1. INTRODUCTION
In India, the Himalayas are the primary habitat for the 85 species of Rhododendron (Ericaceae) [1]. Of these, 36 species are unique to the Himalayan regions of Darjeeling and Sikkim [2].
Even though Rhododendron sp. plants have economic and ethno-medical significance, they are of the least studied plants in India [3,4]. Flowers of this plant have been used to make pickles, juice, jam, syrup, honey, squash, dried items, and treat diarrhea, headache, inflammation, viral, bacterial, and fungal illnesses, and so on [5]. Horticultural values of Rhododendron spp. are internationally known, as well [6].
Many Rhododendron species, including our chosen species, possess ecological, commercial, aesthetic, sacred, and ethno-medicinal and social importance. Anthropogenic disturbances such as deforestation, unsustainable extraction, over-exploitation, and agricultural activities have made many Rhododendron species endangered, rare, and threatened [7], including our chosen species. As a step towards the implementation of ecological preservation measures, determination of genetic diversity is essential. Therefore, it is necessary to know the genetic variability present in a particularly rare species to implement preservation and resource administration [8]. DNA markers are preferred over morpho-anatomical and biochemical ones because of higher specificity and sensitivity, more stability and consistency, unaffected by environmental conditions throughout growth and differentiation, high throughput, and automation of analysis. DNA markers based on polymerase chain reaction (PCR), like random amplified polymorphic DNA (RAPD), have been widely employed to study kinship and variety among plant populations. This technique is very simple and efficient for genomic diversity analysis. Few studies have been published on the use of RAPD molecular markers to examine the genetic variation of Rhododendron [9–11]. The goals of this research were to assess the levels of genetic diversity and genome polymorphism of some rare, threatened Darjeeling Himalayan Rhododendron spp. Among them, there is no report found for assessing genetic diversity by molecular markers. Therefore, this assessment was initially used to determine the genetic relatedness and diversity of selected Darjeeling Himalayan Rhododendron species to utilize effective conservation strategies.
2. MATERIALS AND METHODS
2.1. Collection of Samples
Young and fresh leaf samples (length 1–2.5 cm and breadth 0.5–1 cm) of the 10 Rhododendron species (Fig. 1) were collected for the study. Rhododendron decipiens Lacaita, Rhododendron falconeri Hook.f., Rhododendron fulgens Hook.f., Rhododendron grande Wight, Rhododendron maddenii Hook.f., Rhododendron niveum Hook.f., Rhododendron pendulum Hook.f., Rhododendron setosum D. Don, Rhododendron sikkimense Pradhan & Lachungpa, Rhododendron triflorum Hook.f. were gathered from various elevations in the Darjeeling Hills, beginning in Batasia (2,247 masl) and ending in Sandakphu (3,580 masl) (Table 1 and Fig. 2). The differences in phenotypes between the species were observed and noted in Table 1.
 | Figure 1. Showing plant and a close-up view of flowers of 10 Rhododendron species. [Click here to view] |
2.2. DNA Isolation
Hexadecyl trimethyl ammonium bromide (CTAB) procedure was used to extract DNA from the tender leaves [12]. Initially, 225 mg of leaf sample was crushed using a bowl and stick, and then 60 minutes were spent in an incubation bath (65°C) containing isolation Buffer (600 µl). CTAB (2% wv–1), NaCl (1.40 M), Tris-HCl (100.0 mM, pH 8.0), Polyvinylpyrrolidone (40) (1% wv–1), and 2-mercaptoethanol (1% wv–1) used to make up the isolation buffer. The mixture was extracted using 1:1 chloroformisoamyl alcohol (24:1) after being allowed to cool to room temperature. To separate the phases, the mixture was centrifuged for 10 minutes (15,000 g) at room temperature after being inverted to create an emulsion. After performing an RNase digestion (10 gm l–1 RNase A at 37°C for 60 minutes), a second chloroform-isoamyl alcohol extraction was carried out. Later, by adding a 2/3 volume of cold isopropanol, DNA was separated out of the liquid phase. The pellet was rinsed with 76% (vv–1) ethanol and 0.2 M sodium acetate. The DNA was re-suspended in 50 µl of a buffer containing Tris-HCl (10 mM) and TE-EDTA (1 mM) at pH 8.0. For RAPD analysis, DNA was diluted to 50 ng μl–1 in TATE pH 8.0. DNA purity and concentration were measured using spectrophotometer (Smart-Spec 3000, UV/Vis spectrophotometer, Bio-Rad Laboratories). The OD260/OD230 of the isolated DNA samples was between 1.80 and 2.80, and the OD280/OD280 ratio was 1.7.
2.3. Amplifying DNA and RAPD Procedure
DNA amplifications were carried out using reaction mixtures (25 μl) containing 50 ng template DNA, 2 mM of dNTPs, 2.5 mM MgCl2, 15 ng of degenerate primer, 2.5 μl of 10X PCR buffer, in addition to 1 unit of AmpliTaq-Gold polymerase (Life Technologies; Grand Island, NY). The MJ Mini™ Gradient Thermal from Bio-RAD Laboratories (India) Pvt. Ltd. (PTC-1148G) was used to conduct the PCR. QIAquick PCR Purification Kit was used to clean the PCR products. In the first step, 10 samples were used to test 19 different oligonucleotide primers (10 base pairs) for their ability to produce clear and consistent band patterns. Out of 19, 6 top-performing oligonucleotide primers were chosen for the examination of the current specimens. Six RAPD primers we selected based on their good data reproducibility and finally used for the characterization of ten Rhododendron species. The sequences of 10mer 6 primers are mentioned in Table 2.
2.4. Data Analysis
Data for each band was recorded in Microsoft Excel if either “present” as 1 or “not present” as 0, including monomorphic bands. Genetic data were analyzed by different software. The effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (Hs), total expected heterozygosity (Ht), inbreeding coefficient (Fst), coefficient of gene differentiation (Gst), gene flow (Nm), Shannon’s information index (I), polymorphic percentage (PPL), was calculated by the GenAlEx version 6.502 software [13]. To quantify polymorphic information content (PIC), the 3.25th version of PowerMarker was used [14]. For RAPD-based percentage homology of the samples, pair-wise correlation was performed using the neighbor-joining cluster analysis method produced from Jaccard’s estimate [15]. To create the dendrogram, we used Free Tree Software and the neighbor-joining cluster analysis technique utilizing Jaccard’s estimate [16].
3. RESULTS
3.1. RAPD Polymorphism and Genetic Diversity
The screening was performed using 19 RAPD primers, and 6 of those primers demonstrated amplification across all the selected species. These six primers exhibited dependable and unambiguous banding patterns, with good repeatability and clear band resolution. PCR, using the six-decamer oligonucleotide primers successfully amplified genomic DNA, and the results are summarized in Table 3. A total of 589 bands were detected using six RAPD primers from ten Rhododendron species with an average of 61.10 alleles observed. Using 6 primers among 10 samples, showing a broad range of PPL%, the highest in R. falconeri (93.44%) and lowest in R. setosum (63.50%) and the mean of 79.43% polymorphism. PIC scores were between 0.180 and 0.227 in R. decipiens and R. triflorum, respectively, with an average of 0.209. Observed heterozygosity (Ho) ranges from 0.979 (R. falconeri and R. fulgens) to 0.993 (R. decipiens) which was very low in deference. The results of genetic diversity present within the population (Hs) do not show a significant amount of difference among species. The average variation in a population’s genes (Hs) of the ten Rhododendron species is 0.979. The mean of the whole variation in genes (Ht) among ten Rhododendron species is 0.985. The inbreeding coefficient (Fis) value within individuals was calculated as a negative value except in R. fulgens and R. falconeri. The two species (R. fulgens and R. falconeri) shows neutral Fis result. The degree of gene flow (Nm) ranges from 34.376 (R. triflorum) to 42.080 (R. fulgens) and a mean of 38.505. Shannon’s information index (I) ranges between the lowest 3.944 (R. triflorum) to the highest 4.000 (R. setosum) with an average value of 3.973.
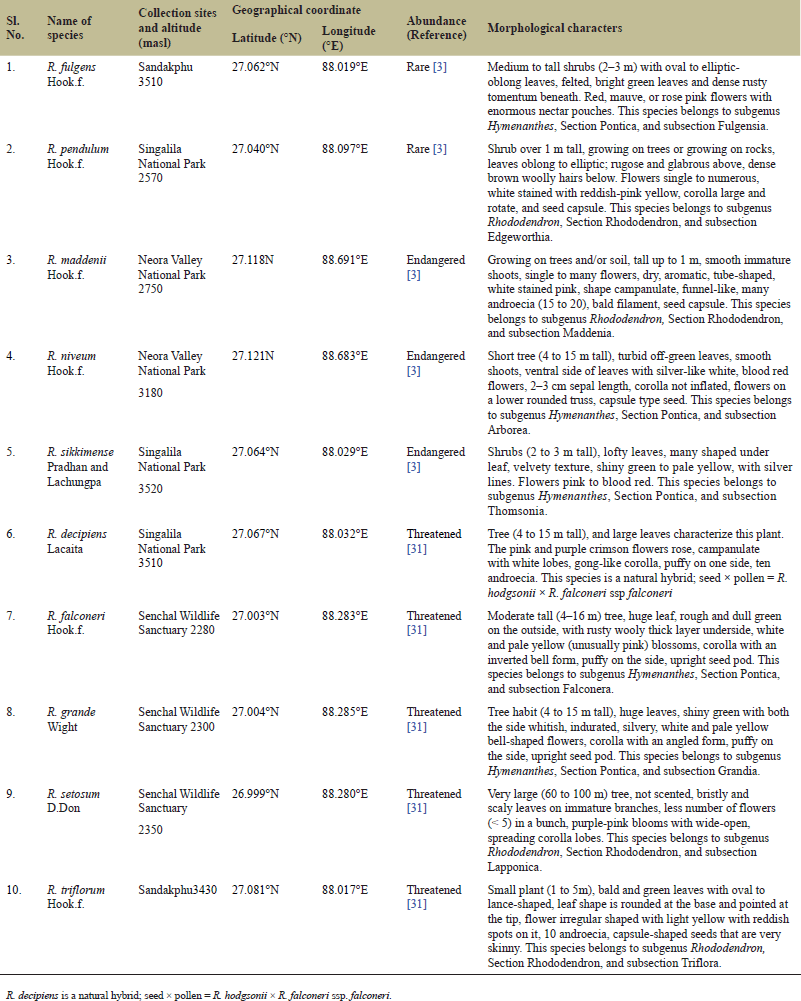 | Table 1. Table showing plant samples, collection site with altitude, abundance status, and the phenotypic differences among the studied species of Rhododendron. [Click here to view] |
 | Figure 2. Geographic locations (in Google map) of the 10 Rhododendron species taken for study. [Click here to view] |
 | Table 2. List of the selected RAPD primers used for study and characterization in 10 Rhododendron species. [Click here to view] |
3.2. Genetic Differentiation and Phylogenetic Tree
The mean inbreeding coefficient (Fst) within subpopulations is 0.006, which is very low. Similarity indices among the ten Rhododendron species based on RAPD analysis of genomic DNA made by Jaccard’s similarity coefficient computer program, are shown in Table 4. The range of similarity correlations was from 0.93827 to 0.45455.
A dendrogram constructed from Jaccard’s estimates in Free Tree through the use of neighbor-joining cluster analysis divided nine (out of ten) Rhododendron species into two main sister groups (I and II). In sister group I, R. niveum, R. fulgens, R. setosum, and R. sikkimense are the most primitive of the taxa under study; whereas R. triflorum and R. maddenii are recent as compared to the previous four taxa, but R. pendulum has most recent origin as indicated by high bootstrap value and relatively smaller branch length. However, sister group II containing R. decipiens and R. falconeri is more recent in origin, than group I due to low bootstrap values (11 and 44) and long branches. Rhododendron grande is out-group taxa (Fig. 3).
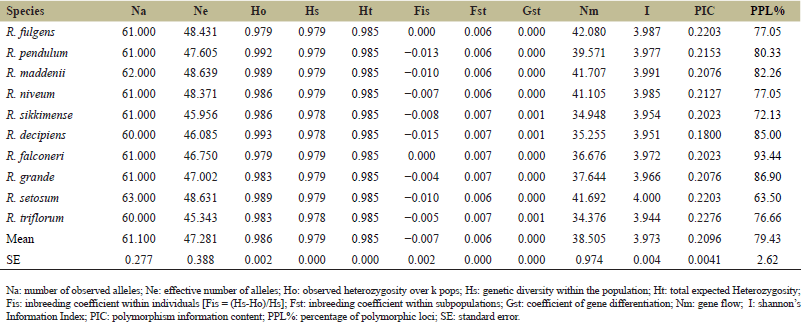 | Table 3. Summary of genetic diversity indices for 10 Rhododendron species used for the study. [Click here to view] |
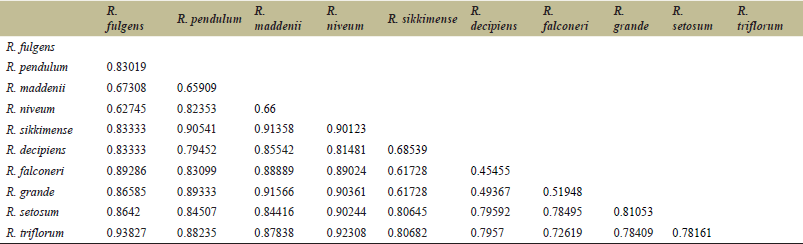 | Table 4. Similarity index or coefficient for RAPD. [Click here to view] |
 | Figure 3. The phylogenetic tree of the 10 Rhododendron genotypes based on RAPD markers utilizing neighbor-joining cluster analysis and Jaccards estimations from Free Tree program. Node values are bootstrapped. The evolutionary position of taxa can be determined by correlating theses values with branch lengths. [Click here to view] |
4. DISCUSSION
Six decamer oligonucleotide random primers were applied for the present investigation. Primers measure moderate to high polymorphism percentage (63.50 to 93.44) with average low Polymorphism Information Content (Mean PIC = 0.2096) of 10 Rhododendron species. Small populations have less genetic variety than big populations due to factors including genetic drift and inbreeding [17]. Due to this, genetic diversity is estimated to be lower in rare and endangered species with restricted geographic ranges than in the same species with wider geographic ranges [18]. We found a high average degree of genetic diversity (percentage of polymorphic loci = 79.43%) in selected Rhododendron species. In general, present findings backed the theory that some rare and endangered organisms may keep their genetic diversity high though their populations are declining [17,19]. High Shannon’s information index (Mean I = 3.973) was calculated among the rare and endangered Rhododendron species. The high genetic diversity was found in the current investigation at the species level (Mean, Na = 61.100, Ho = 0.986, and Hs = 0.979, Ht = 0.985). In earlier studies, it was shown that genetic diversity among endangered plant species is surprisingly high to moderate like Origanum compactum (He = 0.35) a medicinally important plant, Paeonia jishanensis (HE = 0.340), Rhododendron protistum var. giganteum [Nei’s gene diversity (h) = 0.240], Paeonia decomposita (HE = 0.405), and Populus wulianensis (HE = 0.61) [20,21]. Several factors like mating strategy, biological characteristics, and out-breeding could be considered as significant elements that determine increased levels of genetic variety in Rhododendron [17].
The 10 Rhododendron species generate average negative inbreeding coefficients (Fis < 0) (Table 3), illustrating that despite having fragmented habitats, they do not have inbreeding depression [21]. That could be explained by the floral characteristics of the Eastern Himalayan Rhododendron species evolved in such a way that bird pollination is common for this population and weather conditions [22]. On the other hand, increased hybridization (natural or artificial) will elevate in risk of extinction of species or populations [23]. The similarity measurement using Jaccard’s coefficient values varies from 0.45455 to 0.93827 among selected species (Table 4). The genetic diversity among these species clearly indicates that they must have evolved from genetically divergent parents [24].
The RAPD clustering finding revealed a closer link between the Hymenanthes and Rhododendron subgenera. In addition, it demonstrated that certain physical qualities could mirror inherited characteristics. In sister group II, R. decipiens and R. falconeri were gathered together because they shared some common morphological characters (Table 1). Moreover, R. decipiens is a natural hybrid between R. falconeri and R. hodgsonii (Hook. f). In sister group I, R. sikkimense, R. fulgens, and R. niveum were gathered together because they are morphologically small trees or tall shrubs, semi-deciduous species, and associated with the section Pointicum. Rhododendron grande belonging to the same section Pointicum kept as an out-group because it showed little different morphological traits such as trees 4–15 m high, and flowers white to creamy yellow. In Gladiolus plant, the UPGMA cluster analyses method arranged 54 cultivars into four and three primary groups based on their morphological features and RAPD data, while in both cluster studies, “Pusa Lohit” (red-colored flower) branched off from dendrograms, supporting its morphological and genetically uniqueness [25]. Although, R. triflorum, R. setosum, R. pendulum, and R. maddenii were in the subgenus Rhododendron they had a closer relationship with some species of the subgenus Hymenanthes. Therefore, they have been included in the sister group I.
There is a positive correlation between genetic differentiation and Fst value [26]. The average Fst results for the study revealed that the genetic differentiation between species was negligible (Table 3). Gene flow might inhibit differentiation and mitigate the genetic drift when Nm >1 [27]. The gene flow (Mean Nm = 38.505) between Rhododendron species also indicated that gene flow among species is very high. Over gene flow is introgressive and can produce genetic swamping [28]. Through genetic swamping, in which the native organisms have increased the risk of extinction for rare species [29,30].
5. CONCLUSION
The investigation shows that studied Rhododendron species are at a high risk of out-crossing depression, which will lead to a population bottleneck. Therefore, immediate action needs to be taken to implement conservation (in situ and ex situ) measures. Future studies should include more Rhododendron species from the hills to strengthen the comprehensive and generalized conclusions.
6. ACKNOWLEDGMENTS
It is with profound gratitude that the authors acknowledge the Postgraduate Department of Botany at Darjeeling Government College and Hooghly Mohsin College and the Department of Botany at Banwarilal Bhalotia College in Asansol for making this study possible.
7. AUTHOR CONTRIBUTION
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
8. FUNDING
There is no funding to report.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
13. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Tiwari ON, Chauhan UK. Rhododendron conservation in Sikkim Himalaya. Curr Sci 2006;90:532–41.
2. Pradhan UC, Lachungpa ML. Sikkim Himalayan Rhododendrons. Primulaceae Books, Kalimpong, India, 1990.
3. Singh KK, Kumar S, Rai LK, Krishna AP. Rhododendrons conservation in the Sikkim Himalaya. Curr Sci 2003;85:602–6.
4. Balkrishna A, Prajapati UB, Shankar R, Joshi RA. Nutraceutical aspects of Rhododendron (Burans): certainly a need to include some other species for food and beverage production. Int J Sci Res 2022;11:312–21.
5. Kumar V, Suri S, Prasad R, Gat Y, Sangma C, Jakhu H, et al. Bioactive compounds, health benefits and utilization of Rhododendron: a comprehensive review. Agric Food Secur 2019;8:6; CrossRef
6. Sharma M, Gargi A, Bora A. Rhododendron arboreum and its potential health benefit: a review. J Pharm Innov 2022;1:926–33.
7. Menon S, Khan ML, Paul A, Peterson AT. Rhododendron species in the Indian Eastern Himalayas: new approaches to understanding rare plant species distributions. J Am Rhododendron Soc 2012;66:78–84.
8. Izza-Ab-Ghani N, Arifin W, Ismail A. Conservation genetics for managing biodiversity. In: Suratman MN (ed.). Protected area management—recent advances. IntechOpen, London, UK; CrossRef
9. Lanying Z, Yongqing W, Li Z. Genetic diversity and relationship of rhododendron species based on RAPD analysis. Am-Eurasian J Agric Environ 2008;3:626–31.
10. Liu YF, Xing M, Zhao W, Fan RJ, Luo S, Chen X. Genetic diversity analysis of Rhododendron aureum Georgi (Ericaceae) located on Changbai Mountain using ISSR and RAPD markers. Plant Syst Evol 2012;298:921–30; CrossRef
11. Thakur A, Harisha CR, Chauhan S, Kumar A. Genetic characterization of Rhododendron arboreum SM. through RAPD study. Int J Creat Res Thoughts 2020;8:1047–51.
12. Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus 1990;12:13–5.
13. Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012;28:2537–9; CrossRef
14. Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics (Oxford, England) 2005;21:2128–9; CrossRef
15. Legendre P, Legendre L. Numerical ecology. 2nd edition, Elsevier, Amsterdam, The Netherlands, 1998.
16. Pavlícek A, Hrdá S, Flegr J. Free-Tree--freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol (Praha) 1999;45:97–9.
17. Wu FQ, Shen SK, Zhang XJ, Wang YH, Sun WB. Genetic diversity and population structure of an extremely endangered species: the world’s largest Rhododendron. AoB Plants 2014;7:plu082; CrossRef
18. Hamrick JL, Godt MJW. Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL (eds.). Plant population genetics, breeding and genetic resources, Sinauer Associates Inc, Sunderland, UK, pp 43–6, 1989.
19. Swarup S, Cargill EJ, Crosby K, Flagel L, Kniskern J, Glenn KC. Genetic diversity is indispensable for plant breeding to improve crops. Crop Sci 2021;61:839–52.
20. Wang SQ. Genetic diversity and population structure of the endangered species Paeonia decomposita endemic to China and implications for its conservation. BMC Plant Biol 2020;20:510; CrossRef
21. Wu Q, Zang F, Ma Y, Zheng Y, Zang D. Analysis of genetic diversity and population structure in endangered Populus wulianensis based on 18 newly developed EST-SSR markers. Glob Ecol Conserv 2020;24:e01329; CrossRef
22. Huang Z, Song Y, Shuang-Quan H. Evidence for passerine bird pollination in Rhododendron species. AoB Plants 2017;9:plx062; CrossRef
23. Ding JM, Zhang XD, Li GL, Wang J, Huang J, Zhang ZX, et al. Genetic considerations in recovery of endangered plants. Plant Sci J 2018;36:452–58. Available via http://www.whzwxyj.cn/CN/10.11913/PSJ.2095-0837.2018.30452
24. Singh SR, Mir J, Ahmed N, Rashid R, Wani S, Sheikh M, et al. RAPD profile based grouping of garlic Allium sativum germplasm with respect to photoperiodism. J Trop Agric 2011;49:114–7.
25. Pragya P, Bhat KV, Misra RL, Ranjan JK. Analysis of diversity and relationships among Gladiolus cultivars using morphological and RAPD markers. Indian J Agric Sci 2010;80(9):766–72.
26. Wright S. Evolution and the genetics of populations. In: Variability within and among natural populations, Volume 4, University of Chicago Press, Chicago, IL, 1978.
27. Wright S. Evolution in Mendelian populations. Bull Math Biol 1990;52:241–95; CrossRef
28. Fath B. Encyclopedia of ecology. 2nd edition, Elsevier, Amsterdam, The Netherlands, 2018.
29. Todesco M, Pascual MA, Owens GL, Ostevik KL, Moyers BT, Hübner S, et al. Hybridization and extinction. Evol Appl 2016;9:892–908.
30. Buck R, Flores-Rentería L. The syngameon enigma. Plants (Basel) 2022;11:895; CrossRef
31. Singh KK, Rai LK, Gurung B. Conservation of Rhododendrons in Sikkim Himalaya: an overview. World J Agric Sci 2009;5:284–96.