1. INTRODUCTION
Thailand is a highly productive agricultural country, with 47% of the cultivated area comprising rice, rubber, cassava, sugarcane, and palm oil. In Phra Nakhon Si Ayutthaya Province, 89.55% of the land area is under rice cultivation, generating a large amount of rice straw as waste after harvesting [1,2]. Rice straw has high silica and lignin contents, and the C:N ratio (80:1) hampers degradation [3]. Many farmers burn rice straw in the fields as an easier and cheaper disposal option, but this negatively impacts air pollution and public health and reduces nutrients in the organic matter returned to the soil [4-6]. Rice straw is a complex natural polymer and a valuable source of lignocellulosic materials consisting of three major components: cellulose (40–50%), hemicellulose (25–30%), and lignin (15–20%) [7]. Composting rice straw is an effective alternative route for sustainable waste management in Thailand that ensures recycling of the nutrients contained in the residues and also has economic and ecological benefits [8]. Conventional composting requires a long processing period, while the introduction of potent microbial inoculum with specific functions into rice straw compost plays an important role in accelerating composting and improving the conversion of organic matter into nutrients [9,10]. The potential impact of inoculum is mostly generated by mesophilic and thermophilic lignocellulolytic microorganisms with high capacity to produce cellulase, xylanase, and ligninolytic enzymes that degrade cellulose, xylan, and lignin in the rice straw. Lignocellulolytic microorganisms with the capacity to degrade rice straw have been reported from bacteria in the genera Actinobacteria, Bacillus, Clostridium, Cellulomonas, and Pseudomonas with actinobacteria in Actinomycosis bovis, Cellulomonas flavigena, Cellulomonas fimi, Thermobifida fusca, and Xylanimonas cellulosilytica and fungi in Aspergillus niger, Cladosporium cladosporioides, Fusarium spp., Pleurotus ostreatus, Phlebia radiata, and Trichoderma reesei [11]. Bacillus pumilus B37 exhibited optimal lignocellulolytic activities and adaptation to rice straw amended medium [12]. Pleurotus ostreatus T1.1 and Penicillium sp. HC1 played a central role in cellulolytic enzyme production and the ability to use rice straw as a carbon source [13]. The microbial consortium LTF-27 composed of Alcaligenes, Clostridium, Lysinibacillus, Parabacteroides, Sphingobacterium, and uncultured bacteria efficiently degraded rice straw [14], while Firmicutes showed high efficiency in hemicellulose degradation, and Proteobacteria and Bacteroidetes exhibited cellulose and lignin degradation [15]. This is the first study to isolate indigenous mesophilic and thermophilic lignocellulolytic microorganisms from Phra Nakhon Si Ayutthaya Province. Microorganisms with high potential for the production of lignocellulolytic enzymes were screened on agar plates and selected in broth media using specific commercial and rice straw substrates. These microorganisms, belonging to Bacillus licheniformis, Streptomyces sp., Penicillium sp., and Aspergillus sp. were combined into an indigenous microbial consortium to effectively speed up the rice straw composting process. However, efficiencies in rice straw composting need further investigation in field plot experiments. Farmers must be educated about the benefits of rice straw utilization through composting as a sustainable option to avoid environmental pollution through rice straw incineration in the field.
REFERENCES
1. Suebpong P, Ekasingh B, Cramb R. Commercialisation of rice farming in Northeast Thailand. In: R. Cramb, editor. White gold: the commercialisation of rice farming in the Lower Mekong Basin. Singapore: Springer Nature Singapore Pte Ltd; 2020. [CrossRef]
2. Kongjan K, Treewannakul P, Rengkwunkway M. Management of rice stubble and straw of farmers in project of extension for stop burning at agri-area in Phra Nakhon Si Ayutthaya Province. Agricultural Sci J. 2021;52(1):20–31.
3. Kumar A, Gaind S, Nain L. Evaluation of thermophilic fungal consortium for paddy straw composting. Biodegradation. 2008;19:395–402.
4. Tipayarom D, Oanh NTK. Effects from open rice straw burning emission on air quality in the Bangkok metropolitan region. ScienceAsia. 2007;33:339–45.
5. Seglah PA, Wang Y, Wang H, Bi Y, Zhou K, Wang Y, Wang H, Feng X. Crop straw utilization and field burning in Northern region of Ghana. J Clean Prod. 2020;261:121191.
6. Chen L, Gao SYJ, Chen L, Ning H, Hu Z, Lu J, et al. Long-term straw return with reducing chemical fertilizers application improves soil nitrogen mineralization in a double rice-cropping system. Agron. 2002;12(8):1767. [CrossRef]
7. Chaurasia B. Biological pretreatment of lignocellulosic biomass (Water hyacinth) with different fungus for enzymatic hydrolysis and bio-ethanol production resource: advantages, Future work and Prospects. Acta Sci Agric. 2019;3(5):89–96.
8. Taiwo AM. Composting as a sustainable waste management technique in developing countries. J Environ Sci Technol. 2011;4(2):93–102. [CrossRef]
9. Chen Z, Xing R, Yang X, Zhao Z, Liao H, Zhou S. Enhanced in situ Pb (II) passivation by biotransformation into chloropyromorphite during sludge composting. J Hazard Mater. 2021;408:124973.
10. Pan C, Zhao Y, Zhao LI, Wu J, Zhang XU, Xie X, et al. Modified montmorillonite and illite adjusted the preference of biotic and abiotic pathways of humus formation during chicken manure composting. Bioresour Technol. 2021;319:124121. [CrossRef]
11. Benatti ALT, Polizeli MLTM. Lignocellulolytic biocatalysts: the main players involved in multiple biotechnological processes for biomass valorization. Microorganisms. 2013;11(1):162. [CrossRef]
12. Kausar H, Sariah M, Ismail MR, Saud HM, Habib SH, Berahim Z. Development of a potential lignocellulolytic resource for rapid bioconversion of rice straw. Afr J Biotechnol. 2012;11(38):9235–42. [CrossRef]
13. Pedraza-Zapata DC, Sánchez-Garibello AM, Quevedo-Hidalgo B, Moreno-Sarmiento N, Gutiérrez-Rojas I. Promising cellulolytic fungi isolates for rice straw degradation. J Microbiol. 2017;55:711–9. [CrossRef]
14. Zheng G, Yin T, Lu Z, Boboua SYB, Li J, Zhou W. Degradation of rice straw at low temperature using a novel microbial consortium LTF-27 with efficient ability. Bioresour Techno. 2020;304:123064. [CrossRef]
15. Gavande PV, Basak A, Sen S, Lepcha K, Murma N, Rai V, et al. Functional characterization of thermotolerant microbial consortium for lignocellulolytic enzymes with central role of Firmicutes in rice straw depolymerization. Sci Rep. 2021;11:3032. [CrossRef]
16. Sadhu S, Saha P, Mayilraj S, Maiti TK. Lactose-enhanced cellulase production by Microbacterium sp. isolated from fecal matter of zebra (Equus zebra). Curr Microbiol. 2011;62(3):1050–5. [CrossRef]
17. Lueangjaroenkit P, Teerapatsakul C, Chitradon L. Morphological characteristic regulation of ligninolytic enzyme produced by Trametes polyzona. Mycobiology. 2018;46(4):396–406. [CrossRef]
18. Irabor A, Ambaga MT. Evaluation of selected bacterial endophytes for biocontrol potential against Phytophthora blight of bell pepper (Capsicum Annuum L.). J Plant Pathol Microbiol. 2017;8(10):1000424. [CrossRef]
19. Singh V, Haque S, Singh H, Verma J, Vibha K, Singh R, et al. Isolation, screening, and identification of novel isolates of actinomycetes from India for antimicrobial applications. Front Microbiol. 2016;7:1921. [CrossRef]
20. Malviya MK, Pandey A, Trivedi P, Gupta G, Kumar B. Chitinolytic activity of cold tolerant antagonistic species of Streptomyces isolated from glacial sites of Indian Himalaya. Curr Microbiol. 2009;59(5):502–8. [CrossRef]
21. Jagadeesh U. and Muthuraju R. Isolation and screening of potential lignocellulolytic microorganisms from different ecosystems. Mysore J Agric Sci. 2022;56(4):201–8.
22. Photphisutthiphong Y, Vatanyoopaisarn S. Dyadobacter and Sphingobacterium isolated from herbivore manure in Thailand and their cellulolytic activity in various organic waste substrates. Agr Nat Resour. 2019;53(2):89–98.
23. Harnvoravongchai P, Singwisut R, Ounjai P, Aroonnual A, Kosiyachinda P, Janvilisri T, et al. Isolation and characterization of thermophilic cellulose and hemicellulose degrading bacterium, Thermoanaerobacterium sp. R63 from tropical dry deciduous forest soil. PLoS One. 2020;15(7):e0236518. [CrossRef]
24. Sakpetch P, H-Kittikun A, Chandumpai A. Isolation and screening of potential lignocellulolytic microorganisms from rubber bark and other agricultural residues. Walailak J Sci Technol. 2017;14(12):953–67.
25. Saini A, Aggarwal NK, Yadav A. Cellulolytic potential of actinomycetes isolated from different habitats. Bioeng Biosci. 2016;4(5):88–94. [CrossRef]
26. Zyani M, Mortabit D, Mostakim M, Iraqui M, Haggoud A, Ettayebi A, et al. Cellulolytic potential of fungi in wood degradation from an old house at the Medina of Fez. Ann Microbiol. 2009;59:699–704.
27. Bhardwaj N, Kumar B, Agrawal K, Verma P. Current perspective on production and applications of microbial cellulases: a review. Bioresour Bioprocess. 2021;8:95. [CrossRef]
28. Do TH, Dao TK, Nguyen KHV, Le NG, Nguyen TMP, Le TL, et al. Metagenomic analysis of bacterial community structure and diversity of lignocellulolytic bacteria in Vietnamese native goat rumen. Asian Austral J Anim Sci. 2018;31(5):738–47. [CrossRef]
29. Huang J, Weng L, Zhang X, Long K, An X, Bao J, et al. Trypoxylus dichotomus gut bacteria provides an effective system for bamboo lignocellulose degradation. Microbiol Spectr. 2022;10(5):e02147–22. [CrossRef]
30. Sasmitaloka KS, Arif AB, Juniawati, Winarti C, Hayuningtyas M, Ratnaningsih, et al. Xylan production from corn cobs for isolation of xylanase-producing bacteria. IOP Conf Ser Earth Environ Sci. 2019;309:012066. [CrossRef]
31. Niwane S, Lal R, Kuhad RC. Isolation of three xylanase-producing strains of actinomycetes and their identification using molecular methods. Curr Microbiol. 2006;53(3):178–82. [CrossRef]
32. Sunna A, Antranikian G. Xylanolytic enzymes from fungi and bacteria. Crit Rev Biotechnol. 1997;17(1):39–67. [CrossRef]
33. Saini S, Sharma KK. Fungal lignocellulolytic enzymes and lignocellulose: a critical review on their contribution to multiproduct biorefinery and global biofuel research. Int J Biol Macromol. 2021;15(193):2304–19. [CrossRef]
34. Sethi S, Datta A, Gupta BL, Gupta S. Optimization of cellulase production from bacteria isolated from soil. ISRN Biotechnol. 2013;19:985685. [CrossRef]
35. Shakoori FR, Shokat M, Saleem F, Riaz T. Screening, optimization and characterization of xylanase by locally isolated bacteria. Punjab Uni J of Zool. 2015;30(2):65–71.
36. Oliveira PL, Duarte MCT, Ponezi AN, Durrant LR. Purification and partial characterization of manganese peroxidase from Bacillus pumilus and Paenibacillus sp. Braz J Microbiol. 2009;40(4):818–26. [CrossRef]
37. Irfan M, Mushtaq Q, Tabssum F, Shakir HA, Qasi JI. Carboxymethyl cellulase production optimization from newly isolated thermophilic Bacillus subtilis K-18 for saccharification using response surface methodology. AMB Express. 2017;7:29. [CrossRef]
38. Joshi JB, Priyadharshini R, Uthandi S. Glycosyl hydrolase 11 (xynA) gene with xylanase activity from thermophilic bacteria isolated from thermal springs. Microb Cell Factories. 2022;21:62. [CrossRef]
39. Mingardon F, Bagert JD, Maisonnier C, Trudeau DL, Arnold FH. Comparison of family 9 cellulases from mesophilic and thermophilic bacteria. Appl Environ Microbiol. 2011;77(4):1436–42. [CrossRef]
40. Kulkarni CP, Maurya CB. Characterization of the cellulase enzyme produced by actinomycetes isolated from the mangrove coastal areas. Biosci Biotechnol Res Asia. 2017;14(2):685–90. [CrossRef]
41. Niladevi KN, Prema P. Mangrove actinomycetes as the source of ligninolytic enzymes. Actinomycetologica. 2005;19(2):40–7.
42. Chaudhary N, Prabhu S. Thermophilic actinomycetes from hot water spring capable of producing enzymes of industrial importance. Int J of Res Stud Biosci. 2016;4(6):29–35.
43. Taha N, Kadali KK, AL-Hothaly K, Smith AT, Ball AS, Adetutu EM. An effective microplate method (Biolog MT2) for screening native lignocellulosic-straw-degrading bacteria. Ann Microbiol. 2015;65:2053. [CrossRef]
44. Li JX, Zhang F, Jiang DD, Li J, Wang FL, Zhang Z, et al. Diversity of cellulase-producing filamentous fungi from Tibet and transcriptomic analysis of a superior cellulase producer Trichoderma harzianum LZ117. Front Microbiol. 2020;11:1617. [CrossRef]
45. Agrawal N, Verma P, Singh RS, Shahi SK. Ligninolytic enzyme production by white rot fungi Podoscypha elegans strain FTG4. Int J Curr Microbiol Appl Sci. 2017;6(5):2757–64.
46. Zabed HM, Akter S, Yun J, Zhang G, Awad FN, Qi X, Sahu JN. Recent advances in biological pretreatment of microalgae and lignocellulosic biomass for biofuel production. Renew Sust Energ Rev. 2019;105:105–28. [CrossRef]
47. Melati R, Shimizu FL, Oliveira G, Pagnocca F, Souza W, Sant’Anna C, et al. Key factors affecting the recalcitrance and conversion process of biomass. Bioenergy Res. 2019;12:1–20. [CrossRef]
48. Mohamed AH, Youseif SH, El-Mageed FHB, Heikal NZ, Moussa TAA, Saleh SA. Production of cellulase, exoglucanase and xylanase by different microorganisms cultivated on agricultural wastes. Res J of Pharm Biol Chem Sci. 2017;8(4):435–52.
49. Li Y, Zhang X, Xiong L, Mehmood MA, Zhao X, Bai F. On-site cellulase production and efficient saccharification of corn stover employing cbh2 overexpressing Trichoderma reesei with novel induction system. Bioresour Technol. 2017;238:643–9. [CrossRef]
50. Chukwuma OB, Rafatullah M, Tajarudin HA, Ismail N. Lignocellulolytic enzymes in biotechnological and industrial processes: a review. Sustainability. 2020;12(18):7282. [CrossRef]
51. Abo-State MAM, Ghaly MF, Abdellah EM. Production of cellulases and xylanase by thermophilic and alkaliphilic bacterial strains isolated from agricultural wastes. World Appl Sci J. 2013;22(11):1603–12. [CrossRef]
52. Irdawati I, Sofiyyana A, Advinda L, Fiffendy M, Salvia S, Syamsuardi S, et al. Optimization of agricultural waste substrate as an alternative medium for xylan in producing xylanase enzymes by thermophilic bacteria. J Phys Conf Ser. 2021;1940:012052. [CrossRef]
53. Kumar B, Bhardwaj N, Alam A, Agrawal K, Prasad H, Verma P. Production, purification and characterization of an acid/alkali and thermotolerant cellulase from Schizophyllum commune NAIMCC-F-03379 and its application in hydrolysis of lignocellulosic wastes. AMB Express. 2018;8(1):173. [CrossRef]
54. Silva MLC, Souza VB, Santos VS, Kamida, HM, Vasconcellos-Neto JRT, Góes-Neto A, et al. Production of manganese peroxidase by Trametes villosa on unexpensive substrate and its application in the removal of lignin from agricultural wastes. Adv Biosci Biotechnol. 2014;5(14):1067–77. [CrossRef]
55. Nahrowi M, Susilowati A, Setyaningsih R. Screening and identification of lignolytic bacteria from the forest at Eastern slope of Lawu mountain. AIP Conf Proc. 2002;1:020042. [CrossRef]
56. Bettache A, Azzouz Z, Boucherba N, Bouiche C, Hamma S, Maibeche R, et al. Lignocellulosic biomass and cellulolytic enzymes of actinobacteria. Scholarena J Biotechnol. 2018;5(2):203.
57. Song J, Weon HY, Yoon SH, Park DS, Go SJ, Suh JW. Phylogenetic diversity of thermophilic actinomycetes and Thermoactinomyces spp. isolated from mushroom composts in Korea based on 16S rRNA gene sequence analysis. FEMS Microbiol Lett. 2001;202(1):97–102. [CrossRef]
58. Hassine M, Aydi-Ben-Abdallah R, Jabnoun-Khireddine H, Daami-Remadi M. Soil-borne and compost-borne Penicillium sp. and Gliocladium spp. as potential microbial biocontrol agents for the suppression of anthracnose-induced decay on tomato fruits. Egypt J Biol Pest Control. 2022;32:20. [CrossRef]
59. Haas D, Lesch S, Buzina W, Galler H, Gutschi AM, Habib J, et al. Culturable fungi in potting soils and compost. Med Mycol J. 2016;54(8):825–34. [CrossRef]
60. Xie S, Lai Z, Xia H, Tang M, Lai J, Liu Q, et al. A case report of brainstem hemorrhage due to Rhizopus delemar-induced encephalitis diagnosed by metagenomic next-generation sequencing (mNGS). BMC Infect Dis. 2023;23:235. [CrossRef]
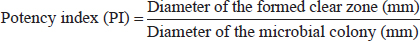
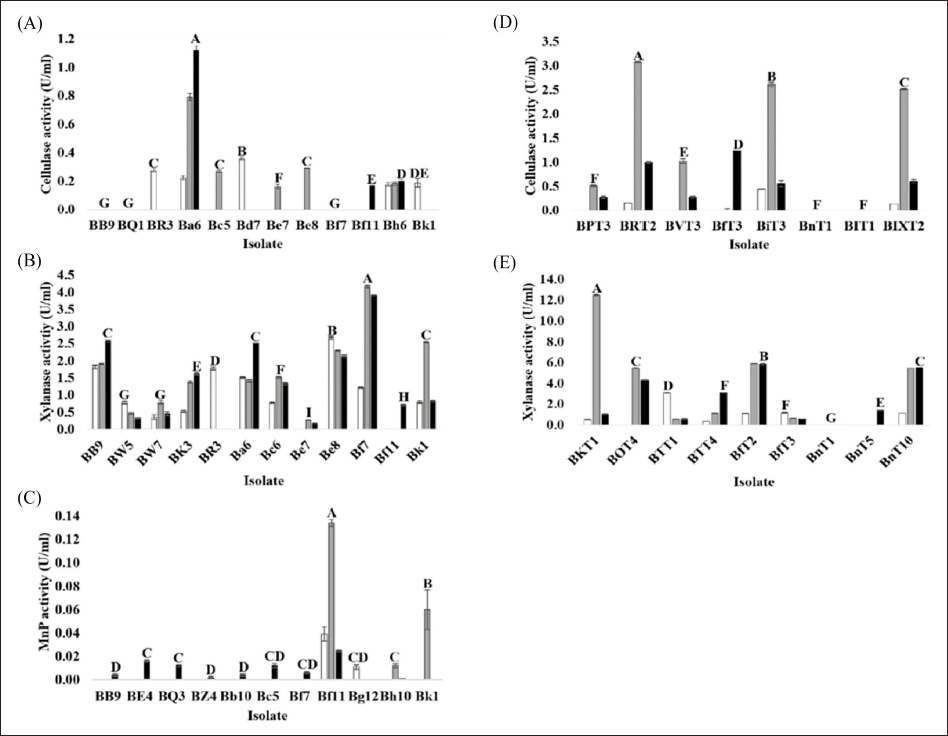
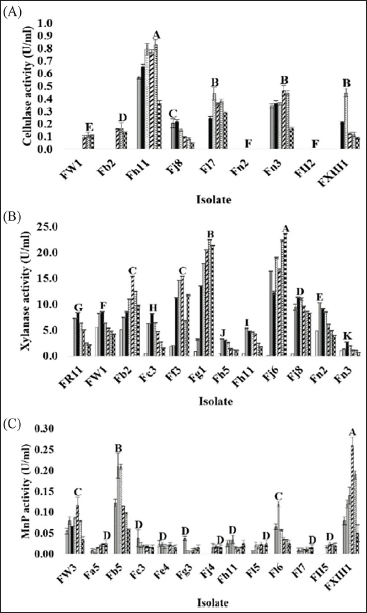
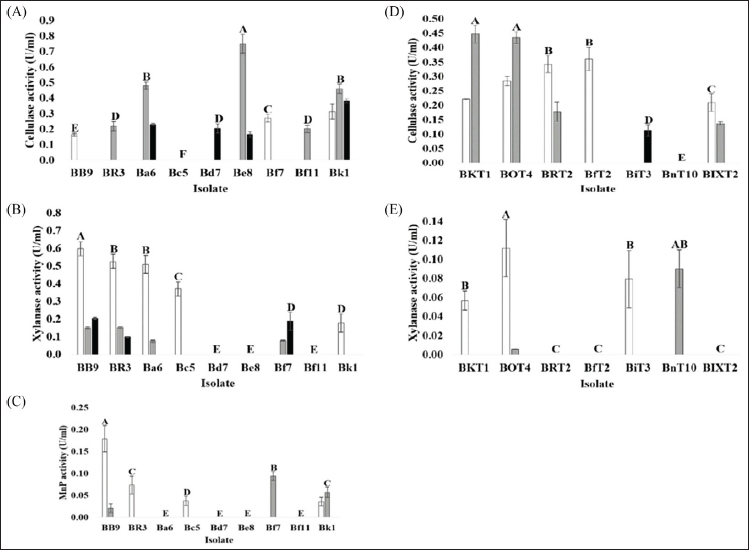
 ), 2 (
), 2 ( ), and 3 (
), and 3 ( ) using a commercial substrate for cultivation.
) using a commercial substrate for cultivation.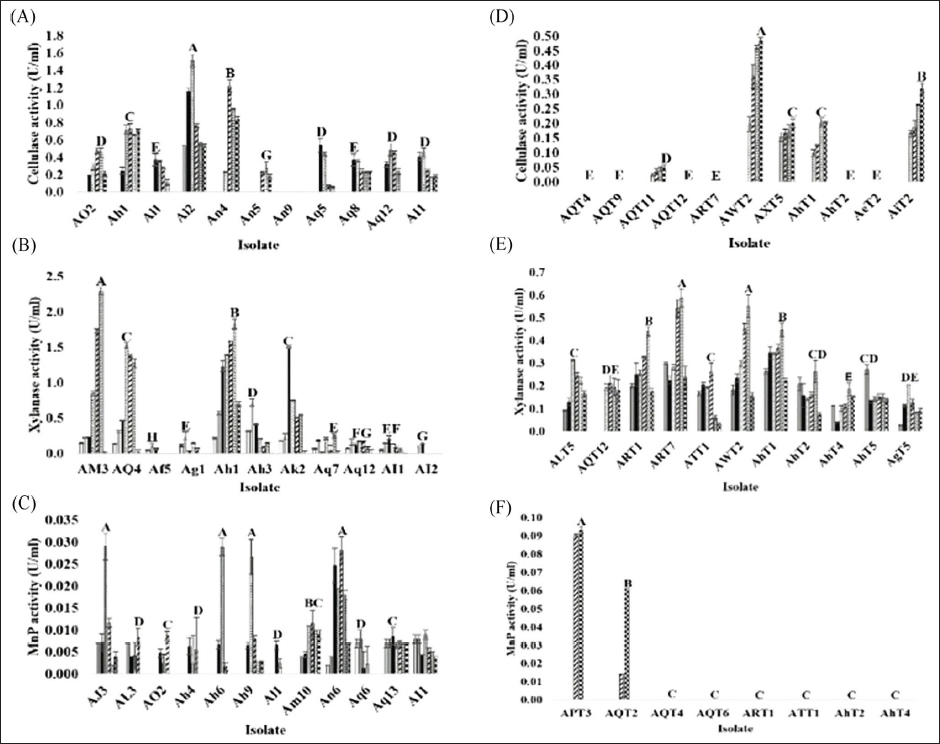
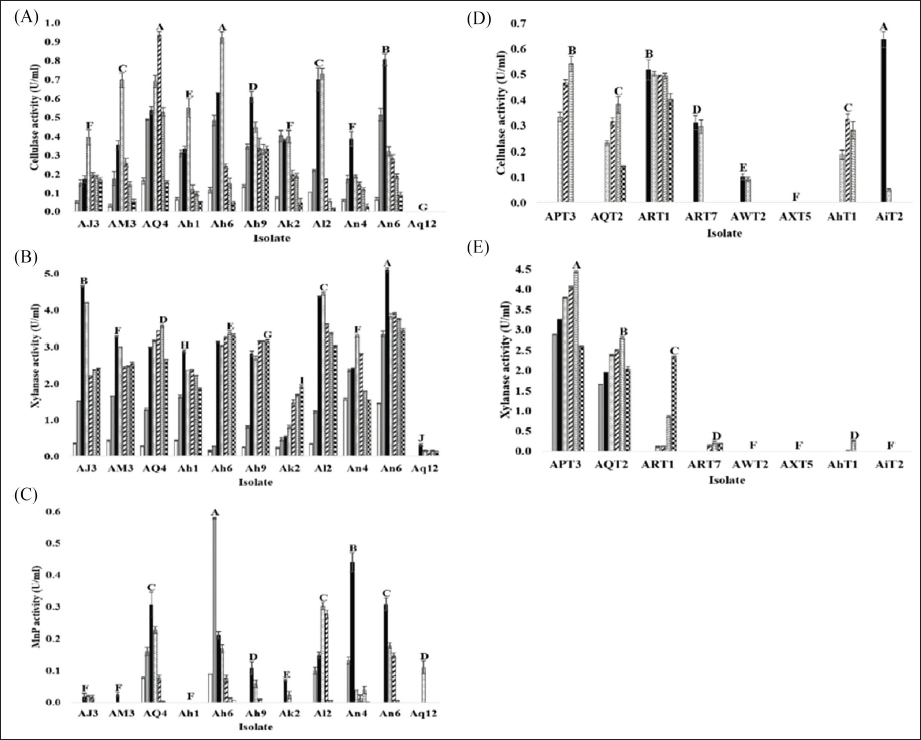
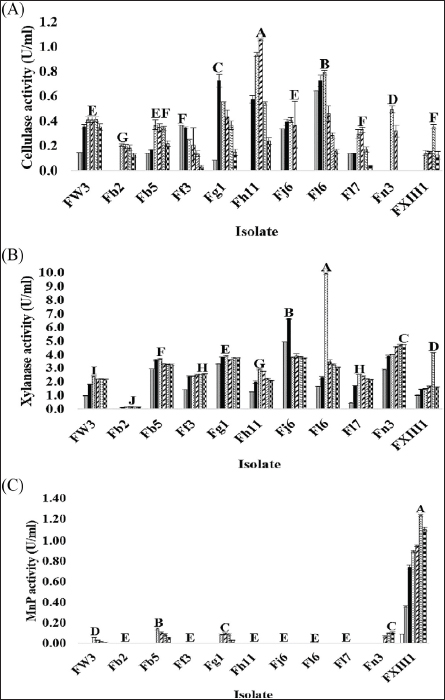
 ), 5 (
), 5 ( ), 6 (
), 6 ( ), and 7 (
), and 7 ( ) using a commercial substrate for cultivation.
) using a commercial substrate for cultivation.