1. INTRODUCTION
Fungal endosymbionts are rich reservoirs for a wide variety of phytochemicals and phytohormones [1]. Endophytes residing inside plant systems contribute to the production of numerous novel compounds which have significant bioactive potential such as anti-cancerous, anti-inflammatory, anti-leishmanial, anti-microbial, and anti-malarial properties [2-4]. Several of these compounds synthesized under biotic and abiotic stress conditions during interdependent interaction between plants and their endophytes increases the survival value of both organisms. The fungus Talaromyces was first reported by mycologist Chester Ray Benjamin in 1955 under Trichocomaceae. The genus exhibits tightly interwoven hyphae with soft, cottony, yellowish fruit bodies (ascocarp) encompassing granules [5]. Talaromyces has been reported to synthesize several important secondary metabolites, including Huperzine A, a potent acetylcholine esterase inhibitor [6], and amestolkaloids, known for anti-inflammatory properties [7].
Whole genome sequencing (WGS) of fungi followed by genome annotation offers an attractive option for the identitification of novel and unexplored secondary metabolites and their biosynthetic genes. Bioinformatic tools such as Funannotate and SMuRF assist researchers in genome pipeline assemblies, annotations and natural product discovery through in silico strategies with precise, non-laborious, and rapid analysis. An important tool to identify Biosynthetic gene clusters (BGCs) in fungal genomes is Antibiotics and Secondary Metabolite Analysis Shell (AntiSMASH), which has been widely used to analyze several fungal genomes for secondary metabolites. Several compounds from Talaromyces spp. have been extensively studied for their bioactivities. However, genome mining of the species has not been carried out so far. In the present work, endophytic Talaromyces spp. isolated from S. samarangense was subjected to WGS and genome mining using AntiSMASH to identify any novel putative bioactive compounds.
2. MATERIALS AND METHODS
2.1. Isolation and Sub-culturing of Endophytic Fungal Strain-YS-1
The explant material (S. samarangense) leaves and stems were collected from Vajamangala village, Mysore district. The explant was thoroughly washed under running tap water. The leaf and stem bits were surface sterilized using 70% ethanol for 4 min, followed by sterile distilled water wash and treated with 1% sodium hypochlorite solution for 4 min. They were again washed with sterile distilled water for 2–3 times, and the leaf bits were placed on solidified potato dextrose agar (PDA) media supplemented with ampicillin 100 mg/mL. The Petri dishes were sealed with parafilmTM and incubated at 25 ± 2°C for 15 days with alternate light and dark cycles. Endophytic fungal mycelia which were observed on leaf and stem explants, were subcultured by point inoculation on PDA and potato dextrose broth media. Samples were monitored for further mycelial growth and were subjected to staining with lactophenol cotton blue in their sporulation state. Among the four endophytic fungal strains isolated from S. samarangense, strain YS-1 was selected for the present study.
2.2. Genomic DNA isolation and WGS of YS1 strain
DNA was isolated from the fungal mycelia by CTAB method [8]. Isolated DNA was eluted with 50 µL of TE buffer and stored at −20°C. The purity of isolated DNA was determined using NanoDrop™ 2000C spectrophotometer (Thermo Fisher Scientific). Genomic DNA samples were electrophoresed on 1% agarose gel to check purity and integrity. The high-quality DNA of strain YS1 was sent for whole genome sequence analysis to Hi-Gx360® Himedia Laboratories Pvt. Ltd., Mumbai, India. The sample analysis was as follows: 250 ng of total DNA was used as input for library preparation using QIASeq FX DNA kit to fragment and obtain adapter ligated and indexed library as per manufacturer’s instructions. The indexed library was sequenced on an Illumina Miseq by paired-end chemistry of 300 cycles. The parameters used to check the raw data quality are included in a MultiQC report and are used to understand the quantity of data obtained for each of the paired read files for an individual sample [9]. The fastp tool (v0.12.4) was used to remove adapter contamination [10].
2.3. Genome Assembly and Annotations
A de novo de-brujin graph-based assembly was carried out to assemble the short reads into larger stretches of DNA called contigs. These contigs are the starting material used for executing a genome annotation that assigns functions to various regions of the genome of the organism in question. Quality assessment for genome assemblies generated by Spades, bioSpades, and Megahit assemblers was performed using the Quast tool [11-13]. It was noticed that the assembly generated by the megahit assembler had a comparatively higher number of contigs with sizes >10kb. Hence, assembly generated in bioSpades mode used for phylogenomics analysis and assembly generated using megahit assembler was masked and further used for annotations and natural product discovery.
A new pipeline carried out fungal gene structural and functional annotation, Funannotate, which uses several gene predictors such as glimerHMM, Augustus, SNAP, and Coding-Quarry to improve its prediction [14]. Proteins, transcripts, and nucleic acid annotation were executed by using software such as MEROPS (version 12), UniProt (version 2022_03), dbCAN (version 11), Pfam (version 35), RepeatsDB (version 1), GO (version 2022-07-01), MIBiG (version 1.4), InterPro (version 90), gene2product (version 1.8).
2.4. Phylogenomic Analysis
The Phylogenomic analysis was carried out by using the Benchmarking Universal Single-Copy Orthologs (BUSCO) quantitative genome analysis tool which basically compares different genomic data sets containing whole, identical and disintegrated genes based on gene prediction strategies [15].
2.5. Genome Mining for Natural Product Discovery
AntiSMASH tool: Fungal version 6.1.1 pipeline was used for the identification of potent secondary metabolite BGCs [16].
3. RESULTS AND DISCUSSION
3.1. YS-1 Strain Identification
The cultured fungal organism showed yellow-colored cottony mycelial mat with white dots covering the entire plate after being incubated for 15 days at 25 ± 2°C. Annotation and phylogenetic analysis results showed that the organism belongs to Talaromyces spp. and the stereomicroscopic view of strain YS-1 was examined [Figure 1a and b].
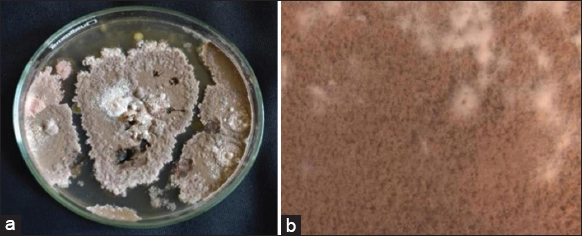 | Figure 1: (a) Mycelial growth on potato dextrose agar plate. (b) Stereomicroscopic view of YS-1 strain. [Click here to view] |
3.2. Genome Assembly and Sequencing
The genome sequence of strain YS-1 was deposited in the NCBI GenBank database [Accession No. PRJNA953512], and the Genome assembly of Talaromyces spp. was accessed by assemblers such as Biosynthetic spades, Spades, and Megahit. These assemblers were compared and analyzed using Quast (Quality assessment tool for genome assemblies). Megahit was found to be an effective assembler that provided data on an organism’s complex metagenomic assemblies. The whole genome size of Talaromyces spp. was found to be 30.5Mb, consisting of 605 contigs (≥0 bp), 372 contigs (≥1000 bp), 208 contigs (≥5000 bp), 168 contigs (≥10000 bp), 149 (≥25000 bp), 125 contigs (≥50,000 bp) and the largest contig was found to be 1112360. The GC content was 46.74%, N50 was 3.33Mb, N75 was 1.86Mb. L50 value was 30 and the L75 value was 65. The Funannotate analysis indicated the presence of 114tRNA, 12722 functional mRNA, 12721 CDS transcripts, and 12721 protein-coding sequences in Talaromyces spp. YS-1 strain. The completeness of genome assembly, as measured by BUSCO, gave a score of 92.9%.
3.3. Phylogenetic Analysis and Phylogenomic Tree
The phylogenetic analysis of the Talaromyces spp. sequenced in the present study (YS-1 Strain) was compared with the isolates available in the database. Three groups could be identified in the phylogenetic tree, with the Talaromyces YS-1 strain forming a close relationship with Talaromyces amestolkiae, Talaromyces marneffei, and Talaromyces stipitatus in the first group. The second group consisted of Talaromyces proteolyticus and Talaromyces rugulosus. The other species Talaromyces atroroseus, formed a separate group [Figure 2].
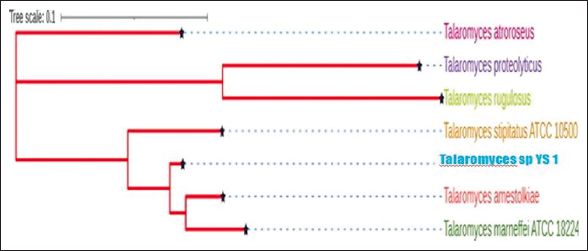 | Figure 2: Phylogenomic tree of fungal Talaromyces strain YS-1. [Click here to view] |
3.4. Genome Mining for Natural Product Discovery
AntiSMASH provides information about the secondary metabolite genes by comparing the query gene cluster with the BGC library. Potential gene clusters of certain secondary metabolites such as terpenes, alkaloids, and phenolic compounds having industrial applications in pharmaceuticals and drug discovery can be analyzed using this comprehensive analysis tool for the fungal genome.
AntiSMASH analysis showed the presence of 76 secondary metabolite BGCs, which include 28 T1Polyketide synthases, 10 Nonribosomal peptide synthetases (NRPSs), 9 terpene, 1 Indole, 12 NRPs like 4 T1 Polyketide synthase (T1 PKS) and NRPs like, 2 T1 PKS Indole, 1 NRPs like terpene, 3 NRPs T1 PKS, 1 Indole NRPs, 2betalactone 1 phosphonate, 1 fungal-RiPP T1 PKS and 1 Other type [Table 1]. Over 40% of these BGCs indicated gene homologies with known clusters in the MIBiG database. Several clusters responsible for coding bioactive compounds such as Pyranonigrin-A, Alternariol, Asperterenoid-A, Naphthopyrone, Clavaric acid, Melanin, and Fusarin showed 100% similarity.
Table 1: Predicted secondary metabolite biosynthetic gene clusters of strain YS-1.
| Region | From | To | Type | Most similar known cluster |
|---|---|---|---|---|
| Region 1.1 | 27,682 | 357,538 | T1PKS | Pyranonigrin E (100%) |
| Region 1.2 | 710,748 | 273,103 | T1PKS | Unknown |
| Region 2.1 | 126,776 | 425,900 | T1PKS | Terretonin (50%) |
| Region 2.2 | 498,524 | 46,806 | NRPS-like | Unknown |
| Region 2.3 | 681,748 | 149,220 | T1PKS, NRPS-like | Zearalenone (18%) |
| Region 3.1 | 291,287 | 267,110 | T1PKS, indole | Trypacidin (35%) |
| Region 3.2 | 454,298 | 424,526 | T1PKS | Viriditoxin (22%) |
| Region 3.3 | 736,454 | 271,795 | T1PKS | Tricholignan A (50%) |
| Region 4.1 | 5,132 | 141,371 | T1PKS | Unknown |
| Region 5.1 | 169,403 | 349,214 | T1PKS | Alternariol (100%) |
| Region 5.2 | 358,147 | 224,543 | NRPS-like, terpene | Squalestatin S1 (60%) |
| Region 5.3 | 689,574 | 171,848 | NRPS-like | Unknown |
| Region 6.1 | 641 | 74,424 | NRPS | Unknown |
| Region 6.2 | 316,319 | 122,148 | T1PKS | Unknown |
| Region 6.3 | 605,092 | 179,953 | NRPS, T1PKS | Emericellamide A/emericellamide B (40%) |
| Region 7.1 | 86,321 | 155,050 | T1PKS, indole | Unknown |
| Region 8.1 | 108,656 | 127,159 | T1PKS | Unknown |
| Region 8.2 | 429,656 | 35,955 | indole | Fumigaclavine C (45%) |
| Region 9.1 | 117,817 | 133,974 | NRPS-like | Unknown |
| Region 10.1 | 114,007 | 51,896 | NRPS-like | Unknown |
| Region 10.2 | 208,461 | 48,783 | NRPS-like | Unknown |
| Region 10.3 | 531,819 | 174,717 | NRPS-like | Unknown |
| Region 10.4 | 598,279 | 61,197 | other | Unknown |
| Region 11.1 | 257,617 | 139,510 | NRPS-like | Unknown |
| Region 11.2 | 491,748 | 72,951 | T1PKS | Azanigerone A (26%) |
| Region 13.1 | 268,179 | 210,071 | Terpene | Asperterpenoid A (100%) |
| Region 14.1 | 64,395 | 71,552 | NRPS | Unknown |
| Region 14.2 | 515,735 | 185,810 | Betalactone | Unknown |
| Region 15.1 | 350,690 | 147,843 | Terpene | Unknown |
| Region 16.1 | 411,349 | 176,667 | NRPS | Unknown |
| Region 17.1 | 277,101 | 48,170 | NRPS | Unknown |
| Region 18.1 | 1 | 154,701 | NRPS | Unknown |
| Region 18.2 | 113,116 | 43,527 | T1PKS | Unknown |
| Region 20.1 | 152,443 | 119,563 | NRPS, T1PKS | curvupallide-B (11%) |
| Region 20.2 | 298,332 | 19,116 | T1PKS | Griseofulvin/epidechlorogriseofulvin/norlichexanthone/dehydrogriseofulvin/4-desmethylgriseofulvin/griseoxanthone B (14%) |
| Region 21.1 | 225,214 | 49,633 | T1PKS | Citrinin (31%) |
| Region 21.2 | 382,522 | 82,811 | Indole, NRPS | Clapurines (27%) |
| Region 22.1 | 1613 | 76,023 | T1PKS | Unknown |
| Region 22.2 | 87,957 | 45,887 | T1PKS | Azanigerone A (26%) |
| Region 22.3 | 219,037 | 48,863 | T1PKS | Ankaflavin/monascin/rubropunctatine/monascorubrin (29%) |
| Region 22.4 | 390,717 | 46,696 | NRPS | Unknown |
| Region 24.1 | 206,270 | 31,408 | T1PKS | Unknown |
| Region 28.1 | 99,615 | 357,538 | T1PKS | Fujikurin A/fujikurin B/fujikurin C/fujikurin D (66%) |
| Region 29.1 | 302,469 | 273,103 | T1PKS | Unknown |
| Region 31.1 | 179,642 | 425,900 | NRPS | Pyranonigrin E (100%) |
| Region 33.1 | 123,646 | 46,806 | T1PKS | Unknown |
| Region 34.1 | 52,652 | 149,220 | Terpene | Unknown |
| Region 37.1 | 78,840 | 267,110 | NRPS-like | Unknown |
| Region 38.1 | 128,355 | 424,526 | NRPS, T1PKS | Unknown |
| Region 39.1 | 111,254 | 271,795 | NRPS-like | Unknown |
| Region 40.1 | 105,708 | 141,371 | Terpene | Unknown |
| Region 41.1 | 1 | 349,214 | Phosphonate | Unknown |
| Region 41.2 | 90,852 | 224,543 | NRPS-like | Unknown |
| Region 47.1 | 27,647 | 171,848 | Betalactone | Unknown |
| Region 48.1 | 2189 | 74,424 | T1PKS | Naphthopyrone (100%) |
| Region 49.1 | 67,645 | 122,148 | fungal-RiPP, T1PKS | Ankaflavin/monascin/rubropunctatine/monascorubrin (20%) |
| Region 54.1 | 13,165 | 179,953 | T1PKS | Pyranonigrin E (100%) |
| Region 54.2 | 116,565 | 155,050 | Terpene | Unknown |
| Region 57.1 | 19,253 | 127,159 | T1PKS, NRPS-like | Unknown |
| Region 59.1 | 196,661 | 35,955 | Terpene | Unknown |
| Region 60.1 | 49,877 | 133,974 | Terpene | Sordarin (11%) |
| Region 63.1 | 163,422 | 51,896 | Terpene | Clavaric acid (100%) |
| Region 67.1 | 92,953 | 48,783 | T1PKS | Chrysoxanthone A/chrysoxanthone B/chrysoxanthone C (8%) |
| Region 68.1 | 132,828 | 174,717 | NRPS-like | Unknown |
| Region 69.1 | 4601 | 61,197 | T1PKS | Melanin (100%) |
| Region 69.2 | 105,785 | 139,510 | T1PKS | Fusarin (100%) |
| Region 71.1 | 502 | 72,951 | NRPS | Unknown |
| Region 71.2 | 73,873 | 210,071 | NRPS | Unknown |
| Region 73.1 | 1 | 71,552 | Terpene | Unknown |
| Region 97.1 | 1 | 185,810 | T1PKS | Sorbicillin (71%) |
| Region 98.1 | 21,324 | 147,843 | T1PKS, NRPS-like | Shearinine D (9%) |
| Region 107.1 | 27,684 | 176,667 | T1PKS | Chrodrimanin B (100%) |
| Region 127.1 | 3370 | 48,170 | NRPS-like | Unknown |
| Region 129.1 | 3888 | 154,701 | T1PKS | Depudecin (33%) |
| Region 130.1 | 1 | 43,527 | NRPS | Unknown |
| Region 139.1 | 1 | 119,563 | T1PKS, NRPS-like | Fusarin (100%) |
AntiSMASH analysis of Talaromyces spp. strain YS-1indicated that the gene clusters in the regions 1.1, 31.1, and 54.1 has significant similarity with the BGCs of Pyranonigrin E from Aspergillus niger ATCC 1015 (GenBank: ACJE01000019.1.) [Figure 3a]. Pyranonigrin- E, isolated from Aspergillus niger, is a pyranopyrrole having effective anti-oxidant and radical scavenging activities [17]. BGC region 5.1 noted significant BLAST hits with Alternariol from Parastagono sporanodorum SN15 (GenBank: KP941080.1) [Figure 3b]. Alternariol is reported to be an effective anti-cancerous and antibacterial metabolite [18]. BGC 13.1 exhibited high similarity with known clusters implicated in the synthesis of Asperterpenoid-A from Talaromyces wortmannii (GenBank: MK140602.1) [Figure 3c]. Asperterpenoid-A has effective anti-inflammatory and antibacterial activities [19]. BGCs found in region 48.1 of the YS-1 strain showed similarity with Naphthopyrone BGC from Aspergillus nidulans FGSC A4 (Gen Bank: BN001302.1) [Figure 3d]. Li et al. [20] have reported that naphthopyrone is a potential antiproliferative compound produced from A. niger. A notable BLAST hit for the BGC region 63.1 was similar to BGC from Hypholomas ublateritium (Gen Bank EU665687.1) for synthesis of Clavaric acid, a potential anti-tumor compound [21] [Figure 3e]. Biosynthetic cluster region 69.1 showed homology with Melanin BGC [Figure 3f]. Melanin is found predominantly in PKS clusters of Fungi and is useful for adjusting to different environmental conditions [22]. The fungal YS-1 strain in regions 69.2 and 139.1 encodes BGC with BLAST hits for Fusarin BGC from Fusarium fujikuroi (Gen Bank: JX308619.1) [Figure 3g]. Fusarin BGCs were been identified in Fusarium spp., along with different polyketide gene clusters [23]. BGC region 107.1 of fungal YS-1 strain presented 100% similarity with BGC from Talaromyces verruculosus (Gen Bank: LC422696.1) meant for biosynthesis of secondary metabolite chrodrimanin B [Figure 3h]. Different classes of secondary metabolites and a new monoterpenoid Chrodrimanin-T have been reported from T. amestolkiae [24]. Chrodrimanin-B- B isolated from Talaromyces funiculosus showed a broad range of anti-bactericidal activity against Staphylococcus aureus, Mycobacterium smegmatis, Mycobacterium phlei, Micrococcus tetragenus and Escherichia coli [25]. In addition, several other bioactives and Antibiotics such as Terretonin, Zearalenone, Trypacidin, Viriditoxin, Tricholignan -A, Squalestatin -S, Emericellamide -A,B, Fumigaclavine -C, Azanigerone -A, Curvupallide -B, Griseofulvin, Citrinin, Clapurines, Fujikurin A,B,C,D, Ankaflavin, Sordarin, Chrysoxanthone -A,B,C, Sorbicillin, Shearinine -D, and Depudecin have been identified in the present study.
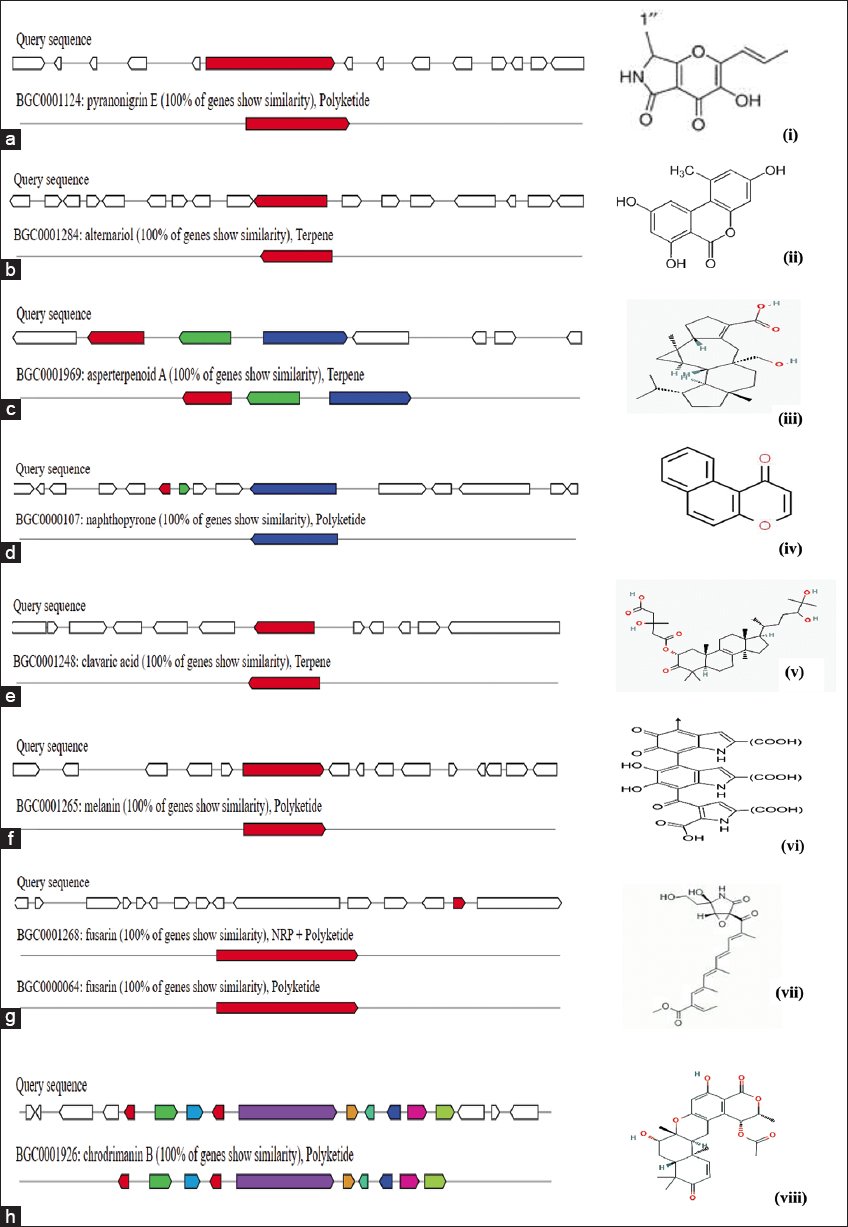 | Figure 3: Comparison of BGCs in Talaromyces spp. YS-1 (a-h) with identified BGCs for biosynthesis of various bioactive compounds (i-Viii). [Click here to view] |
Earlier reports have indicated that Talaromyces species produce several important bioactive compounds. Quinoid compounds such as erythroskirin, islandicin, skyrin, luteoskyrin, regulosin and anthroquinone exhibiting antitumor and antioxidant activities were identified in Talaromyces islandicus [26]. This species also produces mycotoxins and cyclochlorotine, with hepatotoxic and mutagenic activities. Several isocoumarin derivatives and benzofurones identified in the endophytic fungus T. amestolkiae exhibited antibacterial and α-glucosidase inhibitory activities [27,28].
Genome fungiSMASH analysis of T. islandicus detected 19 different types 1 PKS, anthrol reductase BGCs involved in the synthesis of potent alkaloid anthroquinone [29]. Talaropeptins A&B synthesizing NRPs gene cluster was identified in a marine isolate of Talaromyces purpureogenus CX11 [30]. These compounds had antifungal activity. Li et al. [31] have reported the presence of Talaromycolides in Talaromyces pinophilus, which showed antibacterial activity toward S. aureus resistant to methicillin.
In the present study, S. samarangense, a medicinal plant with known pharmacological activities such as anti-oxidant, anti-inflammatory, anti-ulcer, anti-trypanosomal, and hepatoprotective activity, was used as a source for isolation of fungal endophytes [32]. It is suggested that mutualistic interaction between medicinal plants and their endophytes may lead to the development of highly diverse metabolomes with compounds such as alkaloids, terpenoids, flavonoids, coumarins, and tannins [33]. Endophytic Talaromyces spp. was shown to be a reservoir of many bioactive compounds. The systematically organized gene annotation and prediction of BGCs revealed the presence of novel bioactives like clapurines and pyranonigrins, which were reported for the first time in Talaromyces species. The collection of different genes reported in the present analysis provides information about several known, unknown secondary metabolites for drug discovery. Identification of secondary metabolite pathways and their corresponding key gene clusters through in silico analysis assists in developing strategies for targeting these genes through cloning and expression studies. This approach will result in the production of novel metabolites with pharmacological significance.
4. CONCLUSION
Endophyte Talaromyces isolated from medicinal plants is a fascinating reservoir of various metabolites with diverse bioactivities. WGS followed by AntiSMASH analysis indicated the presence of 76 BGCs for the production of like several classes of terpenoids, alkaloids, PKS-NRPSs, and antibiotics. A diverse congregate of genes reported in the present analysis provides information about known and unrevealed metabolites, which can be used further for gene targeting and drug discovery.
5. ACKNOWLEDGMENT
The authors are grateful to the Institution of Excellence (IOE), University of Mysore, for infrastructural facilities and financial assistance to PSN.
6. AUTHOR CONTRIBUTIONS
SS and KRK conceptualized and designed the study. MM, LMP and GN collected and isolated the samples. PSN and SB carried out the experiments and collected the data. PSN, SB, SS and KRK analysed the data and finalized the manuscript. All authors have read and approved the final manuscript.
7. CONFLICT OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
9. DATA AVAILABILITY
The data generated and analyzed are included within this research article.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Schulz B, Boyle C, Draeger S, Römmert AK, Krohn K. Endophytic fungi:A source of novel biologically active secondary metabolites. Mycol Res 2002;106:996-1004. [CrossRef]
2. Ferreira MC, Cantrell CL, Wedge DE, Gonçalves VN, Jacob MR, Khan S, et al. Antimycobacterial and anti-malarial activities of endophytic fungi associated with the ancient and narrowly endemic neotropical plant Vellozia gigantea from Brazil. Mem Inst Oswaldo Cruz 2017;112:692-7. [CrossRef]
3. Toghueo RM. Anti-leishmanial and anti-inflammatory agents from endophytes:A review. Nat Products Bioprospect 2019;9:311-28. [CrossRef]
4. Suresh G, Kokila D, Suresh TC, Kumaran S, Velmurugan P, Vedhanayakisri KA, et al. Mycosynthesis of anticancer drug taxol by Aspergillus oryzae, an endophyte of Tarenna asiatica, characterization, and its activity against a human lung cancer cell line. Biocatal Agric Biotechnol 2020;24:101525. [CrossRef]
5. Benjamin CR. Ascocarps of Aspergillus and Penicillium. Mycologia 1955;47:669-87. [CrossRef]
6. Cao D, Sun P, Bhowmick S, Wei Y, Guo B, Wei Y, et al. Secondary metabolites of endophytic fungi isolated from Huperzia serrata. Fitoterapia 2021;155:104970. [CrossRef]
7. Chen S, Ding M, Liu W, Huang X, Liu Z, Lu Y, et al. Anti-inflammatory meroterpenoids from the mangrove endophytic fungus Talaromyces amestolkiae YX1. Phytochemistry 2018;146:8-15. [CrossRef]
8. Singh P, Sharma A, Bordoloi M, Nandi SP. Molecular identification of endophytic fungi isolated from medicinal plant. Biointerface Res Appl Chem 2020;10:6436-43. [CrossRef]
9. Ewels P, Magnusson M, Lundin S, Käller M. MultiQC:Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016;32:3047-8. [CrossRef]
10. Chen S, Zhou Y, Chen Y, Gu J. Fastp:An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884-90. [CrossRef]
11. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes:A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455-77. [CrossRef]
12. Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST:Quality assessment tool for genome assemblies. Bioinformatics 2013;29:1072-5. [CrossRef]
13. Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT:An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015;31:1674-6. [CrossRef]
14. Lau YL, Lee WC, Xia J, Zhang G, Razali R, Anwar A, et al. Draft genome of Brugiapahangi:High similarity between B. pahangi and B. malayi. Parasites Vectors 2015;8:1-14. [CrossRef]
15. Waterhouse RM, Seppey M, Simao FA, Manni M, Ioannidis P,Klioutchnikov G, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol 2018;35:543-8. [CrossRef]
16. Medema MH, Blin K, Cimermancic P, De Jager V, Zakrzewski P, Fischbach MA, et al. AntiSMASH:Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 2011;39(Suppl 2):339-46. [CrossRef]
17. Riko R, Nakamura H, Shindo K. Studies on pyranonigrins-isolation of pyranonigrin E and biosynthetic studies on pyranonigrin A. J Antibiot (Tokyo) 2014;67:179-81. [CrossRef]
18. Palanichamy P, Kannan S, Murugan D, Alagusundaram P, Marudhamuthu M. Purification, crystallization and anticancer activity evaluation of the compound alternariol methyl ether from endophytic fungi Alternaria alternata. J Appl Microbiol 2019;127:1468-78. [CrossRef]
19. Yan C, Liu W, Li J, Deng Y, Chen S, Liu H. Bioactive terpenoids from:Santalum album derived endophytic fungus Fusarium sp. YD-2. RSC Adv 2018;8:14823-8. [CrossRef]
20. Li DH, Han T, Guan LP, Bai J, Zhao N, Li ZL, et al. New naphthopyrones from marine-derived fungus Aspergillus Niger 2HL-M-8 and their in vitro antiproliferative activity. Nat Prod Res 2016;30:1116-22. [CrossRef]
21. Godio RP, Martín JF. Modified oxidosqualene cyclases in the formation of bioactive secondary metabolites:Biosynthesis of the antitumor clavaric acid. Fungal Genet Biol 2009;46:232-42. [CrossRef]
22. Jia SL, Chi Z, Chen L, Liu GL, Hu Z, Chi ZM. Molecular evolution and regulation of DHN melanin-related gene clusters are closely related to adaptation of different melanin-producing fungi. Genomics 2021;113:1962-75. [CrossRef]
23. Desjardins AE, Proctor RH. Molecular biology of Fusarium mycotoxins. Int J Food Microbiol 2007;119:47-50. [CrossRef]
24. Fu Y, Li C, Zhu J, Zhang L, Wang Y, Chen Q, et al. A new meroterpenoid from endophytic fungus Talaromyces amestolkiae CS-O-1. Biochem Syst Ecol 2020;93:104186. [CrossRef]
25. Guo J, Ran H, Zeng J, Liu D, Xin Z. Tafuketide, a phylogeny-guided discovery of a new polyketide from Talaromyces funiculosus Salicorn 58. Appl Microbiol Biotechnol 2016;100:5323-38. [CrossRef]
26. Schafhauser T, Wibberg D, Rückert C, Winkler A, Flor L, van Pée KH, et al. Draft genome sequence of Talaromyces islandicus (“Penicillium islandicum“) WF-38-12, a neglected mold with significant biotechnological potential. J Biotechnol 2015;211:101-2. [CrossRef]
27. Chen S, Liu Y, Liu Z, Cai R, Lu Y, Huang X, et al. Isocoumarins and benzofurans from the mangrove endophytic fungus Talaromyces amestolkiae possess α-glucosidase inhibitory and antibacterial activities. RSC Adv 2016;6:26412-20. [CrossRef]
28. El-Elimat T, Figueroa M, Raja HA, Alnabulsi S, Oberlies NH. Coumarins, dihydroisocoumarins, a dibenzo-α-pyrone, a meroterpenoid, and a merodrimane from Talaromyces amestolkiae. Tetrahedron Lett 2021;72:153067. [CrossRef]
29. Singh SK, Mondal A, Saha N, Husain SM. Identification and characterization of an anthrol reductase from:Talaromyces islandicus (Penicillium islandicum) WF-38-12. Green Chem 2019;21:6594-9. [CrossRef]
30. Zhou C, Cao X, Ge Y, Wu X, Zhang Z, Ma Y, et al. Talaropeptins A and B, tripeptides with an N- trans-cinnamoyl moiety from the marine-derived fungus Talaromyces purpureogenus CX11. J Nat Prod 2022;85:2620-5. [CrossRef]
31. Li CX, Zhao S, Zhang T, Xian L, Liao LS, Liu JL, et al. Genome sequencing and analysis of Talaromyces pinophilus provide insights into biotechnological applications. Sci Rep 2017;7:490. [CrossRef]
32. Rashied RM, Abdelfattah MA, El-Beshbishy HA, ElShazly AM, Mahmoud MF, Sobeh M. Syzygium samarangense leaf extract exhibits distinct antidiabetic activities:Evidences from in silico and in vivo studies:Syzygium samarangense leaf extract exhibits distinct antidiabetic activities. Arab J Chem 2022;15:103822. [CrossRef]
33. Kaul S, Gupta S, Ahmed M, Dhar MK. Endophytic fungi from medicinal plants:A treasure hunt for bioactive metabolites. Phytochem Rev 2012;11:487-505. [CrossRef]