1. INTRODUCTION
Fish and other aquatic animals are susceptible to changing environments and suitable efforts are needed to replenish the species. A lack of appropriate management strategies may lead to further critical decline for the species in the years to come [1,2]. Clarias magur (Hamilton, 1822), commonly called walking catfish (magur), is an important freshwater clariid species native to India and neighboring nations with promising aquaculture prospects. A variety of known and unknown factors have contributed to a severe drop in the availability of this species, hence, it is categorized as “Endangered (A3cde + 4acde)” in IUCN Red List Status [3]. This species has gained attention and considerable investigations were carried out in the field of aquaculture, conservations, genomics, and so on, including the development of bacterial artificial chromosomes (BAC) genomic resources, to replenish this species in a sustainable aquaculture system.
BACs are DNA constructs used to clone large DNA fragments in bacteria. They are designed to carry and replicate large inserts of DNA, typically ranging from 100,000 to 300,000 base pairs in size. BACs are widely used in molecular biology and genomics research to study and manipulate genes and genomes. They are particularly useful for projects involving sequencing large stretches of DNA. BAC insert DNA can be used for high-throughput BAC end (BE) sequencing, chromosomal mapping, genome sequencing, and other related studies [4,5]. Numerous investigations, such as chromosome mapping using BAC-FISH [6,7] chromosomal investigations using Giemsa, C-banding, Ag-NOR, CMA3 staining, and 18S as well as 5S rDNA-FISH [7], molecular studies, like genome sequencing [8], transcriptome analysis [9], thermal stress studies [10], and so on, were undertaken to better characterize and improve this species. A BAC library of 55,141 clones constructed from C. magur genome is maintained at ICAR-National Bureau of Fish Genetic Resources, Lucknow, India. Moreover, the whole genome assembly of C. magur (GenBank ID: GCA_013621035.1) is also available [8]. The present study was focused on analyzing the BAC clones using their end sequences and whole genome information for gene and simple sequence repeats (SSR) mining and pathway analysis which could aid in genomic selection programs or genetic diversity studies and complement aquaculture production. BAC clone DNA probe that hybridizes on an individual chromosome could be used to identify that particular chromosome. This approach can also be utilized to resolve chromosomal identity in ambiguous morphological conditions. The study will be helpful to identify overlapping clones, select clones for restriction endonuclease fingerprints, identify appropriate clones for FISH mapping, select clone(s) containing genes of interest, and so on. It will also support United Nations’ sustainable development goals (SDGs), particularly SDG 14 of promoting research on life below water, and SDG2 on achieving food security and promoting sustainable agriculture by species conservation and complementing aquaculture production.
2. MATERIALS AND METHODS
2.1. BAC Clone Revival and Culture
The BAC library of C. magur genome consisting 55,141 clones stored in 144 plates of 384-well was utilized in this study. One 384-well plate (ID: 012) was randomly selected and revived. The revived clones were further sub-divided into 96-well plate format for ease of maintenance as well as handling and stored at −80°C for (BE) sequencing and other use. One 96-well plate was randomly selected and thawed after which, 15 μl of each clone was transferred in 15.0 ml centrifuge tubes for revival, culture, and isolation of insert DNA, as per protocol [6,7].
2.2. BAC DNA Isolation and BE Sequencing
Selected clones were subjected to BAC insert DNA isolation, utilizing an alkaline lysis protocol [6]. Briefly, the E. coli culture was incubated overnight and centrifuged at 3,300 rcf for 10 minutes at room temperature. The supernatant was decanted and tubes containing pallets were inverted to remove traces of media. The remaining pellets were pooled in an 8 ml culture tube and used for insert DNA isolation. The isolated BAC inserts DNA were purified with 25 µl magnetic beads using 96-well format recommended protocol of polymerase chain reaction purification procedure of Agencourt AMPure XP, Beckmann Coulter (https://www.beckmancoulter.com/wsrportal/techdocs?docname=B37419). The quality and quantity of isolated insert DNA were determined in a transilluminator with 0.4% agarose gels and in NanoDrop2000 spectrometer (Thermo Fisher Scientific, USA). Both ends of the clones were sequenced in Genetic Analyzer ABI 3500 (Thermo Fisher Scientific, USA) using T7 forward (5′-TAATACGACTCACTATAGGG-3′) and pbRP1 reverse (5′-CTCGTATGTGTGGAATTGTGAGC-3′) primers (http://www.epibio.com). The sequence quality was checked and good-quality sequences of BEs were used in analysis.
2.3. BE Mapping, Gene Mining, and Functional Annotation of Mapped Genes
The qualified BE sequences were mapped on the genome assembly of C. magur) using BLASTN tool with 10−5 e-value. Custom Perl scripts were used for identifying the end-to-end sequence length of BAC insert DNA fragments. The clone-wise mapped end-to-end sequences on the scaffolds were then retrieved and a fasta file of the same was created. Gene prediction, gene mining, and annotation of complete sequences of clones were carried out using OmicsBox platform, wherein InterPro mapping, and annotation using UniProt and NR databases were used. SSRs present in the clone sequences were mined from using MISA tool [11].
2.4. Probe Labelling and BAC-FISH
One µg of isolated BAC inserts DNA from K10 and A23 well clones of 012 plate, containing a comparatively higher number of genes as determined bioinformatically from whole genome data, were taken for physical chromosome mapping. The DNA of K10 and A23 clones were labeled with green fluorescein-12-dUTP (Fermentas, Vilnius, Lithuania) and red tetramethyl-rhodamine-5-dUTP (Roche, Basel, Switzerland) fluorophores using nick translation technique. FISH was carried out to hybridize probes on metaphase chromosomes for 2 to 3 days, following the protocol of Kumar [6]. After hybridization, the chromosomes were counterstained for an hour using VectraShield mounting media (Vector Labs, Burlingame, CA) containing DAPI and antifade. Metaphase spreads hybridized with DNA probes were then examined under a fluorescence microscope (Leica, Wetzlar, Germany) under 3 band filters, viz. DAPI filter (340–380 nm excitation, 461 nm emission) for chromosome visualization in blue, N2.1 filter (515–560 nm excitation, 595–605 nm emission) for rhodamine-labeled probe visualization in red, and I3 filter (450–490 nm excitation) for fluorescein-labeled probe visualization in green color. Chromosomes (visualized in DAPI) and probes (visualized in N2.1 or I3 filters) images were superimposed using Karyo4000 software (Leica, Germany) to determine and visualize the location of the probes’ signals on specific chromosomes. Individual hybridized metaphase spreads were then manually karyotyped and a consensus karyotype was generated.
2.5. Comparative Genomics and Functional Enrichment Analysis
The synteny analysis of annotated genes, present on clones, was performed by searching them against NCBI and Ensembl databases and generating detailed information of the genes using genomes of Danio rerio and Ictalurus punctatus as a query. The complete scaffolds with ID numbers, containing annotated genes of C. magur genome and genes discovered in D. rerio and I. punctatus but present on different chromosomes, were considered for synteny. To make clear image visualization, only one gene of the scaffold was considered for comparisons, though the rest genes were also located on the same scaffold. Circos program [12] was used in synteny visualization.
Functional enrichment analysis of the nodes within the cluster network was carried out using PANTHER gene ontology (GO) tool [13]. GO framework was employed to annotate gene sets, offering a comprehensive structure for functional categorization. Additionally, the KEGG pathway database [14] was applied to provide information on molecular interactions and network reactions. The PANTHER gene list analysis involved a statistical over-representation test, which was employed to identify functionally enriched gene networks within the GO Biological Process category. This statistical test relied on binomial statistical comparison to determine over-represented biological functions associated with the gene set.
2.6. Phylogenetic Analysis
Phylogenetic analysis was carried out for the annotated genes present on undertaken clones of C. magur and genes commonly found in I. punctatus and D. rerio genomes. Neighbour-joining (NJ) methods based on p-distances approach were carried out using MEGA 7 software [15]. The phylogenetic topology of DNA sequences for predicting evolutionary lineages was presented using the interactive tree of life v5.5 tool [16].
2.7. Protein–Protein Interaction (PPI) Network Analysis
The PPI network analysis of annotated genes was carried out using STRING 12.0 [17] and visualized using Cytoscape v3.6.0 tool [18]. The network was built based on a stringent confidence score at a threshold value of 0.04, signifying that only interactions with a high level of confidence were considered for inclusion in the network.
2.8. Functional Annotation
GO-based functional annotation of identified genes was performed through BLAST using UniProtKB/Swiss-Prot database (http://www.uniprot.org/). The most significant BLAST hits were selected which met the following criteria: E-value <1e–5 and similarity >80%.
3. RESULTS AND DISCUSSION
3.1. BEs Mapping, Gene Mining, and Functional Annotation
A few BAC libraries have been established as genomic resources in fishes [19–24]. BAC libraries and their characterization using end sequencing will provide insights into the genome. The isolated plasmid DNA quality was good which was used for forward and reverse-end sequencing. Out of 96 clones revived, good-quality DNA was obtained in 36 clones which were further used for end sequencing. A sufficient amount of BAC insert DNA is required for the BE sequencing of clones. The sequence quality check resulted in 32 clones, out of which 18 clones were sequenced with both ends, and the rest had either forward or reverse end sequence only. Both end sequences of 18 clones were used for further bioinformatic analysis.
The lengths of forward and reverse end sequences of 18 clones ranged from 154 bp (forward end sequence of clone ID: 012B11) to 914 bp (reverse end sequence of clone ID: 012H6) and most of the end sequences were over 500 bp in size (Table 1), indicating good size. Bioinformatic mapping of BE sequences on magur genome revealed their distribution on 17 scaffolds. The mapped 17 scaffolds covered 30,945,289 bp of magur genome and their size ranged from 195,099 bp (Scaffold882) to 6,784,174 bp (Scaffold3) (Table 1). The sizes of the clones were bioinformatically predicted by aligning both the end sequences of the clones on the scaffolds of the magur genome, which ranged from 90.615 kb (012A5 present on Scaffold118) to 150.071 (012H19 present on Scaffold170). A total of 38 genes were present on these clones and identified, annotated, and characterized (Table 1, Supplementary Table 1). A total of 1974 SSRs were identified in 18 clones’ sequences using MISA tool (Table 1). The clone ID: 012A6 contained maximum SSRs (310), while ID: 012H12 contained minimum SSRs (47).
Making use of the genome sequence’s advantages and BAC library of magur, a BAC-based partial physical map of C. magur genome was constructed [25]. There are several databases of BE sequences developed for numerous model species, including human, rice, mouse, and sea urchin [26]. In several studies, BE sequences were generated and utilized for genomic studies [25,27,28] indicating BACs as an important source of live genetic material in different biological contexts. BE sequences are also used for comparative analysis with other model genomes. This will help in determining the sequence orientation of a large genome region on the chromosome for gene discovery.
Metaphase chromosomal complements of fair quality were generated manually with a diploid chromosome (2n) count of 50. Based on morphology and chromosome number, the karyotype is 14m+20sm+8st+8t, with a fundamental arm number of 90 (Fig. 1a and b). A similar karyotype has been obtained by Indian workers in C. magur [25,29].
BAC-FISH is an important tool in physical chromosome mapping, which includes many steps, namely selecting an appropriate clone and obtaining high-quality chromosomal spread for probe signal detection and distinguishing chromosomes based on their morphology [25] employed BAC-FISH to establish its utility as a cytological marker for identifying individual chromosomes. In the present study, clone 012K10, labeled with green color fluorophore, fluorescein 12-dUTP, contained 4 genes (Table 1) and was mapped on 11th pair of sub-metacentric chromosomes (Fig. 1a), while clone 012A23, labeled with a red color fluorophore, rhodamine-5-dUTP, contained 4 genes (Table 1) and were mapped on the 12th pair of sub-metacentric chromosomes (Fig. 1b), with gene descriptions presented in Supplementary Table 1. The genes were well-defined and localized on the chromosomes, and compared with channel catfish and zebrafish in terms of gene localization and their pathways (Fig. 2).
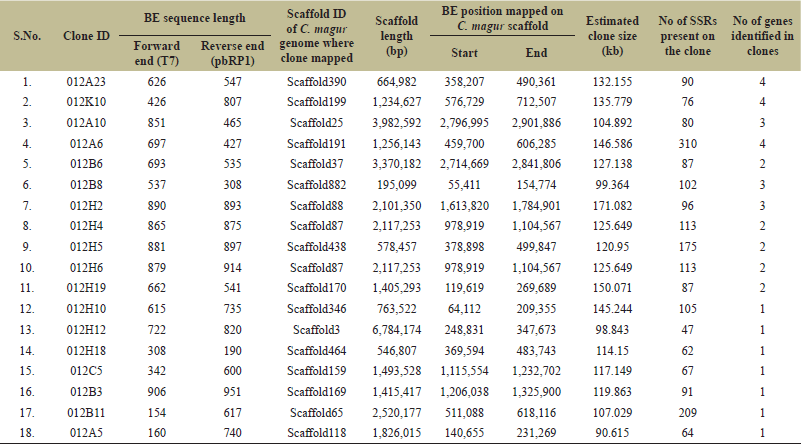 | Table 1. Details of BE mapping on the scaffolds of C. magur genome. [Click here to view] |
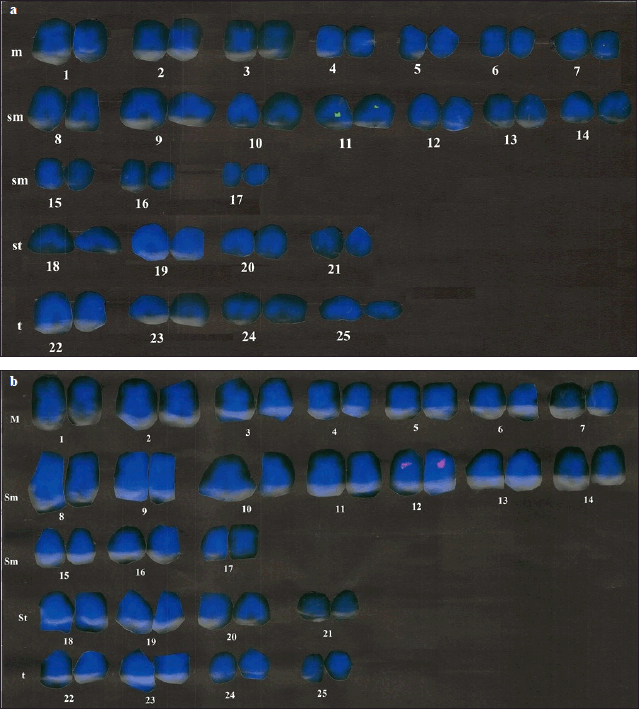 | Figure 1. (a) 012K10 clone signal on 11th submetacentric chromosome. (b) 012A23 clone signal on 12th submetacentric chromosome. [Click here to view] |
3.2. Functional Gene Enrichment
Gene enrichment analysis illuminates a diverse array of biological processes and molecular functions linked to the studied gene set. The cellular anatomical entity (GO: 0110165) has the maximum Go term, involving 13 genes, under the cellular component. Enrichment, such as cellular anatomical entity and protein-containing complex, under cellular components, points towards a focus on cellular structures and protein interactions. Higher-order processes, like “multicellular organismal process” and “response to stimulus”, hint at responses to environmental cues and organismal functions. Notably, the enrichment in “binding” and “catalytic activity” suggests significant involvement in these functions within the studied context. Additionally, “transcription regulator activity” and “ATP-dependent activity” underscore their regulatory and energy-related roles. In biological processes, the enrichment in “cellular process”, “biological regulation”, and “metabolic process” emphasizes their importance in various cellular activities and regulatory pathways (Fig. 3). However, the presence of unclassified entries suggests potential novel functions requiring further investigation.
Four pathways, providing insights into potential cellular mechanisms, were predicted using PANTHER tool. Nicotinic acetylcholine receptor signaling pathway (P00044), PI3 kinase pathway (P00048), p53 pathway (P00059), and Ubiquitin proteasome pathway (P00060) offered glimpses into synaptic transmission, cellular signaling, stress response, and protein turnover, respectively (Fig. 4). However, a significant portion of unclassified entries indicates either a lack of classification or the presence of potential novel pathways, necessitating further exploration. Overall, these analyses offer comprehensive insights into the biological processes, molecular functions, and potential pathways associated with the studied gene set, paving the way for a deeper understanding of their roles in cellular activities and interactions.
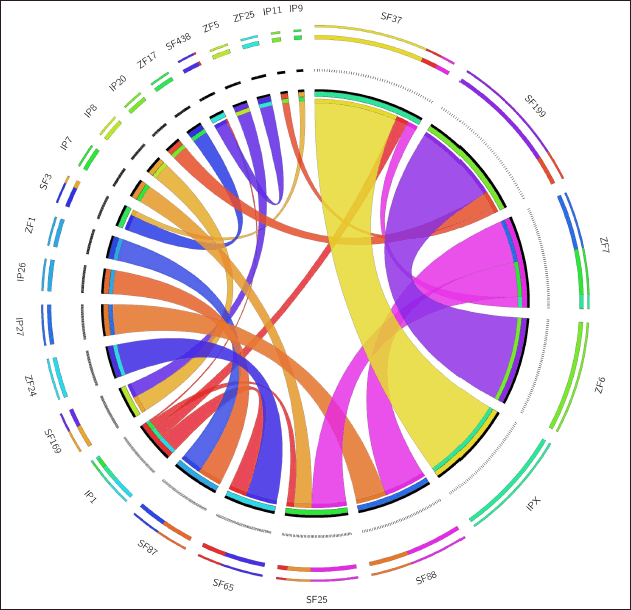 | Figure 2. Circos plot showing synteny of 38 genes present on 17 genome scaffolds of C. magur with I. punctatus and D. rerio. SF represents scaffold of C. magur, IP represents chromosome of I. punctatus and ZF represents chromosome of D. rerio on which gene were present. [Click here to view] |
3.3. Comparative Genomics and Genetic Diversity
By aligning end sequences of 18 clones representing 38 genes (Supplementary Table 1) mapped on the C. magur genome, 9 genes were synchronized with D. rerio and I. punctatus chromosomes. Circos graphic displays gene information and their locations on scaffolds and chromosomes in three species (Fig. 2). Remaining 29 genes present on clones were either present in chromosomes of either of the two species (I. punctatus or D. rerio), hence, were not considered for synteny visualization. The 38 genes located on 18 clones of magur genome were annotated with gene id, protein id, amino acid (AA), and gene (exon) size in bp. Herc1 gene, present on clone 012A6, showed a maximum 3919 AAs, while DAT39_0118 gene, present on clone 012K10, was the smallest with 61 AAs (Supplementary Table 1).
Four NJ-phylogenetic circular unrooted trees were constructed based on the p-distances for clear visualization of 20 common genes found on the chromosomes of I. punctatus, D. rerio, and on C. magur genome scaffolds (Fig. 5 a–d). The Scly gene of I. punctatus did not cluster with D. rerio and C. magur. The NJ-phylogeny of genes present on scaffolds SF_390, SF_390, SF_199, SF_199, SF_25, and SF_191 of C. magur with zebrafish and I. punctatus generated 3 clusters (Fig. 5a), while SF_191, SF_37, SF_882, SF_88, and SF_87 generated 4 clusters (Fig. 5b). Likewise, SF_88, SF_87, and SF_438 grouped into 3 clusters (Fig. 5c) and SF_170, SF_3, SF_159, SF_169, SF_65, and SF_118 generated into 4 clusters with I. punctatus and D. rerio (Fig. 5d).
3.4. Protein-Protein Interaction
A network-based approach was undertaken to identify and prioritize candidate genes in clone-mapped C. magur scaffolds with emphasis on PPI network and its role in unraveling the functional relationships between the genes or proteins. The gene co-occurrence can computationally be predicted by examining genomic datasets from various samples or environments including gene expression profiles and genomic sequencing data. The constructed PPI network revealed two types of interactions among 14 nodes (genes) and between 17 edges (adamts5, cth1, mta2, nos2a, odc1, slc7a5, taf6l, TIMM21, herc1, kif18a, rb1cc1, scly, uba2, adamts1, crocc2, smyhc3, and tpma) (Fig. 6a). These interactions were assessed based on various parameters, including their source in the database, experimental evidence, co-expression patterns and text mining, with 0.004 confidence score and 0.00415 PPI enrichment value (p-value). The clustering analysis of 17 genes in C. magur resulted in 3 distinct clusters represented by red, green, and blue colors (Fig. 6b). The red color cluster contained largest number of 7 genes (adamts5, cth1, mta2, nos2a, odc1, slc7a5, and taf6l), green color cluster had 6 genes (TIMM21, herc1, kif18a, rb1cc1, scly, and uba2) and blue color cluster consisted of 4 genes (adamts1, crocc2, smyhc3, and tpma). The genes exhibiting interaction include uba2-scly and mta2-cth1. Computational inferences of gene neighborhood, fusions, and co-occurrence can reveal some hypothesized interactions and provide insights into putative gene interactions.
 | Figure 3. Panther database analysis showing GO terms associated with molecular function, biological process and cellular components. [Click here to view] |
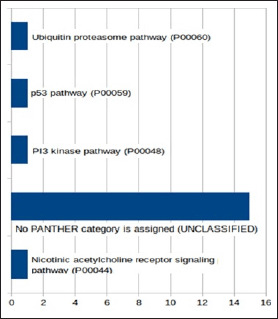 | Figure 4. Bar chart showing results of protein class of genes present on 18 clones in Panther analysis. [Click here to view] |
3.5. Gene Ontology
The genes discussed exhibit diverse functions and locations within cells. The listed genes encompass a diverse array of biological functions and cellular localizations (Table 2). The gene MFSD7 encodes a protein with RNA binding activity primarily localized in the membrane. NCBP1 is involved in various mRNA processing and capping processes, localized in the nucleus, specifically in the nuclear cap-binding complex and nucleus. Nuclear cap-binding proteins NCBP1 and NCBP3 have been reported in some eukaryotes to form a complex that binds to elements of the mRNA processing machinery, enhancing poly(A) RNA production in higher eukaryotes [30]. RPLX gene is primarily localized in the large ribosomal subunit and participates in translation processes with RNA binding activity. SCLY, involved in lyase activity, is primarily found in the cytosol. SLC7A5 has been reported to be essential for mTORC1 protein complex activation during fin regeneration in zebrafish, with leucine and glutamine signaling enhancing cell proliferation and regeneration [31]. MTA2 regulates DNA-templated transcription and is located in the nucleus. CTH1 participates in mRNA destabilization and is found in the cytoplasm and nucleus. TAF6L, involved in transcription initiation, is localized in the membrane and transcription factor complexes. TAF6L has been implicated in the regulation of transcription, cell cycle, cell division, and stem cell maintenance, potentially interacting with c-MYC to activate target gene expression in humans [32]. GNAO1 is part of the G protein-coupled receptor signaling pathway and is localized in the cytoplasm. It has been reported to be part of signal transduction pathways and has been linked to tumorigenesis in humans [33].
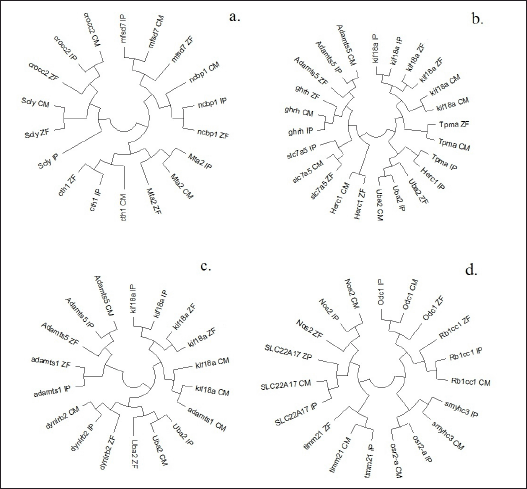 | Figure 5. (a–d) Phylogenetic trees constructed by NJ method based on p-distance of common genes present on chromosomes of I. punctatus and D. rerio with C. magur. [Click here to view] |
 | Figure 6. (a) PPI network and cluster analysis. (b) PPI network of 17 genes present on clones in C. magur resulting in three distinct clusters, denoted by red, green and blue. [Click here to view] |
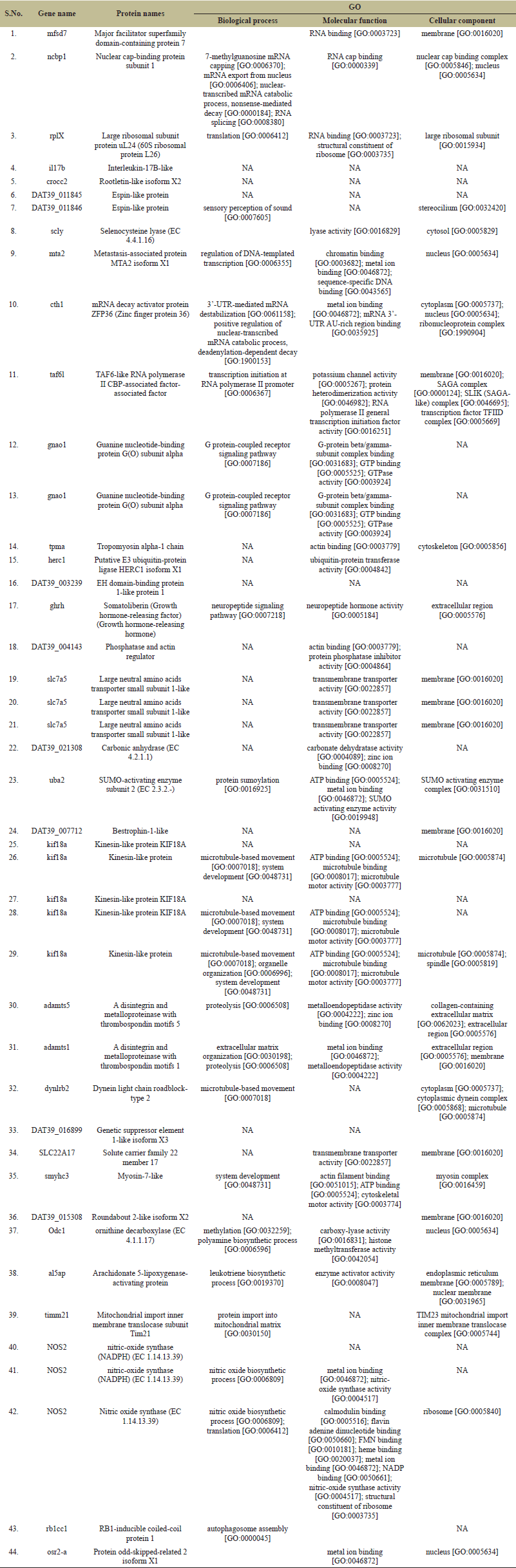 | Table 2. Annotation of genes located on mapped scaffold of C. magur genome. [Click here to view] |
The growth hormone receptor (GHR) is found in various organs of fish, including the liver, brain, and gonads, and plays a role in growth and development [34–37]. GHRH gene, participating in neuropeptide signaling, is primarily found in the extracellular region. SLC7A5 encodes a transmembrane transporter found in the membrane. KIF18A is involved in microtubule-based movement and system development and is primarily localized in the microtubule. AL5AP participates in leukotriene biosynthesis and is located in the endoplasmic reticulum and nuclear membrane. NOS2 participates in nitric oxide biosynthesis and is localized in the ribosome. RB1CC1 is involved in autophagosome assembly. OSR2-A exhibits metal ion binding activity and is localized in the nucleus. OSR2-A exhibits metal ion binding activity and has been implicated in chondrocyte development and fin morphogenesis in D. rerio [38]. Selenocysteine lyase (Scly) is an enzyme that decomposes selenocysteine (Sec) into selenide, providing selenium for the synthesis of new selenoproteins, crucial for various biological functions in mice [39]. The metastasis-associated (MTA) gene family, which includes Mta2 and Mta3, are reported to be involved in D. rerio embryogenesis [40]. Zfcth1 gene in D. rerio, akin to the mammalian TIS11 family, encodes a protein with CCCH zinc fingers and is involved in RNA expression during oogenesis and early embryogenesis, playing a role in cell fate determination and oocyte maturation [41]. ADAMTS5 [42] and ADAMTS1 [43] participate in proteolysis and are localized in the extracellular matrix and membrane, respectively. DYNLRB2 is involved in microtubule-based movement and localized in the cytoplasm and cytoplasmic dynein complex [44] ODC1 is involved in methylation and polyamine biosynthesis and primarily found in the nucleus [45]. TIMM21 is involved in protein import into the mitochondrial matrix, primarily found in the TIM23 mitochondrial import inner membrane translocase complex [46]. Through GO study, genes targeting a diverse array of biological functions could be traced, which may be employed for understanding biological processes to particular stimuli or stress or phenotypes.
4. CONCLUSION
The BAC clone mapping provides a valuable source of identification of genes of interest and is also useful to detect the genes’ order and genetic markers present in the genome. Here, the study provides important information on 38 genes and 1974 SSRs present in the selected 18 clones of C. magur genome. The genes identified in 18 clones using mapping of BE sequences on scaffolds are well-described in terms of gene description, complete gene annotation, protein-protein network analysis, and synteny studies, which are useful resources for undertaking the population dynamics and structural genomics and have application in genomic selection as well as cytogenetic studies. Our study can also lead to the development of the BE sequence and BAC-wise genes database of C. magur, which will be a valuable resource for the formulation of gene-specific constructs for genetic manipulation studies.
5. ACKNOWLEDGMENTS
The present reported work was carried out by the first author at ICAR-NBFGR, Lucknow, for her Ph.D. program. The authors are thankful to the Director, ICAR-National Bureau of Fish Genetic Resources, Lucknow, for providing laboratory facilities and other necessary support to conduct the Ph.D. research work. The authors are also thankful to the DBT, New Delhi, and CABin Scheme of ICAR for providing BAC resource and financial support to the study. The BAC resource utilized here is the outcome of DBT funded project entitled “Whole genome sequencing and development of allied genomics resources in two commercially important fish- Labeo rohita and Clarias batrachus” (Sanction Order BT/PR3688/AAQ/3/571/2011 dated 10.09.2013) and the consumables and other laboratory resources were utilized from the ICAR CABin Scheme sub-project entitled “Construction of physical map of Clarias magur genome” executed at ICAR-NBFGR, Lucknow, under the “Network Project on Agricultural Bioinformatics and Computational Biology” implemented through ICAR-IASRI, New Delhi. The authors are also thankful to Integral University, Lucknow, for providing the necessary guidance and support to the first author. This manuscript is also acknowledged under Integral University Manuscript number- IU/R&D/2024- MCN0002440.
6. LIST OF ABBREVIATIONS
AA, Amino acid; AL5AP, Arachidonate 5-lipoxygenase-activating protein; BAC, Bacterial artificial chromosome; BE mapping, Bac end mapping; C. magur, Clarias magur; CTH1, mRNA decay activator protein ZFP36; D. rerio, Danio rerio; DAPI, 4′,6-diamidino-2-phenylindole; DYNLRB2, Dynein light chain roadblock-type 2; E. coli, Escherichia coli; FISH, Fluorescence in situ hybridization; GO, Gene ontology; GHRH, Growth hormone-releasing hormone; GNAO1, Guanine nucleotide-binding protein G(O) subunit alpha; GSLC7A5, Large neutral amino acids transporter small subunit 1-like; HR, Growth hormone receptor; I. punctatus, Ictalurus punctatus; Kb, Kilobase; KEGG, Kyoto encyclopedia of genes and genomes; KIF18A, Kinesin-like protein KIF18A; MFSD7, Major facilitator superfamily domain-containing protein 7; MTA, Metastasis-associated protein MTA2 isoform X1; MTA2, Metastasis-associated protein MTA2 isoform X1; NCBP1, Nuclear cap binding protein subunit 1; NJ, Neighbour joining; NOS2, Nitric oxide synthase; ODC1, ornithine decarboxylase; OSR2-A, Protein odd-skipped-related 2 isoform X1; PPI, Protein-protein interaction; RB1CC1, RB1-inducible coiled-coil protein 1; Rcf, Relative centrifugal force; RPLX, Large ribosomal subunit protein; SCLY, Selenocysteine lyase; SF, Scaffolds; SLC7A5, Large neutral amino acids transporter small subunit 1-like; SSR, Simple sequence repeats; TAF6L, TAF6-like RNA polymerase II CBP-associated factor-associated factor; TIMM21, Mitochondrial import inner membrane translocase subunit Tim21.
7. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
8. CONFLICTS OF INTEREST
The authors have declared that none of their known financial conflicts or interpersonal connections could have influenced the work presented in this paper.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. SUPPLEMENTARY MATERIAL
The supplementary material can be accessed at the journal's website: Link here [https://jabonline.in/admin/php/uploadss/1297_pdf.pdf].
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
13. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Mishra BM, Khalid MA, Labh SN. Assessment of the effect of water temperature on length gain, feed conversion ratio (FCR) and protein profile in brain of Labeo rohita (Hamilton 1822) fed Nigella sativa incorporated diets. Int J Fish Aquatic Studies 2019;7:06–13.
2. Fricke R, Eschmeyer E, Fong J. Eschmeyer’s catalog of fishes genera/species by family/subfamily. Electronic version accessed 2021;05.02.2021.
3. Vishwanath W. Clarias magur. The IUCN Red List of Threatened Species 2010; e. T168255A6470089; CrossRef
4. Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ, et al. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res 2001;11:483–96.
5. Osoegawa B, Zhu CL, Shu T, Ren Q, Cao GM, Vessere MM. BAC resources for the rat genome project. Genome Res 2004;14:780–5.
6. Kumar R, Baisvar VS, Kushwaha B, Murali S, Singh VK. Improved protocols for BAC insert DNA isolation, BAC end sequencing and FISH for construction of BAC based physical map of genes on the chromosomes. Mol Biol Rep 2020;47:2405–13.
7. Baisvar VS, Kushwaha B, Kumar R, Murali SK, Pandey M, Singh M. Chromosome analysis of walking catfish, Clarias magur (Teleostei, Siluriformes, Clariidae), using staining, FISH with 18S, 5S and BAC probes. J Appl Ichthyol 2021;37:563–71.
8. Kushwaha B, Pandey M, Das P, Joshi CG, Nagpure NS, Kumar R, et al. The genome of walking catfish Clarias magur (Hamilton, 1822) unveils the genetic basis that may have facilitated the development of environmental and terrestrial adaptation systems in air-breathing catfishes. DNA Res 2021;28:dsaa031; CrossRef
9. Banerjee B, Koner D, Hasan R, Bhattacharya S, Saha N. Transcriptome analysis reveals novel insights in air-breathing magur catfish (Clarias magur) in response to high environmental ammonia. Gene 2019;703:35–49.
10. Kumar MS, Baisvar VS, Kushwaha B, Kumar R, Singh M, Mishra AK. Thermal stress induces expression of nuclear protein and Parkin genes in endangered catfish, Clarias magur. Gene 2022;825:146388; CrossRef
11. Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics 2017;33:2583–5.
12. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res 2009;19:1639–45.
13. Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 2019;47:D419–26.
14. Kanehisa M, Sato Y. KEGG mapper for inferring cellular functions from protein sequences. Protein Sci 2020;29:28–35.
15. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870–4.
16. Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 2019;47:256–9.
17. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023;51:638–46.
18. Shannon P, Markiel A, Ozier O, Baliga NS. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504.
19. Thorsen J, Zhu B, Frengen E, Osoegawa K, de-Jong PJ, Koop BF, et al. A highly redundant BAC library of Atlantic salmon (Salmo salar): an important tool for salmon projects. BMC Genomics 2005;4:6–50.
20. Whitaker HA, McAndrew BJ, Taggart JB. Construction and characterization of a BAC library for the European sea bass Dicentrarchus labrax. Anim Genet 2006;37:526.
21. Wang CM, Zhu ZY, Lo LC, Feng F, Lin G. A microsatellite linkage map of Barramundi (Lates calcarifer). Genetics 2007;175:907–15.
22. Wang CM, Lo LC, Feng F, Gong P, Li J, Zhu ZY, et al. Construction of a BAC library and mapping BAC clones to the linkage map of Barramundi, Lates calcarifer. BMC Genomics 2008;25:139–52.
23. Detrich HW, Stuart A, Schoenborn M, Parker SK, Methé BA, Amemiya CT. Genome, enablement of the notothenioidei: genome size estimates from 11 species and BAC libraries from 2 representative taxa. J ExpZool B Mol Dev E 2010;314:369–81.
24. Jang S, Liu H, Su J, Dong F, Xiong F, Liao L, et al. Construction and characterization of two bacterial artificial chromosome libraries of grass carp. Mar Biotechnol 2010;12:261–6.
25. Baisvar VS, Kushwaha B, Kumar R, Kumar MS, Singh M, Rai A, et al. BAC-FISH based physical map of endangered catfish Clarias magur for chromosome cataloguing and gene isolation through positional cloning. Int J Mol 2022;23:15958.
26. Poulsen TS, Johnsen HE, Zhao S, Stodolsky M. BAC end sequencing. In methods in molecular biology. Bacterial artificial chromosomes, library construction, physical mapping and sequencing Humana Press Inc. Totowa 2004;55:157–61.
27. Meyers BC, Scalabrin S, Morgante M. Mapping and sequencing complex genomes: let us get physical. Nat Rev Genet 2004;5:578–89.
28. Anonymous. Annual report of the ICAR-National Bureau of fish genetic resources 2015-2016. ICAR-NBFGR, Lucknow, India, p 103, 2016.
29. Kumar R, Baisvar VS, Kushwaha B. Individual chromosome identification in Clarias magur using BAC-FISH and comparative genes analyses. Nucleus 2021;64:203–9.
30. Yang Y, Zheng L, Li J, Wen B. Identification and validation of N7-methylguanosine-associated gene NCBP1 as prognostic and prognostic immune-associated biomarkers in breast cancer patients. J Cell Mol Immunol 2024;3:1–2.
31. Takayama K, Muto A, Kikuchi Y. Leucine/glutamine and v-ATPase/lysosomal acidification via mTORC1 activation are required for position-dependent regeneration. Sci Rep 2018;8:8278.
32. Seruggia D, Oti M, Tripathi P, Canver MC, LeBlanc L, Di Giammartino DC, et al. TAF5L and TAF6L maintain self-renewal of embryonic stem cells via the MYC regulatory network. Mol Cell 2019;74(6):1148–63.e7; CrossRef
33. Song L, Yu B, Yang Y, Liang J, Zhang Y, Ding L, et al. Identification of functional cooperative mutations of GNAO1 in human acute lymphoblastic leukemia. Blood J Am Soc Hematol 2021;137(9):1181–91; CrossRef
34. Zhu T, Goh EL, Graichen R, Ling L, Lobie PE. Signal transduction via the growth hormone receptor. Cell Signal 2001;13:599–616.
35. Gomez JM, Mourot B, Fostier A, Gac FL. Growth hormone receptors in ovary and liver during gametogenesis in female rainbow trout (Oncorhynchus mykiss). Reproduction 1999;115:275–85.
36. Sakamoto T, Hirano T. Growth hormone receptors in the liver and osmoregulatory organs of rainbow trout: characterization and dynamics during adaptation to seawater. J Endocrinol 1991;130:425–33.
37. Ozaki Y, Fukada H, Tanaka H, Kagawa H, Ohta H, Adachi A, et al. Expression of growth hormone family and growth hormone receptor during early development in the Japanese eel (Anguilla japonica). Comp Biochem Physiol B Biochem Mol Biol 2006;145(1):27–34; CrossRef
38. Lam PY, Kamei CN, Mangos S, Mudumana S, Liu Y, Drummond IA. odd-skipped related 2 is required for fin chondrogenesis in zebrafish. Dev Dyn 2013;242:1284–92.
39. Seale LA, Hashimoto AC, Kurokawa S, Gilman CL, Seyedali A, Bellinger FP, et al. Disruption of the selenocysteine lyase-mediated selenium recycling pathway leads to metabolic syndrome in mice. Mol Cell Biol 2012;32:4141–54.
40. Li X, Jia S, Wang S, Wang Y, Meng A. Mta3-NuRD complex is a master regulator for initiation of primitive hematopoiesis in vertebrate embryos. Blood 2009;114:5464–72.
41. te Kronnie G, Stroband H, Schipper H, Samallo J. Zebrafish CTH1, a C3H zinc finger protein, is expressed in ovarian oocytes and embryos. Dev Genes Evol. 1999;209(7):443–6; CrossRef
42. Obika M, Ogawa H, Takahashi K, Li J, Hatipoglu OF, Cilek MZ, et al. Tumor growth inhibitory effect of ADAMTS1 is accompanied by the inhibition of tumor angiogenesis. Cancer Sci 2012;103:1889–97.
43. Ozler S, Oztas E, Guler BG, Pehlivan S, Kadioglu N, Ergin M, et al. Role of ADAMTS5 in unexplained fetal growth restriction (FGR). Fetal Pediatr Pathol 2016;35:220–30.
44. He S, Gillies JP, Zang JL, Córdoba-Beldad CM, Yamamoto I, Fujiwara Y, et al. Distinct dynein complexes defined by DYNLRB1 and DYNLRB2 regulate mitotic and male meiotic spindle bipolarity. Nat Commun 2023;14:1715.
45. Hogarty MD, Norris MD, Davis K, Liu X, Evageliou NF, Hayes CS, et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res 2008;68:9735–45.
46. Ishihara N, Mihara K. Identification of the protein import components of the rat mitochondrial inner membrane, rTIM17, rTIM23, and rTIM44. J Biochem 1998;123:722–32.