1. INTRODUCTION
Dendrobium with over 1500 species present in the eastern and southeastern countries of the world is the second-largest genus of the orchid family [1]. Since that genus is broad, biologically diversified, and taxonomically complicated, various scientists have divided it into many sections and sub-sections at different times [2]. There are 124 different Dendrobium species in India, making it the country’s second-biggest orchid genus [3]. The widespread distribution, enormous diversity, and great commercial and horticultural importance of Dendrobium species and hybrids all contribute to the plant’s widespread fascination [4]. On the other hand, modern research shows that polysaccharides, alkaloids, amino acids, trace elements, and other active ingredients found in Dendrobium are pharmaceutically important [5]. Due to the huge diversity at different levels of expression, nowadays Dendrobium is considered a model for the study of biodiversity in situ and for biotechnological improvement [6]. Based on morphological important features, Dendrobium species variability and diversity were widely examined. However, the morphological characters are insufficient to clearly separate individuals. Therefore, the best way to measure the variability and diversity within a genus or a population is a resolution by molecular marker, particularly DNA marker. One notable benefit of employing a molecular marker lies in its exceptional purity, as it is unaffected by environmental factors. At present, there exists a wide array of DNA molecular markers. Among these markers, the internal transcribed spacer (ITS) of nuclear rDNA is commonly employed as a molecular tool. This is due to its ability to offer a substantial amount of information for systematic classification [7]. In addition, it is easy to analyze current variations within and among species.
Clusters of hundreds or thousands of tandemly repeated copies of the genes for plant major ribosomal RNA (rRNA) are found on nuclear ribosomal DNA (nrDNA) in the cell nucleus. The ITS sequences are the most important segments of nrDNA and can be used for the identification of biological organisms in lower taxonomic hierarchical levels [8]. The 18S-5.8S and 5.8S-28S regions of ribosomal DNA make up ITS-I and ITS-II, respectively, which are shorter than 300 base pairs. During transcription of rRNA genes, ITS-I and ITS-II are transcribed as RNA transcripts, but they are neither translated as protein nor incorporated as a structural component of the ribosome, so it is free to vary. They seem to be involved in the development of nuclear rRNAs by placing the large and tiny components of the RNAs close together in a processing area [9]. Clements (2003) utilize ITS to do an evolutionary study on the subtribe Dendrobiinae. Numerous studies have used the ITS conserved sequence to characterize and establish the genetic relationships between diverse Dendrobium species [10-15]. Taken together, these results point to a complicated and very diverse range of genetic history at the species level in Dendrobium spp. This article shows the findings of a study of nrDNA sequences from 44 species of Eastern Himalayan Dendrobium. The sequences cover the 5.8S segment and two gaps on either side which are ITS-I and ITS-II. Based on the conserved sequences, the variability of sizes and sequences in a base pair, the percentage of alignment, and the G+C content of the conserved genome of Dendrobium species were studied and usefully exploited to correlate such findings to understand their genetic or phylogenetic relatedness and the probable cladistic position within the selected species of that genus.
2. MATERIALS AND METHODS
Forty-four different epiphytic species of Dendrobium were collected during their new growth time, i.e., sprouting of new and fresh leaves taken from the different elevations of Darjeeling, Sikkim, Arunachal Pradesh, Assam, and Manipur Hills of the Indian part of Eastern Himalaya. The 44 members are derived from nine sections, i.e., Dendrobium (21) Grastidium (1), Breviflores (2), Densiflorum (6), Holochrysa (5), Stachyobium (2), Calcarifera (1), Formosae (5), and Aporum (1) [Table 1]. Bulbophyllum inunctum and Bulbophyllum macranthum xanthine dehydrogenase gene sequences were collected from the gene bank and utilized as outgroups [16] in the analyses. Gene bank reference numbers of the ITS regions of each specimen studied here except the out-group used in the analyses are the outcome of our research work.
Table 1: Sizes (bp), and G+C% of ITS-I, ITS-II (individually), and cumulative ITS-I, 5.8s, and ITS-II of 44 Dendrobium species of Eastern Himalaya.
| S. No. | Accession No. | Name of Plant | Sections | Size (bp) | G+C % | ||||
|---|---|---|---|---|---|---|---|---|---|
| ITS I | ITS II | ITS I-5.8s-ITS II | ITS I | ITS II | ITS I-5.8s-ITS II | ||||
| 1. | KX600499 | Dendrobium aduncum Lindl. | Breviflores | 232 | 242 | 637 | 51.72 | 52.89 | 53.85 |
| 2. | KX600515 | Dendrobium amoenum Wall. ex Lindl. | Dendrobium | 229 | 246 | 638 | 51.96 | 54.47 | 54.7 |
| 3. | KX600514 | Dendrobium aphyllum R. Brown. | Dendrobium | 231 | 244 | 638 | 45.89 | 52.05 | 50.94 |
| 4. | KX600501 | Dendrobium bellatulum Rolfe. | Formosae | 230 | 247 | 640 | 53.04 | 52.63 | 54.37 |
| 5. | KX792018 | Dendrobium bicameratum Lindl. | Breviflores | 230 | 245 | 639 | 51.3 | 52.6 | 53.8 |
| 6. | KX522648 | Dendrobium capillipes Rchb. f. | Holochrysa | 228 | 247 | 638 | 50.88 | 49.39 | 51.72 |
| 7. | KX792016 | Dendrobium cathcartii (Hook. f.) M.A.Clem. and D.L. Jones Dendrobium salaccense Lindl. (Synonym) | Grastidium | 228 | 246 | 637 | 52.1 | 54.4 | 54.7 |
| 8. | KX522638 | Dendrobium chrysanthum Wallich ex Lindley. | Dendrobium | 233 | 241 | 638 | 51.50 | 52.70 | 53.60 |
| 9. | KX522645 | Dendrobium chrysotoxum Lindley. | Densiflorum | 231 | 245 | 640 | 52.81 | 55.10 | 55.31 |
| 10. | KX522644 | Dendrobium crepidatum Lindl. | Dendrobium | 225 | 244 | 632 | 48.44 | 50.41 | 51.42 |
| 11. | KX600510 | Dendrobium cumulatum Lindl. | Calcarifera | 230 | 237 | 631 | 53.48 | 52.32 | 53.72 |
| 12. | KX600511 | Dendrobium denneanum Kerr. Dendrobium clavatum Wall. ex Lindl.(Synonym) | Holochrysa | 233 | 244 | 640 | 49.78 | 50.82 | 52.34 |
| 13 | KX522633 | Dendrobium densiflorum Lindl. ex Wall. | Densiflorum | 232 | 247 | 643 | 50.00 | 52.23 | 53.18 |
| 14. | KX600504 | Dendrobium denudans D. Don. | Stachyobium | 232 | 246 | 641 | 43.53 | 46.75 | 48.05 |
| 15. | KX522643 | Dendrobium devonianum Paxton. | Dendrobium | 233 | 244 | 641 | 44.63 | 49.18 | 49.76 |
| 16. | KX792014 | Dendrobium draconis Rchb. f. | Formosae | 229 | 247 | 640 | 52.4 | 51.0 | 53.0 |
| 17. | KX600498 | Dendrobium falconeri Hook. f. | Dendrobium | 231 | 245 | 639 | 48.48 | 51.02 | 51.96 |
| 18. | KX600516 | Dendrobium farmeri Paxton. | Densiflorum | 229 | 248 | 643 | 47.6 | 50.4 | 50.7 |
| 19. | KX522634 | Dendrobium fimbriatum Hooker. | Holochrysa | 232 | 244 | 640 | 47.41 | 46.31 | 49.53 |
| 20. | KX522642 | Dendrobium formosum Roxb. ex Lindl. | Formosae | 230 | 244 | 638 | 53.48 | 53.69 | 54.70 |
| 21. | KX522636 | Dendrobium gibsonii Paxton. | Holochrysa | 229 | 243 | 635 | 49.78 | 51.44 | 52.44 |
| 22. | KX522646 | Dendrobium gratiosissimum Rchb. f. | Dendrobium | 229 | 246 | 639 | 54.58 | 54.88 | 55.87 |
| 23. | KX600513 | Dendrobium heterocarpum Wall. ex Lindl. | Dendrobium | 232 | 242 | 637 | 51.29 | 50.41 | 52.75 |
| 24. | KX509992 | Dendrobium hookerianum Lindl. | Dendrobium | 216 | 244 | 623 | 48.61 | 50.82 | 52.01 |
| 25. | KX600506 | Dendrobium jenkinsii Wall. ex Lindl. | Densiflorum | 234 | 246 | 643 | 46.58 | 48.37 | 50.23 |
| 26. | KX522640 | Dendrobium lindleyi Steud. | Densiflorum | 228 | 247 | 638 | 48.68 | 48.58 | 51.10 |
| 27. | KX792015 | Dendrobium lituiflorum Lindl. | Dendrobium | 233 | 243 | 639 | 52.7 | 51.4 | 53.9 |
| 28. | KX509991 | Dendrobium longicornu Lindl. | Formosae | 230 | 247 | 640 | 53.48 | 52.23 | 54.22 |
| 29. | KX792017 | Dendrobium moniliforme (L.) Sw. Dendrobium candidum Wall. ex Lindl.(Synonym) | Dendrobium | 233 | 244 | 640 | 52.7 | 51.6 | 53.9 |
| 30. | KX522635 | Dendrobium moschatum Sw. | Holochrysa | 232 | 244 | 640 | 47.41 | 46.31 | 49.53 |
| 31. | KX600497 | Dendrobium nobile Lindl. | Dendrobium | 227 | 246 | 636 | 53.3 | 55.28 | 55.5 |
| 32. | KX509993 | Dendrobium nobile var. pendulum | Dendrobium | 231 | 242 | 636 | 50.22 | 52.89 | 52.99 |
| 33. | KX495131 | Dendrobium nobile var. varginalis | Dendrobium | 231 | 242 | 636 | 49.78 | 52.89 | 52.83 |
| 34. | KX522637 | Dendrobium ochreatum Lindl. | Dendrobium | 231 | 244 | 639 | 45.45 | 52.87 | 51.33 |
| 35. | KX522639 | Dendrobium parishii Rchb. f. | Dendrobium | 229 | 244 | 637 | 46.72 | 54.51 | 52.75 |
| 36. | KX600502 | Dendrobium pendulum Roxb. | Dendrobium | 228 | 246 | 637 | 49.56 | 52.84 | 52.9 |
| 37. | KX600505 | Dendrobium porphyrochilum Lindl. | Stachyobium | 231 | 246 | 640 | 44.15 | 44.31 | 47.19 |
| 38. | KX522641 | Dendrobium primulinum Lindley. | Dendrobium | 231 | 244 | 639 | 45.45 | 52.87 | 51.33 |
| 39. | KX600507 | Dendrobium ruckeri Lindl. | Dendrobium | 230 | 246 | 639 | 52.61 | 54.88 | 55.09 |
| 40. | KX522647 | Dendrobium terminale Par. and Rchb. f. | Aporum | 232 | 248 | 644 | 48.71 | 53.63 | 52.95 |
| 41. | KX600503 | Dendrobium thyrsiflorum Rchb. f. | Densiflorum | 231 | 247 | 641 | 49.35 | 53.44 | 52.89 |
| 42. | KX600508 | Dendrobium transparens Wall. | Dendrobium | 228 | 246 | 637 | 53.07 | 52.84 | 54.47 |
| 43. | KX600500 | Dendrobium wardianum Warner. | Dendrobium | 231 | 242 | 636 | 49.78 | 52.89 | 52.83 |
| 44. | KX522632 | Dendrobium williamsonii J. Day and Rchb. f. | Formosae | 229 | 243 | 635 | 51.96 | 52.26 | 53.38 |
2.1. Genomic DNA Isolation
0.260 mg of young and fresh leaf tissue were collected in triplicate and ground with mortar and pestle. The hexadecyl trimethyl ammonium bromide procedure was used to extract DNA from the tender leaves [17].
2.2. Primer Designing and Polymerase Chain Reaction (PCR) Amplification
The quality of the genomic DNA was analyzed by injecting it into an agarose gel with a concentration of 1.2%. PCR amplification with two bilateral degenerate primers was used to amplify the whole nrDNA ITS region from each species’ genomic DNA. Both the forward and reverse primer sequences are as follows 5’-GGAAGGAGAAGKCGKARCWASG-3’ and 5’-TCCTCCGCTTATWGRTMYKC-3’ for ITS5 and ITS4, respectively. Every cycle of PCR requires separation of the template DNA (94°C), annealing of the primers (52°C), and finally, extension of primers (72°C) to continue up to 35 cycles. The solution mixture for PCR amplification included genomic DNA (1 μL), ITS forward and reverse primers (200 ng each), and dNTPs (2.5 mM each). For the reaction, add 2 μL, ×10 Taq DNA polymerase assay buffer (0.5 μL), Taq DNA polymerase enzyme (3 U/μl) 0.5 μL, water was added, and adjust the total volume to 50 μL. Quality was checked by loading PCR products on 1.2% agarose gel [Figure 1] where the first (L1) is 100 bp and the last lanes (L2) of the gel have 1Kb DNA ladders, and finally bi-directionally sequenced the PCR products.
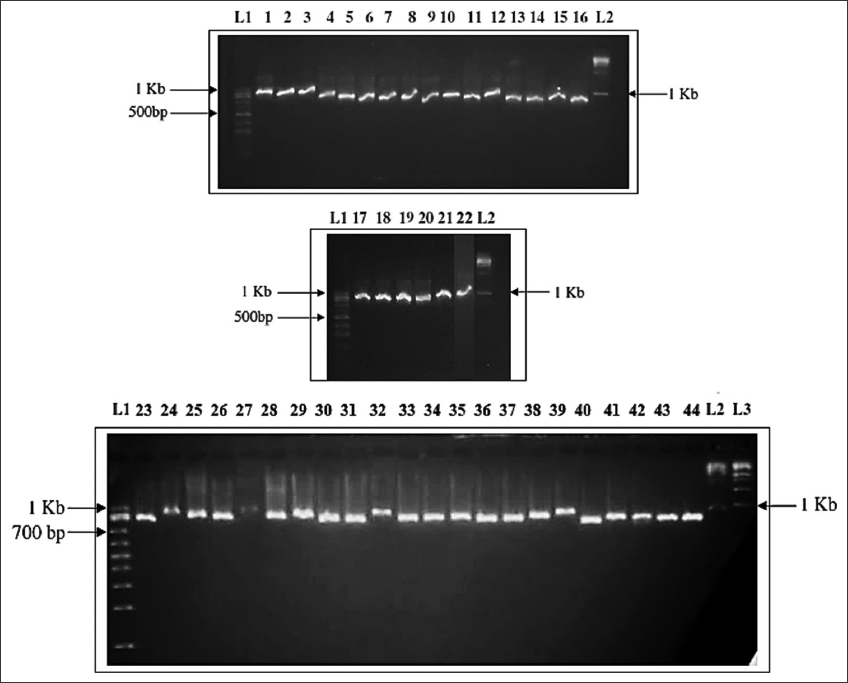 | Figure 1: nrDNA internal transcribed spacer polymerase chain reaction region amplification results of Dendrobium spp. 1. Dendrobium nobile var. varginalis, 2. Dendrobium longicornu, 3. Dendrobium hookerianum, 4. Dendrobium nobile var. pendulum, 5. Dendrobium draconis, 6. Dendrobium williamsonii, 7. Dendrobium densiflorum, 8. Dendrobium fimbriatum, 9. Dendrobium moschatum, 10. Dendrobium gibsonii, 11. Dendrobium ochreatum, 12. Dendrobium chrysanthum, 13. Dendrobium parishii, 14. Dendrobium lituiflorum, 15. Dendrobium primulinum, 16. Dendrobium formosum, 17. Dendrobium devonianum, 18. Dendrobium crepidatum, 19. Dendrobium chrysotoxum, 20. Dendrobium gratiosissimum, 21. Dendrobium terminale, 22. Dendrobium capillipes, 23. Dendrobium nobile, 24. Dendrobium falconeri, 25. Dendrobium aduncum, 26. Dendrobium wardianum, 27. Dendrobium bellatulum, 28. Dendrobium cathcartii, 29. Dendrobium thyrsiflorum, 30. Dendrobium denudans, 31. Dendrobium porphyrochilum, 32. Dendrobium jenkinsii, 33. Dendrobium moniliforme, 34. Dendrobium transparens, 35. Dendrobium lindleyi, 36. Dendrobium cumulatum, 37. Dendrobium denneanum, 38. Dendrobium bicameratum, 39. Dendrobium heterocarpum, 40. Dendrobium aphyllum, 41. Dendrobium amoenum, 42. Dendrobium farmeri, 43. Dendrobium pendulum, 44. Dendrobium ruckeri. L1 = 100bp DNA ladder, L2 = 1kb DNA ladder, DNA ladder run on 1.2% gel electrophoresis. [Click here to view] |
2.3. Sequencing of DNA
The sequencing reaction mixture consisted of Bigdye terminator V.3.1. ready reaction mix (0.4 μL/μL), template (0.1 μg/μL), 2 μL primer (10 pmol/λ), and sterile water (3 μL). For further sequencing, the PCR procedures were initially denaturation at 96°C for 1 min, denaturation continued for 10 s (96°C), hybridization for 5 s (50°C), and finally, elongation was done for 4 min at 60°C. The PCR reaction was achieved as 25 thermal cycles. The PCR outcomes were sequenced (~760 bp) on an ABI 3500 XL Genetic Analyzer.
2.4. Sequencing Data Alignment, Construction of Pairwise Distance Matrix, Phylogenetic Tree, and other Statistical Analysis
The sequence data were aligned by the MUSCLE alignment algorithm. Sequence statistics, nucleotide diversity, selected model-based evolutionary analyses, construction of pairwise distance matrixes, and dendrogram preparation were conducted in MEGA 11 using the Maximum Composite Likelihood model, neighbor-joining algorithm, Tamura-Nei model-based Maximum Likelihood (ML) Technique, Kumar method, and Bayesian-based parsimony method [18]. The neutrality test statistics, specifically Fu and Li’s and Tajima’s D, were computed using the software DnaSP V.5.10.01 [19].
3. RESULTS
3.1. Nucleotide Based Diversity
Purified genomic DNA was amplified using ITS-compatible primer and the quality was checked by loading PCR sequences on 1.2% agarose gel, usually yielding a single band in all cases for each sample [Figure 1]. The ITS fragment (in-between the 18S and 28S rDNA) that was amplified includes segments of the 18S and 5.8S rDNA. The total size of the first internal transcribed spacer (ITS-I) ranged from 216 to 234 base pairs (bp), with a mean of 230.09 bp. The second internal transcribed spacer (ITS-II) had lengths ranging from 237 to 248 bp, with a mean of 244.66 bp. The 5.8S rDNA was 163 bp in length, and this length was shown to be stable and conserved across all species [Table 1]. ITS-I (G+C %) ranged from 43.53 to 54.58 across taxa. ITS-I has an average G+C content of 50.05%. ITS-II had a mean G+C content of 52.11% and a range of 44.31% to 55.28%. The ITS-II region has a comparatively higher percentage (G+C) than the ITS-I region [Table 1] with an overall composition distance of GC% 52.82 [Supplementary Table 1]. Each Dendrobium species was discovered to have a distinct sequence in the ITS-I and ITS-II regions, allowing for easy distinction between them genetically, as this data shows that interspecific DNA sequence variation is relatively strong. Dendrobium is a molecularly natural taxon since the lengths of the ITS-I to ITS-II sections in the 44 Dendrobium species ranged from 623 to 644 bp with an average of 637.7 bp, which is in agreement with earlier research [Supplementary Table 1]. All selected Dendrobium species have 403 polymorphic, 223 conserved, and 337 parsimony informative nucleotides. Single-ton bases number 91 and informative site substitutions average 0.51. Overall composition distance is 0.32, transition and transversion ratio 1.63, and mean evolutionary divergence (d) 0.16 [Supplementary Table 1]. In total, ML fits were performed on 24 nucleotide substitution models. The K2+G model was the only one with a minimum BIC value (13172.666) to be considered the top nucleotide substitution pattern model [Supplementary Table 2]. Molecular evolution among chosen Dendrobium species was calculated by estimating the ML substitution matrix to see the substitution probabilities that led to the most likely model of evolution [Supplementary Table 3]. A tree topology was automatically constructed for Maximum Likelihood (ML) estimation. This calculation yielded the highest log-likelihood of −8102.528. Comparisons of immediate r value (∑r = 100) were performed throughout the assessment and revealed nucleotide percentages of A = 23.70, T = 23.48, G = 29.11, and C = 23.71 [Supplementary Tables 1 and 3] and the transitions exceed transversionsal nucleotide replacements [Table 4]. The total transition/transversion bias (R) is 1.444, with pyrimidine substitution (k2) being more common than purine (k1) [Supplementary Table 1]. Using Maximum Parsimony, we were able to piece together the evolutionary past. All sites and parsimony-informative sites (enclosed in brackets) have indices of 0.5028 (0.4571), 0.6545 (0.6545), and 0.3290 (0.2992), respectively, for consistency, retention, and composite [Supplementary Table 1]. Both ML and Baseyan analysis methods yielded great congruence. Genome sequence substitution patterns were tested for homogeneity. The disparity indices range from 0.00 to 1.89 with a mean of 0.192 which quantifies the present evolutionary differences between sequences [Supplementary Table 1]. The yellow color reading indicated a significant P ≤ 0.05 rejecting the null hypothesis of homogeneous substitution [Supplementary Table 8]. Codon-based neutrality screening was performed among sequences with an estimated mean value (dN/dS) −0.33 [Supplementary Table 1]. As shown in Supplementary Table 9, there is a good chance that ruling out the possibility that dN=dS, the rigorous neutrality null hypothesis (α ≤ 0.05, marked as yellow color). In the same way, an elevated dN–dS ratio and positive readings of the statistic indicate an excessive occurrence of nucleotide base substitutions. Among the species that were selected for analysis, it was observed that the TGT triplet codon (Codon Omega reading) exhibited a notably higher dN–dS value of 4.973, indicating significant (P < 0.05) non-synonymous substitutions [Supplementary Table 10]. The codon use bias of the chosen Dendrobium genomes was determined by calculating relative synonymous codon usage (RSCU) values for 64 synonymous codons. Overrepresented (RSCU average > 1.6) was the codons UUG, CUC, AUC, GUC, UCG, UAU, CAA, GAU, and GAA whereas underrepresented (RSCU average <0.6) was the codons UUC, UUA, CUU, CUA, GUA, UCU, UCA, CCU, ACU, UAC, UAG, CAC, CAG, GAC, GAG, CGG, and AGU [Supplementary Table 11]. The other 38 codes were moderately expressed. The occurrence of triplet codes ending with A and C is more frequent compared to those ending with G and U/T. In addition, the RSCU values (3rd codon of triplet) for the codons C, A, G, and U are 17.65, 16.75, 14.93, and 14.64, respectively. Notably, the codon UCG exhibits the highest RSCU value of 2.68, whereas the codon GAC has the lowest value of 0.16. The codon with the lowest frequency seen is AGU, which occurs at a rate of 0.4. Conversely, the codon with the highest frequency observed is GGC, which occurs at a rate of 7.5 [Supplementary Table 11]. Comparing the relative fixing rates of synonymous and non-synonymous alteration helps explain molecular sequence evolution.
3.2. Comparing Evolutionary Variations
The calculation of genetic distance arises when the dissimilarity between two genes is proportional to the duration of their divergence from a shared ancestral population. The amount of substituted nucleotide per site between sequences is displayed. Calculations used the Maximum Composite Likelihood model. This investigation examined using ITS sequence of 44 Dendrobium species where minimum divergence was calculated (0.000) among Dendrobium moschatum and Dendrobium fimbriatum, Dendrobium ochreatum and Dendrobium primulinum, Dendrobium nobile var. varginalis, and Dendrobium wardianum. The maximum genetic divergence (0.276) was found between Dendrobium aphyllum and Dendrobium denudans [Supplementary Table 12]. Intra-section (within a group) and inter-section (among different groups) genetic differences can be calculated using the Jukes–Cantor pairwise genetic distance analysis algorithm. The Breviflores species group exhibits the highest level of genetic diversity (0.171), whereas the Formosae section of Dendrobiums displays the lowest level (0.045) of genetic diversity [Supplementary Table 4]. The calculation of evolutionary diversity within the taxonomic groups Aporum, Calcarifera, and Grastidium is hindered by the limited number of species available for analysis. The evolutionary distance between Dendrobium and Grastidium group sequences was the smallest (0.117), whereas the distance between Grastidium and Stachyobium group sequences was the greatest (0.285) [Supplementary Table 5]. Species belonging to the Grastidium section exhibit the highest genetic diversity when compared to the Stachyobium and the minimum when compared to the Dendrobium groups. This diversity is reflected in their distinct physical features. These features include elongated and rounded stems, thin and lens-shaped flat leaves, and short peduncles from which inflorescences emerge at leafy stem nodes found in the members of Grastidium [20].
3.3. Analyzing Phylogenetic Tree
The dendrogram [Figure 2] shows that 21 species were included in the section Dendrobium and other members belong to sections Grastidium (1), Breviflores (2), Densiflorum (6), Holochrysa (5), Stachyobium (2), Calcarifera (1), Formosae (5), Aporum (1). The gene bank supplied the outgroup species Bulbophyllum inunctum and Bulbophyllum macranthum for the studies. There are five distinct sub-clades within the Dendrobium section. The first subclade is comprised the species Dendrobium amoenum. Dendrobium nobile makes up the second subclade. The third subclade includes Dendrobium transparens, Dendrobium gratiosissimum, Dendrobium nuckeri, Dendrobium pendulum, and Dendrobium falconeri whereas Dendrobium heterocarpum, Dendrobium nobile var. pendulum, Dendrobium nobile var. varginalis, D. wardianum, D. aphyllum, Dendrobium parishi, D. ochreatum, Dendrobium primilinum, Dendrobium chrysanthum, Dendrobium crepidatum, Dendrobium devonianum, Dendrobium moniliforme, and Dendrobium lutiforum are included in the fourth subclade. The fifth subclade consists of only Dendrobium hookerianum [Figure 2]. The ML-based ancestral tree reveals the presence of four distinct subclades within the Dendrobium section. It is seen that both D. amoenum and D. nobile have a common ancestor. The shared progenitor of the Dendrobium species exhibits the nucleotide “A” in the third position of the codon, a modification that occurred in a more recent ancestor, replacing the nucleotide “C.” In ancient times, the original foundation was referred to as “A” [Supplementary Figure 1]. However, the second subclade of Dendrobium species exhibits a nucleotide substitution from “A” to “C,” which bears similarity to the molecular features observed in section Grastidium. In D. chrysanthum, a notable transition occurs in the nucleotide base from its immediate ancestor, where the base “A” undergoes an abrupt alteration to “C” [Supplementary Figure 1]. Section Grastidium contains a single species, i.e., Dendrobium cathcartii shearing the common ancestor of D. amoenum and D. nobile [Figure 2].
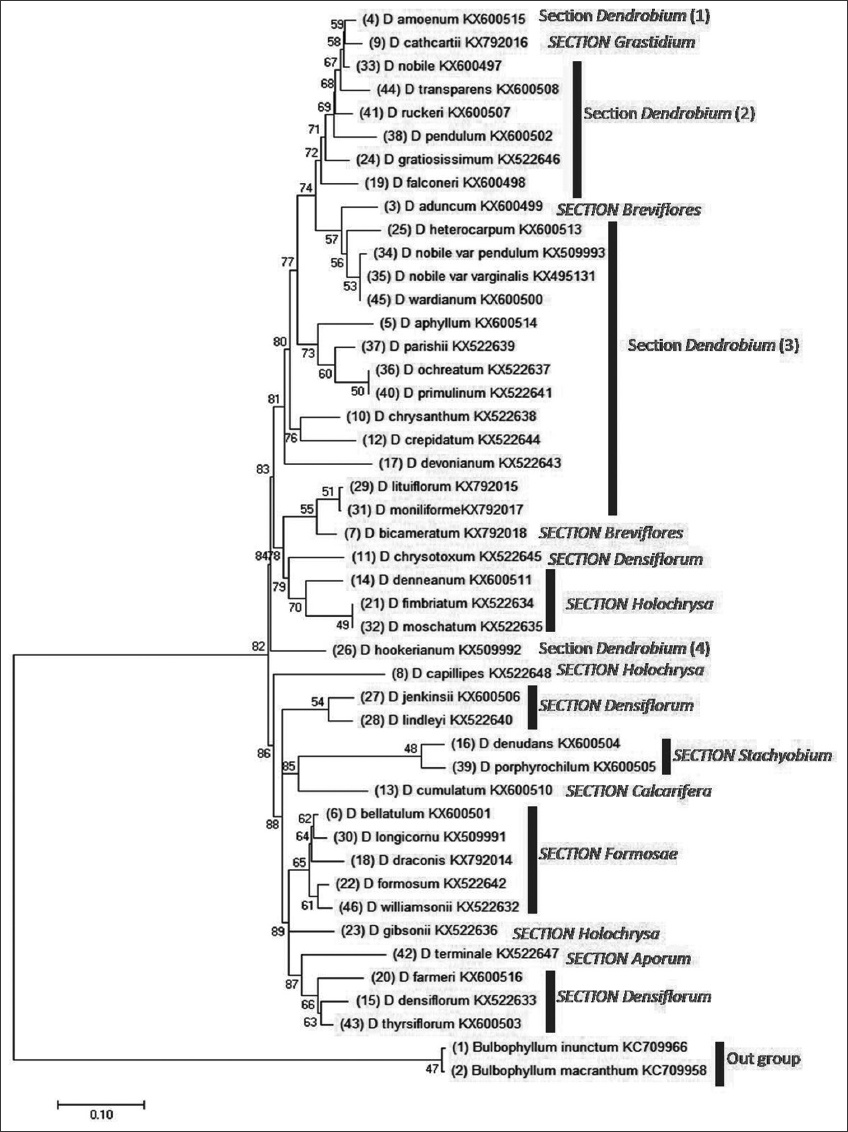 | Figure 2: Analysis of molecular phylogeny using the Tamura-Nei model and the Maximum Likelihood approach. Taxa evolution is represented by the bootstrap consensus tree estimated from 1000 repetitions. Branches collapsed for partitions with fewer than 50% bootstrap replicates. The bootstrap test 1000 replicates a proportion of replicate trees with associated taxa clustered next to branches. The Neighbor-Joining approach was used to build initial tree(s) for the heuristic search from a matrix of pairwise distances computed by the Tamura-Nei model. This investigation examined 44 nucleotides. 1st+2nd+3rd+non-coding were codons. Complete deletion removed all gaps and missing data. The final dataset has 548 positions. MEGA11 performed evolutionary analyses. [Click here to view] |
Section Breviflores consists of two subclades containing a single species in each subclade, i.e., D. aduncum and Dendrobium bicameratum, respectively. Three subclades are noted in the section Densiflorum. Dendrobium chrysotoxum forms a subclade. However, Dendrobium jenkensii and Dendrobium lindeyi constitute together the second subclade. The third subclade includes Dendrobium densiflorum, Dendrobium thyrsiflorum, and Dendrobium farmerii. Section Holochrysa includes three subclades. Dendrobium denneanum, D. fimbriatum, and D. moschatum belong to the first subclade. Dendrobium capillipes is in the second subclade. The third subclade contains a single species, i.e., Dendrobium gibsonii. Section Stachyobium has no subclade in the present investigation and contains only two species D. denudans and Dendrobium porphyrochilum. Section Calcarifera has only the species Dendrobium cumulatum and has no other subclades. Section Formosae includes five species Dendrobium williamsonii, Dendrobium longicornu, Dendrobium bellatum, Dendrobium draconis, and Dendrobium formosum under one subclade. Only one species Dendrobium terminale belongs to section Aporum [Figure 2]. The first, second, and third subclades of the section Dendrobium and section Grastidium are monophyletic. On the other hand, the first subclade of the section Breviflores and D. heterocarpum, D. nobile var. pendulum, D. nobile var. varginalis, D. wardianum of the fourth subclade under section Dendrobium also constitute the second monophyletic group. However, D. aphyllum, D. parishi, D. ochreatum, and D. primilinum of the fourth subclade under section Dendrobium make a separate monophyletic cluster. D. chrysanthum, D. crepidatum. Moreover, D. crepidatum, and D. devonianum under the fourth subclade of the section Dendrobium are polyphyletic. D. moniliforme, D. lutiforum, and D. bicameratum of the second subclade of Breviflores make the fifth monophyletic group. D. chrysotoxum (section Densiflorum), D. denneanum, D. fimbriatum, and D. moschatum (Section Holochrysa) constitute together the sixth monophyletic lineage. Moreover, D. hookerianum is a polyphyletic group. D. capillipes is also a polyphyletic group. D. jenkensii, D. lindeyi, D. denudans, D. pophyrochilium, and D. cumulatum make up the seventh monophyletic group but D. pophyrochilium, and D. cumulatum are polyphyletic. The ninth monophyletic group consists of D. williamsonii, D. longicornu, D. bellatum, D.draconis, and D. formosum, whereas D. gibsonii branches out as a polyphyletic group. D. terminale, D. densiflorum, D. thyrsiflorum, and D. farmerii make them the tenth monophyletic group. It is known that D. capillipes and D. densiflorum do not group with most of the other species in section Dendrobium which they resemble with previous data obtained from ITS. Dendrobium and Grastidium subclades I, II, and III clustered together to create a single phylogenetic unit. This finding demonstrated the close genetic relatedness between groups. Similarly, the subclades I and II of the section Breviflores and Dendrobium were nested together indicating their high phylogenetic relatedness. Monophyletic clustering of D. chrysotoxum of the subclade I of section Densiflorum and species of the first subclade of the section Holochrysa was noted in the dendrogram and they related together with the strong support of the bootstrap value of 79%. D. hookerianum of the fifth subclade of the section Dendrobium was located in the dendrogram in between the first and second subclade of the section Holochrysa as a polyphyletic group [Figure 2]. This species is distantly related to other Dendrobium species [Supplementary Figure 1]. The species under the second subclade of the section Densiflorum are also polyphyletic. Section Calcarifera and Aporum as a paraphyletic group are related to the polyphyletic group of section Stachyobium and Densiflorum, respectively, [Figure 2 and Supplementary Figure 1]. D. longicornu and D. draconis under the section of Formosae are polyphyletic whereas D. longicornu, D. bellatum, D. draconis are monophyletically linked. On the other hand, D. williamsonii and D. formosum are polyphyletic. The selected orchid species, specifically D. gibsonii, belongs to the third subclass within the section Holochrysa and exhibits a direct relationship with the common ancestors of these orchid species [Supplementary Figure 1]. Convergent evolution may have occurred in this particular orchid. D. terminale belonging to section Aporum and the rest of the species under the third subclade of Densiflorum are genetically closely related and form a monophyletic group.
3.4. Tests for Neutrality
According to the neutrality study, Tajima’s D result of −0.1697 was not statistically significant (P > 0.10) among the selected species [Supplementary Table 6], which is consistent with a neutral evolution model. The results of the D*, F*, and Fu’s Fs tests for Fu and Li were −2.80834 (P < 0.05), −2.19041 (0.10 > P > 0.05), and −4.677 (P < 0.01), respectively [Supplementary Table 7]. The findings of Tajima’s D and Fu and Li’s D* test results show a statistical anomaly in the neutral evolution theory. Fu’s Fs is highly sensitive to recent population growth and an excess of alleles, therefore its negative value and statistical significance here make logical sense. A value of D below 0 indicates that natural selection has recently been at work, and the population has grown since the last bottleneck. While there is substantial evidence indicating that the chosen species are undergoing a selective sweep, it is important to consider the significant D* score.
4. DISCUSSION
Many species of Dendrobium are recognized to share a similar appearance. As a result, identifying them dependent on morphological features is challenging, unless they are in bloom. Many Dendrobium morphological features appear homoplasious in several molecular investigations [21]. In this investigation, we observed that the DNA segment of ITS may be used to identify individual species, taxonomical markers, and germplasm preservation in Dendrobium. Most ITS nucleotides are variable and provide extensive systematic data. ITS-I and ITS-II lengths are retained in one population. It was found that the lengths of ITS-I and ITS-II in angiosperms are typically 187–298 base pairs and 187–252 base pairs, respectively, whereas the length of 5.8S is consistently 163 or 164 base pairs and in angiosperm, ITS nucleotide sequence divergence ranges from 1.2% to 10.2% among species and 9.6–28.8% among genera [9]. The lengths of both the ITS area and the 5.8S region in the current study are consistent with the previously reported standard findings. ITS-I and ITS-II were genetically divergent between Dendrobium species, as stated in earlier research and it also indicated better taxonomic identification [22,23]. The ITS sequences have been widely used because of their significance in molecular studies of the orchid family [24]. It is a common tool for tracing the evolutionary history of Dendrobium [11,25] and related families across a wide range of taxonomic groups [26,27]. The result shows 223 (34.97%) conserved bases, 337 (52.85%) parsimony informative sites, and 91 (14.27%) single-ton bases number. The mean percentage of the polymorphic site (63.20%), conserved sites (34.97%), and low mean disparity index (0.192) provide evidence that all species are closely related. However, singleton bases serve as the naturally occurring sites for selection or mutation. The transition (S) and transversion (V) ratio (1.63) shows extremely reactive to saturating mutations and the result differs from conventional Orchid ts/tv ratios ranging from 0.66 to 1.02 [28]. The CI, RI, composite index, and parsimony-informative sites (in bract) make it clear that all species can keep evolving and make changes to their genomes that are either synonymous or non-synonymous to fit in with their new surroundings. A small deviation (R = 1.444) from perfectly balanced transition and transversion contents indicates that transition substitutions are more common than transversion [Supplementary Table 1 and 3], as is usual for advanced taxa, and confirms the amino acid conservative theory [29]. The dN/dS results support the purifying selection (allelic elimination through selective means) of the selected species. The top ML-based DNA models evaluated the K2+G technique as the best nucleotide substitution pattern model because it had the least BIC values among the 24 models [Supplementary Table 2]. The transition-and-transversion-based Tamura-Nei model (K2+G) hypothesized differences between the species. This suggests that transversional substitution rates are similar and purine-purine and pyrimidine-pyrimidine transitions are employed [30]. Mutations detected through multiple sequence alignment supported these findings. Nucleotide composition showed moderate differences (0.32) among sequences [Supplementary Table 1]. However, the base composition may increase GC% due to ambient nitrogen overabundance [31]. These character distributions are optimum for the maximum log-likelihood (−8102.528) phylogenetic tree. The use of particular codons varies greatly not only between species but also between genes and even within genes at different places. Natural codon use variance for UCG (RSCU= 2.68) can be explained by either mutation or natural selection [Supplementary Table 11]. The variability of the GAC codon is comparatively lower inside the ITS sequences. A sensitive indicator of pressure for selection at the amino acid level is provided by Codon Omega results [Supplementary Table 10]. The TGT codon, which exhibits a significant dN–dS value of 4.973 (P < 0.05), is responsible for encoding the amino acid cysteine. Cysteine, encoded by the TGT codon, is an antioxidant that, in Cymbidium goeringii, prevents browning and stimulates rhizome growth by blocking the actions of catalase and polyphenol oxidase [32].
The species of Breviflores exhibit the greatest level of genetic variety (d = 0.171), as indicated in Supplementary Table 4. In addition, the maximum genetic distance (d = 0.285) is observed between the Grastidium and Stachyobium groups [Supplementary Table 5] confirming the finding by their almost opposite morphological characteristics the plant belongs to the group Grastidium and Stachyobium [20]. The study of the data has revealed a high degree of branch support, as evidenced by the presence of a large number of parsimony instructive characters. The resolution of clades was high, and species boundaries were clearly defined. The dendrogram [Figure 2] agrees well with results from previous studies [11,12,14]. In phylogenetic tree analysis, the values at each node are bootstrap. These values can be correlated with branch lengths and conclusions on the evolutionary position of taxa can be drawn. The species of Dendrobium which have high bootstrap values and relatively smaller branch length are considered the most recent origin and advanced. However, those species that have low bootstrap values but long-branch lengths can be correlated as phylogenetically primitive [33]. All of these species (apart from D. falconeri) belong to the first or second subclade of the Dendrobium section, the Grastidium section, and D. chrysanthum has an unstable AC (adenine and cytosine) intermediate as compared to their ancestors [Supplementary Figure 1]. However, the unsteady AC intermediate is extremely rare and the substitution frequencies were highly variable over the two-step procedure, demonstrating that AC is subject to distinct selective forces from those exerted on GU/T which supports compensatory mutation [34]. The monophyletic status of sects. Stachyobium, Grastidium, and Dendrobium are supported in the present study. However, Grastidium was poorly represented due to a lack of data. All reasons viewed, and acknowledging that monophyly is one of the limited objective standards for delimiting genera, it seems better to widen Dendrobium to encompass all previous segregate genera [27]. In contrast, many infra-generic taxa within the genus Dendrobium, namely section Dendrobium, section Holochrysa, and section Densiflora exhibit paraphyly or polyphyly. Consequently, a thorough reassessment and redefinition of their taxonomic classification is warranted. The efficacy of ITS sequencing in Dendrobium taxonomy is still little explored, thus we studied the DNA sequencing methodologies and variability of the ITS-I, 5.8S, and ITS-II locations in 44 wild species and their impact on phylogenetic reconstruction. The present findings corroborate those of other research reporting that current sequence data show that the ITS sections of those species exhibit several differentiations, which match with published sequences. This provides further evidence that Dendrobium species can be distinguished from one another based on the sequence of their ITS-I and ITS-II segments [35]. A negative value of Tajima’s D (−0.1697) denotes an overabundance of low-to-moderate polymorphisms (mean 63.20%), in comparison to expected, signifying the size of the population increase following a bottleneck and the observed nucleotide diversity (π) of 0.1377 [Supplementary Table 6] exhibits a close resemblance to previous research findings in the genus Dendrobium [36]. In summary statistics (Tajima’s D and Fu and Li’s test), probabilities are simulated values, not calculated. Since these p-values are approximate. The Fu’s Fs significant value [Supplementary Table 7] indicates an overabundance of alleles and a possible selective sweep [37]. At present, population growth is the most acceptable explanation for the deviations from neutrality, as supported by the negative values of Tajima’s D, Fu and Li’s D*, F*, and Fu’s Fs.
5. CONCLUSION
This study evaluated the ITS sequences of 44 Dendrobium species. A significant amount of variation is present in the ITS sequences among the selected species of Dendrobium. ML tree analysis strongly supports the possible use of the ITS region for distinguishing morphologically most similar Dendrobium species. In the future, more numbers of Dendrobium species should be analyzed to confirm the suitability of the ITS region for identifying all Dendrobium species and other orchid genera.
6. ACKNOWLEDGMENT
For providing the resources needed to complete this work, the authors would like to express their deep appreciation to the Postgraduate Department of Botany, Hooghly Mohsin College, Hooghly.
7. AUTHOR CONTRIBUTIONS
Animesh Mondal: Sample collection, laboratory work, assessment of data, and early manuscript writing. Kalyan Kumar De: oversaw the experimentation, verification, review, and editing of the updated manuscript.
8. FUNDING
This research was supported by a grant (F.No. 42-988/2013/SR) from the University Grant Commission, New Delhi, India.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
All the raw data regarding the research are available within this manuscript.
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
13. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Tang XG, Yuan YD, Zhang JC. How climate change will alter the distribution of suitable Dendrobium habitats. Front Ecol Evol 2020;8:536339. [CrossRef]
2. Lokho A. Diversity of Dendrobium Sw. Its distributional patterns and present status in the Northeast India. Int J Sci Res 2013;3:1-9.
3. Deori C, Sarma SK, Hynniewte TM. Dendrobium Orchids of Northeast India. Guwahati, Assam, India:Purbayon Publication;2019.
4. Zhang GQ, Xu Q, Bian C, Tsai WC, Yeh CM, Liu KW, et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci Rep 2016;6:19029. [CrossRef]
5. Teixeira da Silva JA, Ng TB. The medicinal and pharmaceutical importance of Dendrobium species. Appl Microbiol Biotechnol 2017;101:2227-39. [CrossRef]
6. Bhattacharyya P, Kumaria S, Tandon P. High-frequency regeneration protocol for Dendrobium nobile:A model tissue culture approach for propagation of medicinally important orchid species. S Afr J Bot 2016;104:232-43. [CrossRef]
7. Szlachetko DL, Kolanowska M, Naczk A, Gorniak M, Dudek M, Rutkowski P, et al. Taxonomy of Cyrtochilum-alliance (Orchidaceae) in the light of molecular and morphological data. Bot Stud2017;58:8.
8. Mort ME, Archibald JK, Randle CP, Levsen ND, O'Leary TR, Topalov K, et al. Inferring phylogeny at low taxonomic levels:Utility of rapidly evolving cpDNA and nuclear ITS loci. Am J Bot 2007;94:173-83. [CrossRef]
9. Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA:A valuable source of evidence on angiosperm phylogeny. Ann Mo Bot Gard 1995;82:247-77. [CrossRef]
10. Yuan ZQ, Zhang JY, Liu T. Phylogenetic relationship of China Dendrobium species based on the sequence of the internal transcribed spacer of ribosomal DNA. Biol Plant 2009;53:155-8. [CrossRef]
11. Xiang XG, Schuiteman A, Li DZ, Huang WC, Chung SW, Li JW, et al. Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol Phylogenet Evol 2013;69:950-60. [CrossRef]
12. Liu YT, Chen RK, Lin SJ, Chen YC, Chin SW, Chen FC, et al. Analysis of sequence diversity through internal transcribed spacers and simple sequence repeats to identify Dendrobium species. Genet Mol Res 2014;13:2709-17. [CrossRef]
13. Moudi M, Go R. Monophyly of four sections of genus Dendrobium (Orchidaceae):Evidence from nuclear ribosomal DNA internal transcribed spacer (ITS) sequences. Int J Bioassays 2015;4:3622-6.
14. Feng S, He R, Yang S, Chen Z, Jiang M, Lu J, et al. Start codon targeted (SCoT) and target region amplification polymorphism (TRAP) for evaluating the genetic relationship of Dendrobium species. Gene 2015;567:182-8. [CrossRef]
15. Wang S, Hou F, Zhao J, Cao J, Peng C, Wan D, et al. Authentication of Chinese herbal medicines Dendrobium species and phylogenetic study based on nrDNA ITS sequence. Int J Agric Biol 2018;20:369-74. [CrossRef]
16. Moudi M, Go R. The comparison between nuclear ribosomal DNA and chloroplast DNA in molecular systematic study of four sections of genus Dendrobium sw. (orchidaceae). Int J Bioassays 2016;5:4944-52.
17. Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus 1990;12:13-5.
18. Tamura K, Glen S, Kumar S. MEGA11:Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022-7. [CrossRef]
19. Librado P, Rozas J. DnaSP v5:A software for comprehensive analysis of DNA polymorphism data. Bioinformatics2009;25:1451-2.
20. Deori C. Morphological diversity within the genus Dendrobium Swartz (Orchidaceae) in Northeast India. Richardiana 2014;16:85-110.
21. Feng S, Jiang Y, Wang S, Jiang M, Chen Z, Ying Q, et al. Molecular identification of Dendrobium Species (Orchidaceae) based on the DNA barcode ITS2 region and its application for phylogenetic study. Int J Mol Sci 2015;16:21975-88. [CrossRef]
22. Wang X, Chen X, Yang P, Wang L, Han J. Barcoding the Dendrobium (Orchidaceae) species and analysis of the intragenomic variation based on the internal transcribed spacer 2. Biomed Res Int 2017;2017:2734960. [CrossRef]
23. Liu H, Fang C, Zhang T, Guo L, Ye Q. Molecular authentication and differentiation of Dendrobium species by rDNA ITS region sequence analysis. AMB Express 2019;9:53. [CrossRef]
24. Nordin FA, Saibeh K, Go R, Mangsor KN, Othman AS. Molecular phylogenetics of the orchid genus Spathoglottis (Orchidaceae:Collabieae) in Peninsular Malaysia and Borneo. Forests 2022;13:2079. [CrossRef]
25. Schuiteman A. Dendrobium (Orchidaceae):To split or not split?Gard Bull Singapore 2011;63:245-57.
26. Manokar J, Balasubramani SP, Venkatasubramanian P. Nuclear ribosomal DNA-ITS region based molecular marker to distinguish Gmelina arborea Roxb. Ex Sm. from its substitutes and adulterants. J Ayurveda Integr Med 2018;9:290-3. [CrossRef]
27. Zhang J, Chi X, Zhong J, Fernie A, Alseekh S, Huang L, et al. Extensive nrDNA ITS polymorphism in Lycium:Non-concerted evolution and the identification of pseudogenes. Front Plant Sci 2022;13:984579. [CrossRef]
28. Salazar GA, Chase MW, Soto Arenas MA, Ingrouille M. Phylogenetics of Cranichideae with emphasis on Spiranthinae (Orchidaceae, Orchidoideae):Evidence from plastid and nuclear DNA sequences. Am J Bot 2003;90:777-95. [CrossRef]
29. Lyons DM, Lauring AS. Evidence for the selective basis of transition-to-transversion substitution Bias in two RNA viruses. Mol Biol Evol 2017;34:3205-15. [CrossRef]
30. Kumar S. Patterns of nucleotide substitution in mitochondrial protein coding genes of vertebrates. Genetics 1996;143:537-48. [CrossRef]
31. Bohlin J, Eldholm V, Pettersson JH, Brynildsrud O, Snipen L. The nucleotide composition of microbial genomes indicates differential patterns of selection on core and accessory genomes. BMC Genomics 2017;18:151. [CrossRef]
32. Huang W, Fang Z. Different amino acids inhibit or promote rhizome proliferation and differentiation in Cymbidium goeringii. Hortic Sci 2021;56:79-84.
33. Hong Y, Luo Y, Gao Q, Ren C, Yuan Q, Yang QE. Phylogeny and reclassification of Aconitum subgenus Lycoctonum (Ranunculaceae). PLoS One 2017;12:e0171038.
34. Kimura M. The role of compensatory neutral mutations in molecular evolution. J Genet 1985;64:7-19. [CrossRef]
35. Saha PS, Sengupta M, Jha S. Ribosomal DNA ITS1, 5.8S and ITS2 secondary structure, nuclear DNA content and phytochemical analyses reveal distinctive characteristics of four subclades of Protasparagus. J Syst Evol 2017;55:54-70. [CrossRef]
36. Roy SC, Moitra K, De Sarker D. Assessment of genetic diversity among four orchids based on ddRAD sequencing data for conservation purposes. Physiol Mol Biol Plants 2017;23:169-83. [CrossRef]
37. Ashfaq M, Paul Hebert DN, Sajjad Mirza M, Arif M, Shahid M, Ghulam S, et al. Genetic diversity indices and neutrality tests (Fu's Fs and Tajima's D) in the mtCOI-5'(barcode) sequences of putative species in Bemisia tabaci complex from Pakistan and India. PLoS One 2014;9(5):e97268.
SUPPLEMENTARY MATERIAL
The supplementary material can be accessed at the journal's website:
Link Here [https://jabonline.in/admin/php/uploadss/1193_pdf.pdf].