Microbial drug resistance is among the most important public health issues in recent time [1], but its implication in public health is not well emphasized in many countries [2]. Several pathogens of public health importance have developed ability to resist many drugs they were originally susceptible to [3]. This is as a result of the frequent use and in addition, abuse of antibiotics in community, hospitals, and relatedsettings [4]. Bacteria capable of resisting different antibiotics have been found in both hospital and community settings; thus, there is now no localization of reservoirs of drug resistance gene to a particular setting [5,6]. The natural driving forces responsible for the dissemination of bacteria with multi-resistance property world-wide include migrant birds, household animals, travellers, as well as movement of commercial food across the globe [7,8].
Geckos (Geckonidae) are reported as reservoirs of zoonotic bacteria and are considered an avenue for infection to humans through fecal contamination [8]. The zoonotic pathogens in geckos are mostly intestinal commensals such as non-typhoidal salmonellae, Citrobacter freundii, Shigella sonnei, Serratia marcescens, Klebsiella pneumonia, and Escherichia coli to mention a few. There are over 100 million cases of enteropathogenic illnesses reported globally each year with significant hospitalizations and mortalities [9]. Geckos constitute a great threat by the indiscriminate littering of human habitat with their droppings. Hence, the contamination of the environment and spread of medically important pathogens could be inevitable sources of infection to humans and animals. Besides, severe clinical problems associated with enteropathogenic bacteria may increase mortality rate especially in developing countries where there are limited therapeutic options for infected patients. In Nigeria only pocket of studies investigated isolation of bacteria in wall geckos [10-12] with no reference to their resistance patterns. Thus, there is a dearth in information on susceptibility pattern of gecko-associated pathogens in this region. This study was, therefore, design to investigate the diversity and antibiotic resistance pattern of medically important bacteria isolated from household gecko in Ilishan-Remo Ogun State.
2. MATERIALS AND METHODS
2.1. Study Areas
The sample collection points for this study were households in Ilishan-Remo. Ilishan-Remo is a small town located within Irepodun district in Ikenne Local Government Area, Ogun State. It is situated in South Western Nigeria within Latitude 6.8932 E and Longitude 3.7105 N.
2.2. Sample Collection and Bacterial Isolation
Wall geckos were caught from different households with nets and were sacrificed within glass jars by the help of chloroform as described elsewhere [13]. The samples were transported to the Department of Microbiology Laboratory, Babcock University, in clean polythene bags for further processing. The body surfaces of the geckos were sterilized with 70% alcohol and dissected to get the intestines. With the help of sterilized laboratory gadgets such as forceps and scissors, the intestines were aseptically obtained and transferred into test tubes containing nutrient broth (Oxoid, UK). The samples were homogenized and then incubated at 37°C for 24 h. Isolation was carried out as described elsewhere [13].
2.3. Antibiotic Susceptibility Test
The antibiogram was performed by agar diffusion and interpreted following criteria recommended by recognized body [14]. Antibiotics used were manufactured by Abtek, Biologicals Ltd which include ceftazidime (30 µg); cefuroxime (30 µg); gentamicin (10 µg); ofloxacin (5 µg); amoxicillin/clavulanate (30 µg); nitrofurantoin (30 µg); cefixime (5 µg); ciprofloxacin (5 µg); ceftriaxone (30 µg); erythromycin (5 µg); and cloxacillin (5 µg).
2.4. Molecular Characterization of Bacterial Isolates
Based on the preliminary tests (selective agar media, Gram staining, and biochemical tests), 12 bacteria were picked for sequencing. Bacterial DNA was extracted using Qick-DNA™ miniprep plus kit (Zymo research, Biolab, USA. Agarose electrophoresis was used to check the quality of the DNA before polymerase chain reaction. The bacterial 16S rRNA was amplified by PCR with forward primer 341: 5’-CCTACGGGAGGCAGCAG-3’and reverse primer R806 5’-GGACTACHVGGGTWTCTAAT-3’. Amplification reaction was achieved using GeneAmp PCR9700 system(Applied Biosystems) with the program set up: Initial denaturation at 94°C for 3 min, denaturation 94°C for 30 s, annealing 50°C for 30 s, extension 68°C for 30 s, and final extension 68°C for 5 min. The program was set for 30 cycles. The amplicons were sent to Inqaba Biotech (South Africa) for sequencing. The sequence reads trimmed and contigs assembled with help of BioEdit (version 7.2.5.0) [15]. Neighbor-Joining method was used to infer the evolutionary history [16]. Jukes-Cantor method was used to calculate the evolutionary distances [17]. Gaps and missing data were eliminated. MEGA6 was used for evolutionary analyses [18].
2.5. Data Analysis
Data (susceptibility) were analyzed descriptively using SPSS Statistics for Windows, Version 23.0 (IBM SPSS Statistics for Windows, Version 23.0. Armonk, NY: IBM Corp).
3. RESULTS AND DISCUSSION
3.1. Occurrence of Bacterial Isolates and Susceptibility Pattern
A total of 163 bacteria were obtained from 51 samples of Hemidactylus frenatus. Molecular characterization, indicated that the bacteria isolated were made up of 5 genera; Hafnia, Klebsiella, Salmonella, Enterobacter, and Cedecea [Table 1]. All the sequenced data have been deposited in GenBank under the accession numbers: MT271752-MT271763. Enterobacter species had the highest occurrence and closely, followed by Hafnia species. Cedecea species had the least occurrence. The phylogenetic relationship between the species identified and those in database is shown [Figure 1]. All the species identified fell within the same cluster with 99% identity except Enterobacter hormaechei subsp. hormaechei (73%).
Table 1: Species distribution and susceptibility pattern of bacteria obtained from intestinal track of wall geckos.
| Antibiotics | Name of bacteria and % resistance to different antibiotics | ||||
|---|---|---|---|---|---|
| Hafnia spp n=46 | Klebsiella spp n=18 | Salmonella spp n=43 | Enterobacter spp n=49 | Cedecea spp n=7 | |
| CAZ (30 µg) | 46 (100) | 18 (100) | 31 (72.1) | 49 (100) | 7 (100) |
| CRX (30 µg) | 46 (100) | 18 (100) | 43 (100) | 49 (100) | 7 (100) |
| GEN (10 µg) | 30 (65.2) | 9 (50) | 28 (65.1) | 46 (93.9) | 3 (42.9) |
| OFL (5 µg) | 46 (100) | 18 (100) | 41 (95.3) | 49 (100) | 7 (100) |
| AUG (30 µg) | 46 (100) | 18 (100) | 43 (100) | 49 (100) | 7 (100) |
| NIT (30 µg) | 46(100) | 0 | 38 (88.4) | 49 (100) | 0 |
| CXM (5 µg) | 46 (100) | 0 | 35 (81.4) | 49 (100) | 0 |
| CPR (5 µg) | 25 (54.3) | 0 | 13 (30.2) | 6 | 0 |
| CTR (30 µg) | 0 | 18 (100) | 0 | 0 | 7 (100) |
| ERY (5 µg) | 0 | 18 (100) | 0 | 0 | 7 (100) |
| CXC (5 µg) | 0 | 14 (77.8) | 0 | 0 | 7 (100) |
n: Sample size, CAZ: Ceftazidime, CRX: Cefuroxime, GEN: Gentamicin, OFL: Ofloxacin, AUG: Amoxicillin/clavulanate, NIT: Nitrofurantoin, CXM: Cefixime, CPR: Ciprofloxacin, CTR: Ceftriaxone, ERY: Erythromycin, CXC: Cloxicillin, 0: Not tested
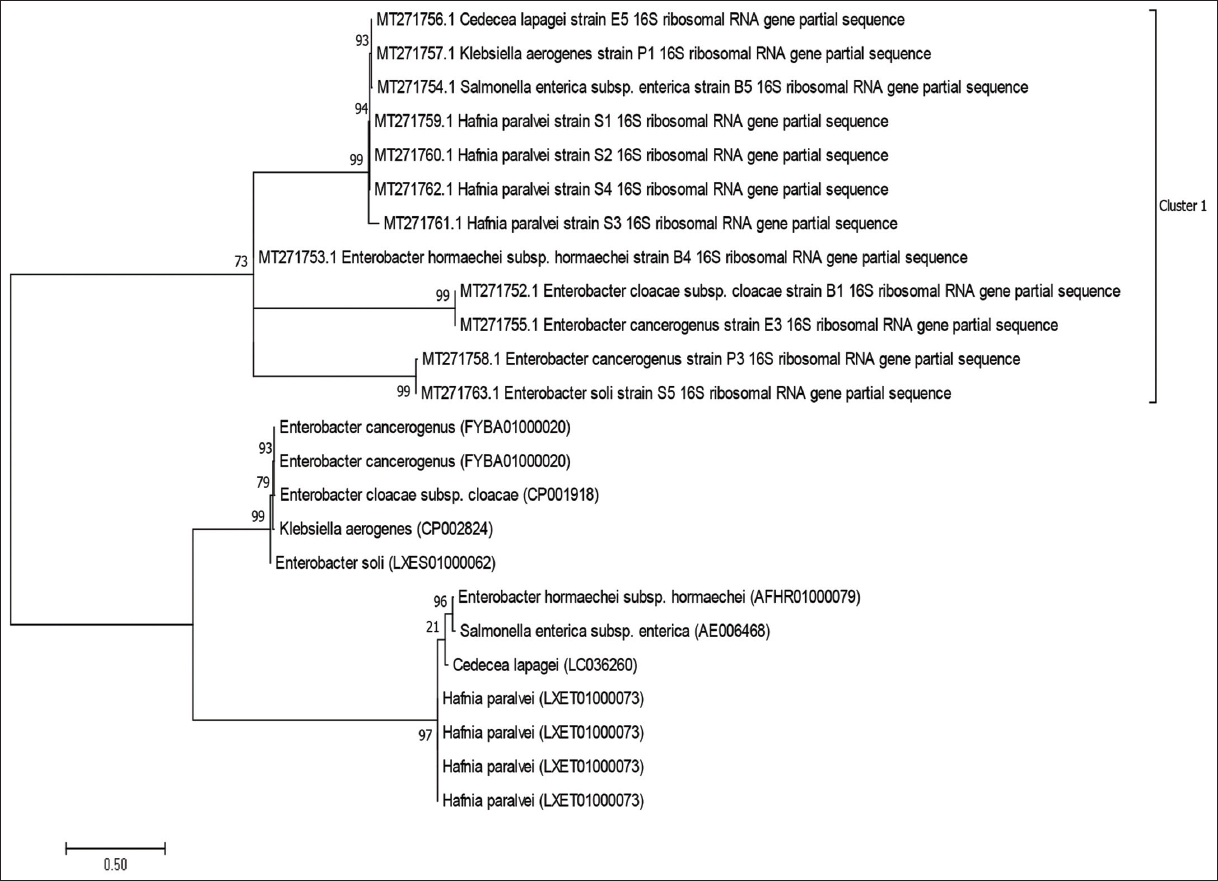 | Figure 1: Phylogenetic relationship of the species identified with close relatives in database. The evolutionary history was inferred using the Neighbor-Joining method and distances were computed using the Jukes-Cantor method. Cluster 1: Cedecea lapagei strain E5, Klebsiella aerogenes strain P1and Salmonella enterica subsp. enterica strain B5 had 93% identity, but were 94% close to Hafnia paralvei strain S1, H. paralvei strain S2, H. paralvei strain S3, and H. paralvei strain S4 (99%). Close family: Enterobacter hormaechei subsp. hormaechei strain B4, E. cloacae subsp. cloacae strain B1, Enterobacter cancerogenus strain E3, E. cancerogenus strain P3 and E. soli strain S5 were 73% identical to the aforementioned species. [Click here to view] |
The species isolated showed varying susceptibility patterns to the selected antibiotics. Hafnia species were resistant to ceftazidime, cefuroxime, ofloxacin, amoxicillin/clavulanate, nitrofurantoin, and cefixime with 65.2% (30) and 54.3% (25) of the organisms resistant to gentamicin and ciprofloxacin, respectively [Table 1]. Klebsiella species were not different in resistance pattern as they were resistant to ceftazidime, cefuroxime, ceftriaxone, erythromycin, ofloxacin, and amoxicillin/clavulanate with half of the species resistant to gentamicin [Table 1]. The trend in resistance pattern of Enterobacter and Cedecea species was similar to other species described above. Enterobacter species isolated were resistant to ceftazidime, cefuroxime, ofloxacin, amoxicillin/clavulanate, nitrofurantoin, and cefixime. Likewise, Cedecea species were resistant to ceftazidime, cefuroxime, ceftriaxone, erythromycin, cloxicillin, ofloxacin, and amoxicillin/clavulanate. More than 50% of Salmonella species were resistant to seven antibiotics tested [Appendix 1-5].
The family Enterobacteriaceae is predominantly diverse within the intestinal tract of geckos. The phylogenetic tree revealed that all the species obtained were within this family with close species having 99% identity, thus explaining the reason for one cluster group obtained. A review by Janda and Abbott [19] suggested that 16S rRNA gene sequencing provides genus identification in most cases (>90%) but less so with regard to species (65 to 83%). A more stringent boundary for species delineation was proposed to increase the accuracy of identification [20]. Pairwise nucleotide similarity values for the species in this study were within the acceptable range for species identification, thus, stressing the importance of sequencing method in microbial taxonomy.
Studies on geckos in indicated that they are reservoirs for bacteria such as Salmonella, Citrobacter, Erwinia, Shigella, Edwardsilla, Enterobacter, Serratia, Proteus, Klebsiella, and Escherichia coli [11,21]. This study has indeed strengthened the fact that geckos pose serious risk to human population especially the rural dwellers. The isolation of pathogenic bacteria in this study (Hafnia species ,Klebsiella species, Cedecea species, Salmonella species, and Enterobacter species) was consistent with previously studies on geckos and closely related species [21,22]. Food contamination due to non-typhoidal Salmonella is a significant reason for both irregular gastroenteritis and epidemic universally. In sub-Saharan Africa, community-acquired infection due to Salmonella enterica, Salmonella Typhi, and non-typhoidal Salmonella has been reported [23]. Salmonella causes intestinal-associated sicknesses such as diarrhea and vomiting with graded fever as well as life-threatening septicemia [24]. The infection is severe especially in elderly, children, and immunocompromised patients. The species of Salmonella relevant to human infections are the S. enterica, (S. ser. Typhimurium and S. ser. Enteritidis).
Antimicrobial drug resistance in Salmonella infections has been a growing concern in the treatment and management of patients. In this study, Salmonella species were multidrug-resistant (>50%) and this was comparable to 76% report by Elkenany et al. [25]. According to EU summary report on antimicrobial resistance, Salmonella species were among the multidrug-resistant zoonotic pathogens [26]. The importance of Salmonella infections cannot be overemphasized. Salmonella infection was estimated to be 185 diseases per 100,000 populace for each year in Australia [27]. Specifically, S. enterica infection results in 450 death with an estimate of 24, 000 hospitalization annually within United States [28].
The isolation of medically important bacteria from geckos such a Hafnia species is of great concern. For instance, Hafnia alvei has been implicated as cause of various diseases such as bacteremia, pneumonia, gastroenteritis, meningitis, nosocomial wound infections, endophthalmitis, and a buttock abscess [29]. Diarrhea-associated H. alvei has been reported to cause acute gastroenteritis in children [30,31]. Acute gastroenteritis and diarrhea were reported in 16% of Finnish tourists who visited Morocco [32].
The multi-drug resistance observed among the H. alvei in this study may complicate patients’ recovery leading to high mortality in the event of infection. However, previous report showed that this species were highly susceptible to most antibiotics [29]. The cause of the disparity in susceptibility pattern of this species in this study compared to the former is unknown. However, it might not be unconnected with increasing antibiotic pressure that is prevalent in the environment in recent times. It is can also be postulated that gene exchange might have been occurring among the intestinal microbiota in the host. Besides, geographical location, differential power of techniques, batches of antibiotics used and strains encountered might have contributed significantly to this variation.
Wall geckos are “innocent” in appearance, but “guilty” as they are reservoirs of zoonotic pathogens such as Enterobacter, Klebsiella, and Cedecea species. Enterobacter species causes serious infection in patients, particularly to individuals on mechanical ventilation [32]. Two species: E. hormaechei and Enterobacter cloacae are most encountered in human infections. E. cloacae was reported as the most widely recognized Enterobacter species causing nosocomial diseases and scores of data regarding their resistance to antibiotics have been highlighted [33]. The pathogenic instruments contributing the sickness related to E. cloacae are unclear. However, its capacity to form biofilms and to produce different cytotoxins is significant for its virulence potentials. While this species remain as commensal microflora in the intestinal tracts of living creatures and pathogenic in plants and creepy crawlies, notable nosocomial infections (bacteremia, endocarditis, septic joint pain, osteomyelitis, and skin/delicate tissue diseases), as well as lower respiratory tract-urinary tract and intra-abdominal infections have been reported [34]. E. cloacae in general taints different medical gadgets [35,36] and there has been its recurrent in neonatal units with a few episodes of disease reported [37,38]. The two species of Enterobacter reported in this study have been associated with infections. E. hormaechei, for example, is commonly considered a causative pathogen for human infection and it does not usually cause diseases in animals. However, it was first found to be associated with respiratory disease in unweaned calves in China as well as respiratory and blood stream infections among premature infants in intensive care nursery at the Hospital of the University of Pennsylvania [39,40].
Among the species reported in this study, Cedecea has less prevalence. However, it is an opportunistic pathogen commonly isolated from immunocompromised patients and has been linked to infections such as bacteremia, scrotal abscess, chronic renal, and heart diseases, pneumonia [41-45]. The first case fatality due to Cedecea lapagei infection was reported in a 52-year-old Mexican who developed septic shock and multiple organ failure [46].
Klebsiella species are universally widespread in nature and presumably have two regular natural surroundings (surface water, sewage, soil, and plants) and the mucosal surfaces of animals (horses, humans, and swine), for habitation [47]. Nosocomial infections associated with Klebsiella species include chronic pulmonary obstruction, neonatal sepsis, urinary tract infections, septicemia, and wound infections [47,48]. In this study, nearly all the Klebsiella species were resistant to antibiotics used. The previous cases of Klebsiella aerogenes in different hospitals were reported to be multidrug-resistant strains [49,50] and studies indicated that its resistance against carbapenem involves over-expression of AmpC or ESBLs enzymes together with mutations that disrupt membrane permeability [33].
4. CONCLUSION
This study revealed that household geckos harbor pathogenic bacteria with spectrum of resistance to different antibiotic. Hence, exposure to fecal droppings of wall geckos could be a possible source of bacterial infection that may be difficult to treat. This study recommends that general public should maintain high level of domestic hygiene to avoid contamination of food and food stuffs with fecal droppings of wall geckos.
5. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
6. FUNDING
There is no funding to report.
7. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. ETHICAL APPROVALS
Not applicable.
9. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. World Health Organization. 2014. Antimicrobial Resistance, Global Report on Survellance. Geneva: World Health Organization; 2014. Available from: http://www.apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1. [Last accessed on 2020 Apr 13].
2. Mouiche MM, Moffo F, Akoachere JT, Okah-Nnane NH, Mapiefou NP, Ndze VN, et al. Antimicrobial resistance from a one health perspective in Cameroon: A systematic review and meta-analysis. BMC Public Health 2019;19:1135. [CrossRef]
3. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mo Biol Rev 2010;74:417-33. [CrossRef]
4. Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog Glob Health 2015;109:309-18. [CrossRef]
5. Levy SB, Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat Med 2004;10:S122-9. [CrossRef]
6. Munita JM, Arias CA. Mechanism of antibiotic resistance. Microbiol Spect 2016;4:1-37. [CrossRef]
7. Vila J. Multidrug-resistant bacteria without borders: Role of international trips in the spread of multidrug-resistant bacteria. J Travel Med 2015;22:289-91. [CrossRef]
8. Damborg P, Broens EM, Chomel BB, Guenther S, Pasmans F, Wagenaar JA, et al. Bacterial zoonoses transmitted by household pets: State-of-the-art and future perspectives for targeted research and policy actions. J Comp Pathol 2016;155:S27-40. [CrossRef]
9. Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness and death in the United States. Emerg Infect Dis 1999;5:607-25. [CrossRef]
10. Oboegbulem SI, Iseghimhen AU. Wall gekos Gekonidae as reservoirs of Salmonella in Nigeria: Problem for epidemiology and public health. Int J Zoonoses 1985;12:228-32.
11. Gugnani HC, Oguike JU, Sakazaki R. Salmonellae and other enteropathogenic bacteria in the intestines of wall geckos in Nigeria. Antonie Leeuw J Microbiol 1986;52:117-20. [CrossRef]
12. Orji MU, Onuigbo HC, Mbata TI. Isolation of Salmonella from poultry droppings and other environmental sources in Awka, Nigeria. Int J Infect Dis 2005;9:86-9. [CrossRef]
13. Singh BR, Singh V, Ebibeni N, Singh RK. Maternal transfer of bacteria to eggs of common house gecko (Hemidactylus frenatus). J Microbiol Res 2014;4:78-85.
14. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard. 12th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2018. p. M02-A13.
15. Hall RM, Collis CM, Kim MJ, Partridge SR, Recchia GD, Stokes HW. Mobile gene cassettes and integrons in evolution. Ann N Y Acad Sci 1999;870:68-80. [CrossRef]
16. Saitou N, Nei M. The neighbour-joining method: A new method for reconstructing phylogenetic trees. J Mol Biol Evol 1987;4:406-25.
17. Jukes TH, Cantor CR. Evolution of protein molecules, In: Mammalian Protein Metabolism. Vol. 3. New York: H.N. Munro, Academic Press; 1969. p. 21-132. [CrossRef]
18. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. Molecular evolutionary genetics analysis version 6.0. J Mol Biol Evol 2013;30:2725-9. [CrossRef]
19. Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J Clin Microbiol 2007;45:2761-4. [CrossRef]
20. Stackebrandt E, Ebers J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006;33:152-5.
21. Singh BR, Singh V, Ebibeni N, Singh RK. Antimicrobial and herbal drug resistance in enteric bacteria isolated from faecal Droppings of common house Lizard/gecko (Hemidactylus fenatus). Int J Microbiol 2013;2013:340848. [CrossRef]
22. Ajayi JO, Ogunleye AO, Happi AN, Okunlade AO. Bacteria isolated from the oral and cloaca swabs of lizards co-habitating with poultry in some poultry farms in Ibadan, Oyo State, Nigeria. Afr J Biomed Res 2015;18:211-5.
23. Msemo OA, Mbwana J, Mahende C, Malabeja A, Gesase S, Crump JA, et al. Epidemiology and antimicrobial susceptibility of Salmonella enterica bloodstream isolates among febrile children in a rural district in Northeastern Tanzania: A cross-sectional study. Clin Infect Dis 2019;68:S177-82. [CrossRef]
24. Bjelland AM, Sandvik LM, Skarstein MM, Svendal L, Debenham JJ. Prevalence of Salmonella serovars isolated from reptiles in Norwegian zoos. Acta Vet Scand 2020;62:3. [CrossRef]
25. Elkenany R, Elsayed MM, Zakaria AI, El-sayed SA, Rizk MA. Antimicrobial resistance profiles and virulence genotyping of Salmonella enterica serovars recovered from broiler chickens and chicken carcasses in Egypt. BMC Vet Res 2019;15:124. [CrossRef]
26. European Food Safety Authority. EU summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. EFSA J 2015;13:4036. [CrossRef]
27. Kirk M, Ford L, Glass K, Hall G. Foodborne illness, Australia, circa 2000 and circa 2010. Emerg Infect Dis 2014;20:1857-64. [CrossRef]
28. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States-major pathogens. Emerg Infect Dis 2011;17:7-15. [CrossRef]
29. Giinthard H, Pennekamp A. Clinical significance of extraintestinal Hafnia alvei isolates from 61 patients and review of the literature. Clin Infect Dis 1996;22:1040-5. [CrossRef]
30. Albert MJ, Alam K, Islam M, Montanaro J, Rahman AS, Haider K, et al. Hafnia alvei, a probable cause of diarrhea in humans. Infect Immun 1991;59:1507-13. [CrossRef]
31. Albert MJ, Faruque SM, Ansaruzzaman M, Islam MM, Haider K, Alam K, et al. Sharing of virulence associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J Med Microbiol 1992;37:310-4. [CrossRef]
32. Ridell J, Siitonen A, Paulin L, Mattila L, Korkeala H, Albert MJ. Hafnia alvei in stool specimens from patients with diarrhea and healthy controls. J Clin Microbiol 1994;32:2335-7. [CrossRef]
33. Davin-Regli A, Pages JM. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 2015;6:392. [CrossRef]
34. Fata F, Chittivelu S, Tessler S, Kupper Y. Gas gangrene of the arm due to Enterobacter cloacae in a neutropenic patient. Southern Med J 1996;89:1095-6. [CrossRef]
35. Dugleux G, Le Coutour X, Hecquard C, Oblin I. Septicemia caused by contaminated parenteral nutrition pouches: The refrigerator as an unusual cause. J Parenter Enteral Nutr 1991;15:474-5. [CrossRef]
36. Wang SA, Tokars JI, Bianchine PJ, Carson LA, Arduino MJ, Smith A, et al. Enterobacter cloacae bloodstream infections traced to contamined human albumine. Clin Infect Dis 2000;30:35-40. [CrossRef]
37. Fernandez-Baca V, Ballesteros F, Hervas JA, Villalon P, Alberti S. Molecular epidemiological typing of Enterobacter cloacae isolates from a neonatal intensive care unit: three-year prospective study. J Hosp Infect 2001;49:173-82. [CrossRef]
38. Pestourie N, Garnier F, Barraud O, Bedu A, Ploy MC, Mounier M. Outbreak of AmpC b-lactamase-hyper-producing Enterobacter cloacae in a neonatal intensive care unit in a French teaching hospital. Am J Infect Control 2014;42:456-8. [CrossRef]
39. Wenger PN, Tokars JI, Brennan P, Samel C, Bland L, Miller M, et al. An outbreak of Enterobacter hormaechei infection and colonization in an intensive care nursery. Clin Infect Dis 1997;24:1243-4. [CrossRef]
40. Wang Z, Duan L, Liu F, Hu Y, Leng C, Kan Y, et al. First report of Enterobacter hormaechei with respiratory disease in calves. BMC Vet Res 2020;16:1. [CrossRef]
41. Bae BH, Sureka SB. Cedecea davisae isolated from scrotal abscess. J Urol 1983;130:148-9. [CrossRef]
42. Dalamaga M, Pantelaki M, Karmaniolas K, Matekovits A, Daskalopoulou K. Leg ulcer and bacteremia due to Cedecea davisae. Eur J Dermatol 2008;18:204-5.
43. Abate G, Qureshi S, Mazumder SA. Cedecea davisae bacteremia in a neutropenic patient with acute myeloid leukemia. J Infect 2011;63:83-5. [CrossRef]
44. Peretz A, Simsolo C, Farber E, Roth A, Brodsky D, Nakhoul F. A rare bacteremia caused by Cedecea davisae in patient with chronic renal disease. Am J Case Rep 2013;14:216-8. [CrossRef]
45. Ginn PS, Tart SB, Sharkady, SM, Thompson DK. Urinary catheter colonization by multidrug-resistant Cedecea neteri in patient with benign prostatic hyperplasia. Case Rep Infect Dis 2018;2018:7520527. [CrossRef]
46. Herrera VR, De Silva MF, Alcaraz HO, Espiritu GC, Peña KC, Melnikov V. Death related to Cedecea lapagei in a soft tissue bullae infection: A case report. J Med Case Rep 2018;12:328. [CrossRef]
47. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998;11:589-603. [CrossRef]
48. Bergogne-Berezin E. Nosocomial pathogens: New pathogens, incidence, prevention. Presse Med 1995;24:89-97.
49. Salso S, Culebras E, Andrade R, Picazo JJ. Outbreak of TEM-24-producing Enterobacter aerogenes in a Spanish hospital. J Clin Microbiol 2003;9:299-305. [CrossRef]
50. Malek A, McGlynn K, Taffner S, Fine L, Tesini B, Wang J, et al. Next-generation-sequencing-based hospital outbreak investigation yields insight into Klebsiella aerogenes population structure and determinants of carbapenem resistance and pathogenicity. Antimicrob Agents Chemother 2019;63:e02577-18. https://doi.org/10.1128/AAC.02577-18