]. The extracted lipids’ ability to be used in the transesterification process to create biodiesel was validated by the presence of FFAs in those lipids [70]. Lipid sample from crude oil isolates was extracted and it contained a variety of FFAs, according to the GC-MS analysis, with oleic acid being the most frequently found FFA followed by n-hexadecanoic acid and octadecanoic acid [52]. In the present study, GC-MS of the extracted and esterified lipid sample revealed the presence of hexadecenoic acid, heptadecenoic acid, octadecadienoic acid, octadecenoic acid, and octadecatrienoic acid [Figure 4]. These are polyunsaturated and monounsaturated fatty acids which are suitable for biodiesel production.
 | Figure 4: Gas chromatography–mass spectrometry spectrum. [Click here to view] |
3.4.2. Insight through FT-IR
The absorption spectrum by FT-IR is demonstrated in Figure 5. C-H stretching can be confirmed by absorption bands between 3010 and 2854 cm-1 [61]. In our sample, we got peaks at 2853.69, 2923.19, and 3007.00 indicating C-H stretching. The RCOOR0’ carbonyl group (C=O) is identified by the distinctive absorption band at 1743 cm-1 [61]. Peak obtained in the range 1741–1750 cm-1 depicts the presence of fatty acid methyl ester [71]. The existence of RCOOR’ carbonyl group was confirmed by us from the distinctive absorption band at 1741.83 cm-1, which is signature for methyl ester. The vibrations of symmetric and asymmetric stretching of C-H alkane groups are indicated by the peaks at 2850.41 cm-1 and 2924.32 cm-1, respectively. They may be methyl (CH3) or methylene groups in the ester chains of the biodiesel, and they need more energy to create stretching vibrations within their bond than typical C-H bending vibrations of alkene groups, which are visible at low energy and frequency ranges [71,72]. We got peaks at 2853.59 and 2923.19 which are indications of symmetric and asymmetric stretching vibrations of C-H alkane groups, respectively. The bending vibration of C-H methyl groups in the fuel is responsible for the band region between 1377.23 and 1465.03 cm-1 [71]. We found alkane bending and primary alcohol stretching by peaks at 1460.76 cm-1.
 | Figure 5: Fourier-transform infrared spectroscopy measurements. [Click here to view] |
3.4.3. NMR unveils biodiesel
Two characteristic peaks by 1H NMR spectrum are indication of the existence of methyl ester in FAME [73]. The signal for the -CH2- group next to the carbonyl group, which is utilized as an internal benchmark to roughly measure the other groups in the biodiesel molecules, is at 2.27 ppm and the signal for methyl ester is at 3.63 ppm [74-76]. In the 1H NMR spectrum obtained from Bacillus transesterified lipids, the methoxy proton shown as a singlet at 3.65 ppm and an alpha –CH2 protons shown at 2.26 ppm are characteristic of methyl esters. 1.28 ppm is for the aliphatic -CH2, and neither the ester group nor the double bonds have an impact on their chemical shifts [76]. In our result, backbone CH2, especially –(CH2)n was confirmed by proton shift at 1.22–1.42 ppm [Figure 6a]. The carbon resonance from triglycerides was shown by 13C NMR spectrum. The signals for the terminal chain methyl (-CH3) carbon are at 13.9 ppm, and the signals for the aliphatic methylene (-CH2-s) carbons are in the range of 34 to 27 ppm [76]. In our study, peak observed at 14.3 ppm suggests the presence of terminal carbon of methyl groups. Moreover, bands between 22.0 and 34.0 ppm are due to carbon of methylene group (-CH2-)n. The signals for the unsaturated (--CH=CH-) carbons and the outer carbons of the nun-conjugated (-CH=CH-CH2-CH=CH-) are at 129.8 ppm and 127.9 ppm, respectively [76]. The observed peaks between 120 and 135 ppm suggest the presence of sp2 -C in the present study. At 174.0 ppm, the carbonyl carbon signal (-COO-) is detectable [76]. In our study, we observed peak at 173 ppm which is assigned to carbonyl carbon of ester groups [Figure 6b].
 | Figure 6: Nuclear magnetic resonance (NMR) spectroscopy measurements. (a) 13C NMR spectrum. (b) 1H NMR spectrum. [Click here to view] |
3.5. Biodiesel Revealed: Unraveling Physicochemical Properties of Biodiesel
Biodiesel properties were tested by the American Society for Testing and Materials (ASTM) along with the recommended values for biodiesel ASTM-D6751 which are presented in Table 2. One of the most important properties of fuel is viscosity as it causes changes in the operation of fuel injector and it is affected by temperature. Bacillus fuel possessed 4.8 Mm2s-1 viscosity, which can be considered in normal range given by ASTM standards. Higher cloud point and pour point are indications of fuel having more polyunsaturated fats and less viscosity. Moreover, these properties are better suited for varying weather conditions. Biofuel from test organism showed 0.8823 g cm-3 density at 40°C, which is also in the range of ASTM standards. The product showed specific gravity slight higher with comparison to standards of ASTM, which is 0.8895 g cm-3 at 40°C. The temperature at which biodiesel starts turning into wax form is known as cloud point while pour point can be defined as the temperature at which fuel remains in fluid form. The cloud point and pour point of the FAME were 4°C and 14.4°C, respectively, which are also in the range of standard biodiesel. Minimum temperature at which vaporization can be done is known as flash point. The flash point for the test sample is 106.5°C which falls in the normal range for standard biofuel. Cetane number for the biodiesel obtained in our study is 61. Cetane number measures the quality or performance of diesel fuel. The higher the number, the better the fuel burns within the engine of a vehicle.
Table 2: Properties of biodiesel produced by Bacillus paralicheniformis (OQ202112).
| Properties | Standard Biodiesel | Biodiesel from Bacillus paralicheniformis (OQ202112) | Units |
|---|---|---|---|
| Kinematic viscosity @ 40°C | 1.9–6.0 | 4.8 | Mm2s-1 |
| Density @ 40°C | 0.86–0.90 | 0.8823 | g cm-3 |
| Specific gravity @ 40°C | 0.88 | 0.8895 | g cm-3 |
| Pour point | −15–16 | -14.0 | °C |
| Cloud point | −3–12 | 4.0 | °C |
| Flash point | 100–170 | 106.5 | °C |
| Acid number | 0.5 | 0.66 | mgKOH/g |
| Iodine number | - | 115.54 | %(m/m) |
| Cetane number | 47 minimum | 61 | - |
4. CONCLUSION
B. paralicheniformis (OQ202112) isolated from seawater sample was found to be very effective in utilizing groundnut husks as an agro waste and accumulating PUFAs. The organism accumulated PUFA using groundnut husks and these fatty acids are proven effective in FAME formation. Biodiesel produced by the test organism using the agro waste showed suitable properties on comparing with the standards given by ASTM and ASAM. The cetane rating obtained for the biodiesel was in the range that marks good quality of the fuel. Thus, we can conclude that the isolated organism shows promising capacity to be used as a single-cell oil factory and the oil produced by the organism is suitable for biodiesel production. This organism uses agro waste which is harmful for the environment and produces biodiesel that helps in minimizing dependence on non-renewable energy resources. Thus, it can be used as a potential source for future fuel factory.
5. ACKNOWLEDGMENT
Authors would like to acknowledge 1) Mr. Jaimin Parikh, 2) Dr. Ishan Raval, 3) Mr. Malay Pandya, 4) Mr. Dhruvik Kanani, 5) Dr. Keyur Bhatt, 6) Dr. Hardik Shah, 7) Dr. Amruta Desai for their support in this research work. Authors would also like to acknowledge Biotechnology Research Laboratory-MUIS and chemical research laboratory-MUIS for providing with FT-IR and other facilities. Authors are thankful to Gujarat Biotechnology Research laboratory, National Institute of Pharmaceutical Education and Research, and Indrashil Institute of Science and Technology for providing instrument availability for the research.
6. AUTHORS’ CONTRIBUTIONS
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Roshani Chaudhary and Priti Patel. The first draft of the manuscript was written by Roshani Chaudhary and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
7. FUNDING
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Paramjeet S, Manasa P, Korrapati N. Biofuels:Production of fungal-mediated ligninolytic enzymes and the modes of bioprocesses utilizing agro-based residues. Biocatal Agric Biotechnol 2018;14:57-71. [10.1016/j.bcab.2018.02.007]
2. Khot M, Gupta R, Barve K, Zinjarde S, Govindwar S, Kumar AR. Fungal production of single cell oil using untreated copra cake and evaluation of its fuel properties for biodiesel. J Microbiol Biotechnol 2015;25:459-63. [10.4014/jmb.1407.07074]
3. Abomohra AE, Elsayed M, Esakkimuthu S, El-Sheekh M, Hanelt D. Potential of fat, oil and grease (FOG) for biodiesel production:A critical review on the recent progress and future perspectives. Prog Energy Combust Sci 2020;81:100868. [10.1016/j.pecs.2020.100868]
4. Gomez JA, Höffner K, Barton PI. From sugars to biodiesel using microalgae and yeast. Green Chem 2016;18:461-75. [10.1039/C5GC01843A]
5. Bhatnagar A, Chinnasamy S, Singh M, Das KC. Renewable biomass production by mixotrophic algae in the presence of various carbon sources and wastewaters. Appl Energy 2011;88:3425-31. [10.1016/j.apenergy.2010.12.064]
6. Chisti Y. Constraints to commercialization of algal fuels. J Biotechnol 2013;167:201-14. [10.1016/j.jbiotec.2013.07.020]
7. Albers E, Johansson E, Franzén CJ, Larsson C. Selective suppression of bacterial contaminants by process conditions during lignocellulose based yeast fermentations. Biotechnol Biofuels 2011;4:59. [10.1186/1754-6834-4-59]
8. Basso LC, de Amorim HV, de Oliveira AJ, Lopes ML. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res 2008;8:1155-63. [10.1111/j.1567-1364.2008.00428.x]
9. Amorim HV, Lopes ML, de Castro Oliveira JV, Buckeridge MS, Goldman GH. Scientific challenges of bioethanol production in Brazil. Appl Microbiol Biotechnol 2011;91:1267-75. [10.1007/s00253-011-3437-6]
10. Renneberg R, Berkling V, Loroch V. Viruses, antibodies, and vaccines. In:Biotechnology Beginners. Cambridge:Academic Press;2017. 165. [10.1016/B978-0-12-801224-6.00005-9]
11. Gouda MK, Omar SH, Aouad LM. Single cell oil production by Gordoniasp. DG using agro-industrial wastes. World J Microbiol Biotechnol 2008;24:1703-11. [10.1007/s11274-008-9664-z]
12. Alvarez HM, Silva RA, Herrero M, Hernández MA, Villalba MS. Metabolism of triacylglycerols in Rhodococcusspecies:Insights from physiology and molecular genetics. J Mol Biochem 2013;2.
13. Behera AR, Dutta K, Verma P, Daverey A, Sahoo DK. High lipid accumulating bacteria isolated from dairy effluent scum grown on dairy wastewater as potential biodiesel feedstock. J Environ Manage 2019;252:109686. [10.1016/j.jenvman.2019.109686]
14. Wu Y, Li Q, Zhang R, Dai X, Chen W, Xing D. Circulating microRNAs:Biomarkers of disease. Clin Chim Acta 2021;516:46-54. [10.1016/j.cca.2021.01.008]
15. Li Q, Du W, Liu D. Perspectives of microbial oils for biodiesel production. Appl Microbiol Biotechnol 2008;80:749-56. [10.1007/s00253-008-1625-9]
16. Wentzel A, Ellingsen TE, Kotlar HK, Zotchev SB, Throne-Holst M. Bacterial metabolism of long-chain n-alkanes. Appl Microbiol Biotechnol 2007;76:1209-21. [10.1007/s00253-007-1119-1]
17. Rubio-Rodríguez N, Beltrán S, Jaime I, de Diego SM, Sanz MT, Carballido JR. Production of omega-3 polyunsaturated fatty acid concentrates:A review. Innov Food Sci Emerg Technol 2010;11:1-2. [10.1016/j.ifset.2009.10.006]
18. Delong EF, Yayanos AA. Biochemical function and ecological significance of novel bacterial lipids in deep-sea procaryotes. Appl Environ Microbiol 1986;51:730-7. [10.1128/aem.51.4.730-737.1986]
19. Adarme-Vega TC, Thomas-Hall SR, Schenk PM. Towards sustainable sources for omega-3 fatty acids production. Curr Opin Biotechnol 2014;26:14-8. [10.1016/j.copbio.2013.08.003]
20. Moi IM, Leow AT, Ali MS, Rahman RN, Salleh AB, Sabri S. Polyunsaturated fatty acids in marine bacteria and strategies to enhance their production. Appl Microbiol Biotechnol 2018;102:5811-26. [10.1007/s00253-018-9063-9]
21. Gladyshev MI, Sushchik NN, Makhutova ON. Production of EPA and DHA in aquatic ecosystems and their transfer to the land. Prostaglandins Other Lipid Mediat 2013;107:117-26. [10.1016/j.prostaglandins.2013.03.002]
22. Freese E, Rütters H, Köster J, Rullkötter J, Sass H. Gammaproteobacteria as a possible source of eicosapentaenoic acid in anoxic intertidal sediments. Microb Ecol 2009;57:444-54. [10.1007/s00248-008-9443-2]
23. Allen EE, Facciotti D, Bartlett DH. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl Environ Microbiol 1999;65:1710-20. [10.1128/AEM.65.4.1710-1720.1999]
24. Yoshida K, Hashimoto M, Hori R, Adachi T, Okuyama H, Orikasa Y, et al. Bacterial long-chain polyunsaturated fatty acids:Their biosynthetic genes, functions, and practical use. Mar Drugs 2016;14:94. [10.3390/md14050094]
25. Kawamoto J, Kurihara T, Yamamoto K, Nagayasu M, Tani Y, Mihara H, et al. Eicosapentaenoic acid plays a beneficial role in membrane organization and cell division of a cold-adapted bacterium, Shewanella livingstonensis Ac10. J Bacteriol 2009;191:632-40. [10.1128/JB.00881-08]
26. Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 2001;293:290-3. [10.1126/science.1059593]
27. Yazawa K. Production of eicosapentaenoic acid from marine bacteria. Lipids 1996;31:S297-300. [10.1007/BF02637095]
28. Allen EE, Bartlett DH. Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology (Reading) 2002;148:1903-13. [10.1099/00221287-148-6-1903]
29. Shulse CN, Allen EE. Widespread occurrence of secondary lipid biosynthesis potential in microbial lineages. PLoS One 2011;6:e20146. [10.1371/journal.pone.0020146]
30. Griffiths MJ, Harrison ST. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J Appl Phycol 2009;21:493-507. [10.1007/s10811-008-9392-7]
31. Holden E, Gilpin G. Biofuels and sustainable transport:A conceptual discussion. Sustainability 2013;5:3129-49. [10.3390/su5073129]
32. Chen XF, Huang C, Xiong L, Chen XD, Ma LL. Microbial oil production from corncob acid hydrolysate by Trichosporon cutaneum. Biotechnol Lett 2012;34:1025-8. [10.1007/s10529-012-0869-8]
33. Gong Z, Shen H, Wang Q, Yang X, Xie H, Zhao ZK. Efficient conversion of biomass into lipids by using the simultaneous saccharification and enhanced lipid production process. Biotechnol Biofuels 2013;6:36. [10.1186/1754-6834-6-36]
34. Hu C, Wu S, Wang Q, Jin G, Shen H, Zhao ZK. Simultaneous utilization of glucose and xylose for lipid production by Trichosporon cutaneum. Biotechnol Biofuels 2011;4:25. [10.1186/1754-6834-4-25]
35. Kumar M, Rathour R, Gupta J, Pandey A, Gnansounou E, Thakur IS. Bacterial production of fatty acid and biodiesel:Opportunity and challenges. In:Refining Biomass Residues for Sustainable Energy and Bioproducts. Cambridge:Academic Press;2020. 21-49. [10.1016/B978-0-12-818996-2.00002-8]
36. Cherubini F. The biorefinery concept:Using biomass instead of oil for producing energy and chemicals. Energy Convers Manage 2010;51:1412-21. [10.1016/j.enconman.2010.01.015]
37. FitzPatrick M, Champagne P, Cunningham MF, Whitney RA. A biorefinery processing perspective:Treatment of lignocellulosic materials for the production of value-added products. Bioresour Technol 2010;101:8915-22. [10.1016/j.biortech.2010.06.125]
38. Ryan J, Farr H, Visnovsky S, Vyssotski M, Visnovsky G. A rapid method for the isolation of eicosapentaenoic acid-producing marine bacteria. J Microbiol Methods 2010;82:49-53. [10.1016/j.mimet.2010.04.001]
39. Kwolek-Mirek M, Zadrag-Tecza R. Comparison of methods used for assessing the viability and vitality of yeast cells. FEMS Yeast Res 2014;14:1068-79. [10.1111/1567-1364.12202]
40. Hartman TL. The use of Sudan black B as a bacterial fat stain. Stain Technol 1940;15:23-8. [10.3109/10520294009110328]
41. Wei Z, Zeng G, Kosa M, Huang D, Ragauskas AJ. Pyrolysis oil-based lipid production as biodiesel feedstock by Rhodococcus opacus. Appl Biochem Biotechnol 2015;175:1234-46. [10.1007/s12010-014-1305-4]
42. Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 1959;37:911-7. [10.1139/y59-099 https://doi.org/10.1139/o59-099]
43. Thorpe RF, Ratledge C. Fatty acids of triglycerides and phospholipids from a thermotolerant strain of Candida tropicalis grown on n-alkanes at 30 and 40 degrees C. J Gen Microbiol 1973;78:203-6. [10.1099/00221287-78-1-203]
44. Naureen R, Tariq M, Yusoff I, Chowdhury AJ, Ashraf MA. Synthesis, spectroscopic and chromatographic studies of sunflower oil biodiesel using optimized base catalyzed methanolysis. Saudi J Biol Sci 2015;22:332-9. [10.1016/j.sjbs.2014.11.017]
45. Gelbard G, Bres O, Vargas RM, Vielfaure F, Schuchardt UF. 1H nuclear magnetic resonance determination of the yield of the transesterification of rapeseed oil with methanol. J Am Oil Chem Soc 1995;72:1239-41. [10.1007/BF02540998]
46. Knothe G. Fuel properties of highly polyunsaturated fatty acid methyl esters. Prediction of fuel properties of algal biodiesel. Energy Fuels 2012;26:5265-73. [10.1021/ef300700v]
47. Lang X, Dalai AK, Bakhshi NN, Reaney MJ, Hertz PB. Preparation and characterization of bio-diesels from various bio-oils. Bioresour Technol 2001;80:53-62. [10.1016/S0960-8524(01)00051-7]
48. Moawia RM, Nasef MM, Mohamed NH, Ripin A, Farag H. Production of biodiesel from cottonseed oil over aminated flax fibres catalyst:Kinetic and thermodynamic behaviour and biodiesel properties. Adv Chem Eng Sci 2019;9:281-98. [10.4236/aces.2019.94021]
49. Estupiñán M, Hernández I, Saitua E, Bilbao ME, Mendibil I, Ferrer J, et al. Novel Vibriospp. strains producing omega-3 fatty acids isolated from coastal seawater. Mar Drugs 2020;18:99. [10.3390/md18020099]
50. Zhang Q, Li Y, Xia L. An oleaginous endophyte Bacillus subtilis HB1310 isolated from thin-shelled walnut and its utilization of cotton stalk hydrolysate for lipid production. Biotechnol Biofuels 2014;7:152. [10.1186/s13068-014-0152-4]
51. Qadeer S. Utilizing Potential Oleaginous Microbes for Lipid Based Biofuel Production From Food Processing Waste. Doctoral Dissertation. Rawalpindi:PMAS-Arid Agriculture University.
52. Qadeer S, Mahmood S, Anjum M, Ilyas N, Ali Z, Khalid A. Synchronization of lipid-based biofuel production with waste treatment using oleaginous bacteria:A biorefinery concept. Process Saf Environ Prot 2018;115:99-107. [10.1016/j.psep.2017.10.011]
53. Alvarez HM, Kalscheuer R, Steinbüchel A. Accumulation and mobilization of storage lipids by Rhodococcus opacusPD630 and Rhodococcus ruber NCIMB 40126. Appl Microbiol Biotechnol 2000;54:218-23. [10.1007/s002530000395]
54. Zhou Y, Han LR, He HW, Sang B, Yu DL, Feng JT, et al. Effects of agitation, aeration and temperature on production of a novel glycoprotein GP-1 by Streptomyces kanasenisi ZX01 and scale-up based on volumetric oxygen transfer coefficient. Molecules 2018;23:125. [10.3390/molecules23010125]
55. Udeh B. Bio-waste transesterification alternative for biodiesel production:A combined manipulation of lipase enzyme action and lignocellulosic fermented ethanol. Asian J Biotechnol Bioresour Technol 2018;3:1-9. [10.9734/AJB2T/2018/40789]
56. Pocan P, Bahcegul E, Oztop MH, Hamamci H. Enzymatic hydrolysis of fruit peels and other lignocellulosic biomass as a source of sugar. Waste Biomass Valorization 2018;9:929-37. [10.1007/s12649-017-9875-3]
57. Ribeiro BD, De Castro AM, Coelho MA, Freire DM. Production and use of lipases in bioenergy:A review from the feedstocks to biodiesel production. Enzyme Res 2011;2011:615803. [10.4061/2011/615803]
58. Kanakdande AP, Khobragade CN, Mane RS. Utilization of pomegranate waste-peel as a novel substrate for biodiesel production by Bacillus cereus(MF908505). Sustain Energy Fuels 2020;4:1199-207. [10.1039/C9SE00584F]
59. Hadiyanto H, Lestari SP, Widayat W. Preparation and characterization of Anadara granosa shells and CaCo3 as heterogeneous catalyst for biodiesel production. Bull Chem React Eng Catal 2016;11:21-6. [10.9767/bcrec.11.1.402.21-26]
60. Salamatinia B, Mootabadi H, Bhatia S, Abdullah AZ. Optimization of ultrasonic-assisted heterogeneous biodiesel production from palm oil:A response surface methodology approach. Fuel Process Technol 2010;91:441-8. [10.1016/j.fuproc.2009.12.002]
61. Fadhil AB, Ali LH. Alkaline-catalyzed transesterification of Silurus triostegus Heckel fish oil:Optimization of transesterification parameters. Renew Energy 2013;60:481-8. [10.1016/j.renene.2013.04.018]
62. Sinha S, Agarwal AK, Garg S. Biodiesel development from rice bran oil:Transesterification process optimization and fuel characterization. Energy Convers Manage 2008;49:1248-57. [10.1016/j.enconman.2007.08.010]
63. Srivastava A, Prasad R. Triglycerides-based diesel fuels. Renew Sustain Energy Rev 2000;4:111-33. [10.1016/S1364-0321(99)00013-1]
64. Ma F, Hanna MA. Biodiesel production:A review. Bioresour Technol 1999;70:1-15. [10.1016/S0960-8524(99)00025-5]
65. Freedman BE, Pryde EH, Mounts TL. Variables affecting the yields of fatty esters from transesterified vegetable oils. J Am Oil Chem Soc 1984;61:1638-43. [10.1007/BF02541649]
66. Canakci M, Van Gerpen J. Biodiesel production viaacid catalysis. Trans ASAE 1999;42:1203-10. [10.13031/2013.13285]
67. Encinar JM, Gonzalez JF, Rodríguez-Reinares A. Biodiesel from used frying oil. Variables affecting the yields and characteristics of the biodiesel. Ind Eng Chem Res 2005;44:5491-9. [10.1021/ie040214f]
68. Dorado MP, Ballesteros E, López FJ, Mittelbach M. Optimization of alkali-catalyzed transesterification of Brassica carinata oil for biodiesel production. Energy Fuels 2004;18:77-83. [10.1021/ef0340110]
69. Arabolaza A, Rodriguez E, Altabe S, Alvarez H, Gramajo H. Multiple pathways for triacylglycerol biosynthesis in Streptomyces coelicolor. Appl Environ Microbiol 2008;74:2573-82. [10.1128/AEM.02638-07]
70. Soccol CR, Dalmas Neto CJ, Soccol VT, Sydney EB, da Costa ES, Medeiros AB, et al. Pilot scale biodiesel production from microbial oil of Rhodosporidium toruloides DEBB 5533 using sugarcane juice:Performance in diesel engine and preliminary economic study. Bioresour Technol 2017;223:259-68. [10.1016/j.biortech.2016.10.055]
71. Oyerinde AY, Bello EI. Use of Fourier transformation infrared (FTIR) spectroscopy for analysis of functional groups in peanut oil biodiesel and its blends. Br J Appl Sci Technol 2016;13:1-4. [10.9734/BJAST/2016/22178]
72. Hariram V, Bose A, Seralathan S. Dataset on optimized biodiesel production from seeds of Vitis vinifera using ANN, RSM and ANFIS. Data Brief 2019;25:104298. [10.1016/j.dib.2019.104298]
73. Mello VM, Oliveira FC, Fraga WG, do Nascimento CJ, Suarez PA. Determination of the content of fatty acid methyl esters (FAME) in biodiesel samples obtained by esterification using 1H-NMR spectroscopy. Magn Reson Chem 2008;46:1051-4. [10.1002/mrc.2282]
74. Fauconnot L, Robert F, Villard R, Dionisi F. Chemical synthesis and NMR characterization of structured polyunsaturated triacylglycerols. Chem Phys Lipids 2006;139:125-36. [10.1016/j.chemphyslip.2005.11.004]
75. Mantovani AC, Chendynski LT, Galvan D, de Macedo Júnior FC, Borsato D, Di Mauro E. Thermal-oxidation study of biodiesel by proton nuclear magnetic resonance (1H NMR). Fuel 2020;274:117833. [10.1016/j.fuel.2020.117833]
76. Doudin KI. Quantitative and qualitative analysis of biodiesel by NMR spectroscopic methods. Fuel 2021;284:119114. [10.1016/j.fuel.2020.119114]