1. INTRODUCTION
Dexras1 is a small monomeric G protein [1-6], GTP binding signal transducer proteins [7] which get activated by G protein-coupled receptors [8]. It belongs to RAS superfamily proteins that are reported to be involved in various cell signaling processes [4] and is also named Activator of G protein signalling1 (AGS1) [9], which is encoded by the gene RASD1 present in humans. The full-length cDNA of Dexras1 contains a 281-aminoacid and molecular mass of 31,700Da [1]. Dexras1 has all of the conserved domains of the RAS superfamily needed for the binding of guanine nucleotide (Σ1–Σ4) domains, an effector loop [10-13], and a C terminus having a typical CAAX motif [12,14-15] that undergoes prenylation and regulates Dexras1 subcellular localization by promoting the translocation of the RAS protein to the plasma membrane [Figure 1] [15-17].
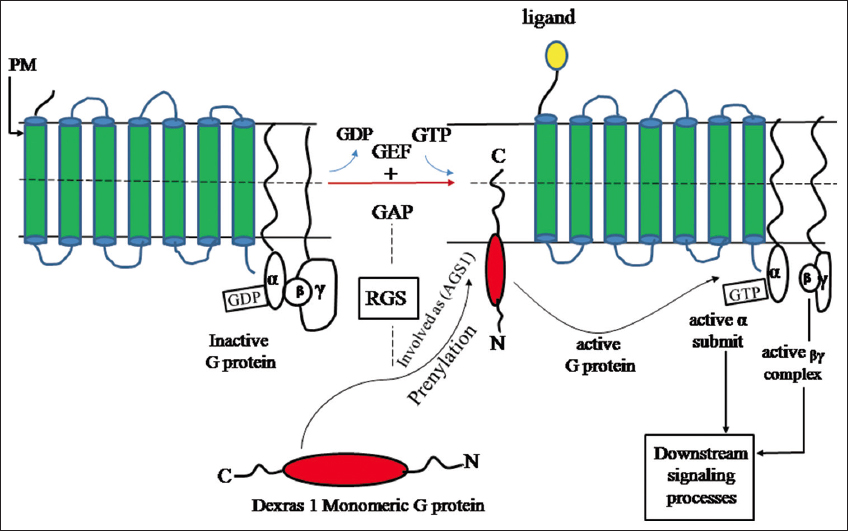 | Figure 1: Dexras1 prenylation and translocation to the plasma membrane: Dexras1 functions as an activator of G protein signaling1 (AGS1) after prenylation, by which inactive GDP bound trimeric G protein transformed to active GTP bound protein, in the presence of GAP and GEF, for further downstream signal transduction process. [Click here to view] |
With its unique characteristics, Dexras1 is reported to be involved in various pathophysiological processes such as cardiovascular diseases, Huntington’s disease, regulation of rhythms, neurotransmitter-mediated behavior, infertility, reproduction, anxiety disorders, obesity, and a medical biomarker in cancer [Table 1] [18-22]. The exact molecular mechanism of signaling through this protein in modulation of various physiological processes is mostly attributed to its C-Terminus but not much is known about the 3D structure of this protein. It is reported in many studies that the identification and characterization of a protein could be a potential biomarkers [23]. Dexras1 could also be explored more on the bases of its structural information, but the Protein Data Bank file of Dexras1 structure is unavailable in any of the databases to date. Out in silico studies have predicted Dexras1 structure by the use of homology modeling [24]. Our present study is in continuation of in silico study. In the present study, experiments were carried out to subclone, express, and purify Dexras1 protein, circular dichroism (CD) technique was used to elaborate on Dexras1 structural determinants.
Table 1: Important milestones and comparative novelty of present work.
| Work done | Outcome and Findings | Year and Author |
|---|---|---|
| Introduced Dexras1, as a new Ras superfamily member. | Dexras1 was discovered as a dexamethasone inducible protein in AtT-20 cell lines. | 1998 Kemppainen and Behrend |
| Dexras1 is involved in the inhibition of 3’,5’- cAMP triggered secretion in AtT-20 corticotroph cells. | Linear protein Structural analyses described the presence of common GTP binding domains and unique N and C terminals in Dexras1. | 2001 Graham et al. |
| Reviewed the previous work done and reported the role of Dexras1 in various pathophysiological processes. | Dexras1 is involved in Cardiovascular Disease, Huntington’s disease, Modulation of N-type calcium channels, Cancer, regulation of circadian rhythms, and hormonal regulations. | 2014 Thapliyal et al. |
| And its role in repeated uterine implantation failure was also reported. | Expression in the uterus and reproductive organs were reported. And its involvement in infertility was also reported. | 2018 Hong and Choi |
| A new type of antiadipogenic agent, and tigliane diterpenoids inhibit GRα-Dexras1 axis in adipocytes. | Dexras1 investigated as an important therapeutic agent in obesity. | 2019 Song et al. |
| Dexras1 protein in silico structural analysis was done. | Dexras1 pretended 3D structure produced by homology modeling, a bioinformatics approach. | 2019 Verma et al. |
| Dexras1 sub-cloning expression and purification protocols were standardized for some structural analysis. | Dexras1 protein is expressed in prokaryotic cells and some preliminary structural determinants are known by CD which will help in further 3D structure determination. | Present work |
2. MATERIALS AND METHODS
The expression of the protein in Escherichia coli strains is affected by a protein and its nature. Most of the recombinant proteins are expressed as soluble proteins and some are expressed in insoluble form aggregates due to their intracellular nature, as membrane-bound proteins. These insoluble forms are called “inclusion Bodies” [25]. Some studies have also reported that recombinant proteins show more excellent temperature stability than native proteins [26]. Dexras1wt (wild type) is a membrane protein and its C terminal might get attached to the membrane during prenylation, which might affect its solubility nature, keeping that in mind, the experiments were conducted for subcloning expression and purification of Dexras1wt protein [Figure 2]. The purified protein was used to analyze some preliminary structural details by a biophysical technique, CD.
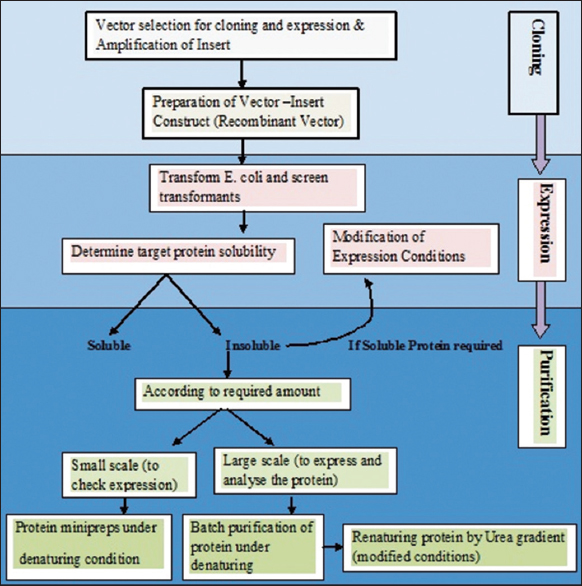 | Figure 2: Strategy of Dexras1wt gene sub-cloning, expression, and purification. [Click here to view] |
All the experiments of gene subcloning were conducted in the Mechanistic Biological Chemistry Lab, Department of Biotechnology, IIT Roorkee with the kind support of Dr. Krishna Mohan Poluri (Associate Prof. Dept. of Biotech., IIT Roorkee). Strains, E. coli (DH5α) and E. coli (BL21), and chemicals used in gene sub-cloning were also provided by the same lab. Primers were designed using OligoPerfect Thermo Fischer Scientific Server and were procured from Macrogen Company. The Dexras1/Ags1 (Accession number: AF498923) wild-type gene in the form of complementary DNA, already inserted in the vector pcDNA3.1, named Dexras1 wtpcDNA3.1, was provided by Dr. Ashish Thapliyal (Dept. of Life Sciences, Graphic Era Deemed to be University) and Dr. Brett A. Adams (Dept. of Biology, Utah State University, USA) [27]. The PCR cleanup/Gel extraction kit (Cat. no. NP-36105) was obtained from Nucleopore, Genetix-Biotech Asia Pvt. Ltd., and Restriction enzymes NdeI (Cat. no. FD0583) and XhoI (Cat. no. FD0694) were procured from Thermo Fischer Scientific. The chemicals and protocols for the protein expression and purification studies were used from QIAexpressionist™ manual (2001–2003) and QIAexpress type IV Kit (Cat. no. 32149, QIAGEN).
2.1. Subcloning of Dexras1 wt Gene
For subcloning, the clone of Dexras1wt pcDNA3.1 gene was amplified using a forward primer of 28 mer 5’GCAACATATGATGAAACTGGCCGCGATG-3’ and a reverse primer of 29 mer5’GCAACTCGAGCTAGCTGATGACGCAGCGC-3’ with the thermal cycle: Hot start at 95°C, 1: 30 min, denaturation at 95°C, 1 min, annealing at 58°C, 30 s, extension at 72°C, 1 min for 35 cycles, and final elongation at 72°C for 5 min. The amplified product of PCR was cleaned up using the PCR cleanup kit and was sub-cloned in a suitable customized Maltose Binding Protein (MBP) vector, derived from a pMAL series vector having a T7 promoter/terminator, 6× His tag site, and TEV protease cleavage site. The vector and gene were both digested with Nde1 and Xho1 restriction endonucleases and extracted from the gel using standard protocols [28]. The gel eluted and precipitated Vector and insert were ligated in 1:3 ratios in a total volume of 20 μL using the T4 DNA ligase (Thermo Fisher Scientific EL0011) enzyme. The recombinant vector thus produced had, 5’T7 Promotor – HisTag – MBP – Vector sequence – TEV Protease Cleavage sequence – Nde-1 Site – Insert Gene(Dexras1wt) ---- Stop-codon -- Xho-1 – vector – T7 Terminator3’, sequence [Figure 3]. The transformation of the ligated product (Dexras1wt-MBP)/recombinant plasmid was done into host competent cells E. coli DH5α cells prepared by the Rubidium Chloride method [29]. The positive transformant colonies were selected based on ampicillin resistance and the recombinant plasmid (positive transformants) was isolated using the alkaline lysis method [30] and the final transformation was done in another strain known as E. coli BL21 for expression of a protein. The positive transformants were selected based on the ampicillin-resistant marker present in the vector and maintained in LB medium for further study.
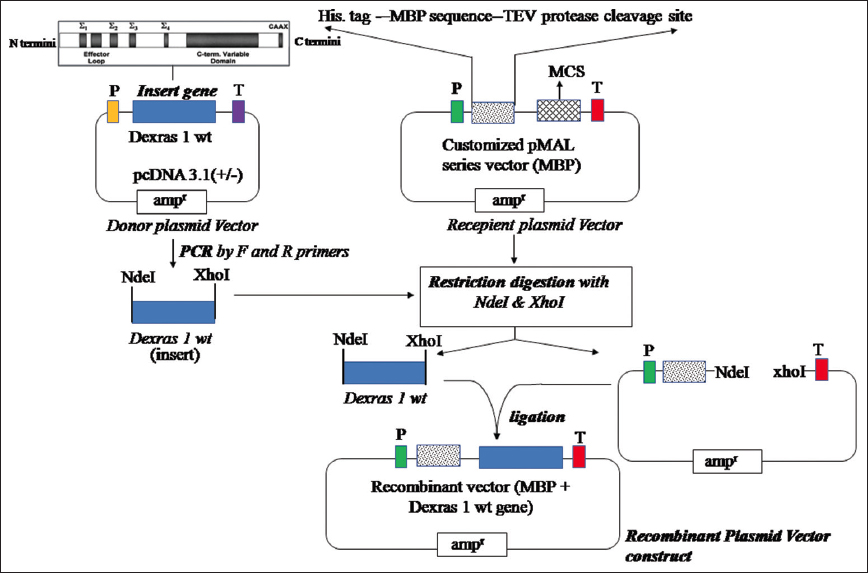 | Figure 3: Construction of recombinant plasmid used for expression of Dexras1wt gene: Donor, pCDNA 3.1 vector containing Dexras1 wt gene insert, Recipient: customized pMAL series MBP vector containing P(T7 polymerase) promoter, 6× Histidine affinity tag sequence, MBP sequence, TEV protease cleavage site, and MCS-multiple cloning site for restriction endonuclease including NdeI&Xho I. The Dexras1wt gene was amplified by PCR with NdeI and XhoI restriction sites added at the ends using F and R primers and subcloned into the recipient vector after digestion and ligation process to generate recombinant plasmid vector construct with Dexras1wt gene insert. [Click here to view] |
2.2. Expression of Protein
The solubility of Dexras1 was analyzed using different expression protocols. For the expression of recombinant Dexras1 wt protein, a single colony of positive transformants of E. coli Bl21 (with MBP vector and Dexras1wt gene) was inoculated in 5mL LB amp (100 μg/mL stock) media and allowed to grow overnight at 37°C. To induce the expression of the protein, 10 mL LB amp media was inoculated with 100 μL overnight culture and, further, incubated for 2–3 h in a shaker at 37°C. As the O.D600nm reached 0.6, 1 mL culture was taken instantly before induction, pelleted out and the pellet re-suspended in 100 μL 1X SDS PAGE gel loading dye, named as an un-induced control sample. The sample was frozen at −20°C until it is needed for SDS-PAGE analysis. The rest of the growing culture was induced by 1 mM Isopropyl β-D-1 thiogalactopyranoside (IPTG, 1M stock) to induce expression. The culture was allowed to grow for the additional 4–5 h. The second 1 mL sample was pellet out and re-suspended in 100 μL 1X SDS-PAGE gel loading dye named as an induced sample and frozen at −20°C for SDS page analysis. The protocol of induction and expression was repeated with different concentrations of IPTG (1 mM, 0.5 mM) and also with different growth conditions (temp.20°C and 30°C and time 3 h, 4 h, 16 h, and 20 h) to analyze the highest expression conditions of Dexras1wt protein. Protein expression was analyzed by SDS-PAGE of 5% stacking, and 12% resolving polyacrylamide gels using the standard protocol. The separate wells were loaded with induced and non-induced protein sample supernatant (30 μL each), along with BSA (66 kD) and lysozyme (14 kD) as a marker protein. Initially, the gel was set to run at 70 volt 1 h and later at 90–100 volt for 3 h.
2.3. Purification of Protein
The purification of protein was done both in the native and denatured form to analyze its soluble or insoluble nature following the protocol of QIAexpressionist™ manual (2001–2003). After checking the solubility of the protein, the large-scale expression and purification of protein were done.
2.3.1. Determination of dexras1wt protein solubility in native form (small scale)
A 100 mL culture of transformants (E. coli BL21 with Dexras1wt gene) was induced with 1 mM IPTG to determine the protein solubility in native conditions. Induced and un-induced 1–1 mL culture was pelleted out for SDS Page analysis and the rest culture was harvested by centrifugation at 13000 rpm for 20 min at 4°C. 10 mL lysis buffer (20 mM tris and 500 mM NaCl) was added and the suspension was left at −80°C overnight. Lysozyme (5 mg/mL stock) and PMS (100 mM stock) were added and cells were sonicated (5 s. on, 10 s. off with 50% power supply for 20 min). After centrifugation, at 14000 rpm for 30 min., the pellet and the supernatant(s) were stored at 4°C. Pre-regenerated Nickel column (with the buffers used sequentially: DW 50 mL, 1M imidazole 50 mL, DW 50 mL, 4M NaCl 50 mL, DW 50 mL, 0.4 M EDTA 50 mL, DW 50 mL, and 100 m M Nickel Sulfate 50 mL) was loaded with supernatant(s) and passed 3–4 times through the column and collected in a fresh tube (stored at 4°C). 50 mL of lysis buffer (20 mM Tris and 400 mM NaCl) passed through the column and the first lysis eluent was stored at 4°C. Again, the column was washed twice with 50 mL buffer (imidazole 10 mM, for wash1 (W1)/50 mM for wash2 (W2), 20 mM tris, and 500 mM NaCl) and eluent collected as W1 W2. The protein was eluted (E) with 50 mL buffer (400 mM imidazole and 20 mM tris and 500 mM NaCl). All the samples were stored at −20°C and used to check the expression of protein in 12% SDS.
2.3.2. Determination of dexras1wt protein solubility in denatured form (small scale)
Dexras1wt protein purification was also done on small scale in denatured conditions using protocols of QIAexpressionist kit manual (QIAexpress type IV Kit, 2001–2003, Cat. no. 32149, QIAGEN) with slide modifications. The 100 mL transformants culture (E. coli BL21 containing Dexras1wt gene) was induced with 1 mM IPTG at 37°C for 4 h and the induced and un-induced pellet of 1–1 mL cultures were pelleted out to analyze SDS PAGE. The rest of the culture was centrifuged and 5 mL/g wet weight pellet was treated with lysis buffer 100 mM NaH2PO4, 10 mM Tris, 8 M urea, pH 8.0. The cells were stirred at room temperature (RT) for 1 h by gentle vortexing. After lysis of cells, the translucent solution was centrifuged at 10000× g for 30 min at RT and supernatant (cleared lysate) was used for batch purification of 6× His tagged protein by Nickel-NTA column chromatography and 20 μL of this clear lysate was also stored at −20°C for SDS-PAGE analysis. For batch purification, 1 mL 50% Nickel-Nitriloacetic acid (Ni-NTA) slurry was mixed with 4 mL clear lysate by gentle shaking at 200 rpm for 1 h. The lysate mixture was loaded onto a column and the effluent was collected. The resin was washed with 4 mL buffer100 mM NaH2PO4, 10 mM Tris, 8 M urea wash buffer pH 6.3 2 times, and the protein was eluted 4 times in 0.5 mL buffer elution buffer 100 mM NaH2PO4, 10 mM Tris, 8 M urea pH 5.9. All the samples were stored at 4°C for analysis of protein expression by SDS-PAGE after purification of protein.
2.3.2.1. Large-scale protein purification
A 1-l culture was set to grow using the optimized protocol for small-scale culture that the expression conditions were kept constant (Temp. 37°C, Induction with 1 mM IPTG for 4 h). Dexras1 protein was purified under denaturing conditions, following the referenced protocol [31] with slight modifications. The pellet was mixed with 6 mL lysis buffer, 100 mM NaH2PO4, 10 mM Tris, 8M urea, and pH 8.0 by gentle stirring at RT for 1 h. The lysate was centrifuged at 10,000 g at 4°C for 30 min and the supernatant was mixed with 2 mL, 50% slurry of Ni-NTA resins, and stirred at RT for 1 h. These resins were loaded into a 5 mL bed volume Ni-NTA column. To facilitate the slow removal of urea, resins were washed with a gradually decreasing urea gradient (8M-0M) in Na-phosphate (100 mM) and Tris-Cl (10 mM) buffer, pH 7.8. The resin was, then, washed with a buffer containing Na-phosphate (100 mM) and Tris-Cl (10 mM) buffer, pH 7.6. The protein was eluted with 250 mM Imidazole in 100 mM Na-phosphate, 10 mM Tris-Cl, and pH 7.0 buffer. The fractions of eluted protein were analyzed on SDS-PAGE and those containing protein were pooled and quantified by Bradford’s assay. The quantified protein was dialyzed with 12–14 MWCO dialysis membrane (Dialysis Membrane-110, Cat. no. LA395, HiMedia) against sodium phosphate buffer (10mM, pH 7.4). The dialyzed sample was treated with TEV protease enzyme (procured from Mechanistic Biological Chemistry Lab, IIT ROR) and passed through the Ni-NTA column to remove the MBP sequence from Dexras1wt protein. The purified protein was concentrated by ultracentricon 10MWCO, provided by Mechanistic Biological Chemistry Lab, IIT ROR, and analyzed by spectrophotometer (BioSpectrometer Basic 230 V/50–60 Hz, Eppendorf) by taking O.D between 230 and 280 nm. The highest O.D observed at 280 nm. The concentration of the protein was calculated using the extinction coefficient of a protein. The concentrated protein was kept in aliquots at 4°C.
2.4. Analyzing Some Structural Determinants by CD Spectroscopy Technique
CD spectroscopy, a biophysical tool, is used to study structural aspects of optically active biomolecules such as protein or DNA [32]. The molar extinction coefficient of Dexras1 protein was calculated based on its amino acid sequence using the formula: nWx5500+nYx1490+nCx125 (Nw = total number of tryptophan, nY = total number of tyrosine, nC = total number of cysteine in the sequence of Dexras1 protein). The concentration of the protein sample was calculated using its absorbance (O.D) at 280 nm and its molar extinction coefficient by the formula: C=A280nm/ε (C = concentration of protein, A = Absorbance at 280 nm, e = Molar extinction coefficient). The sample of Dexras1 wt protein was taken in a quartz cuvette of 1 cm path length. The far-UV CD spectra of native protein were recorded on a CD spectrometer (J-815-450S, JASCO) from a wavelength of 250–200 nm at 20°C in the laboratory of Kusuma School of Biological Sciences, IIT Delhi. The CD spectra were used to interpret the secondary structure of Dexras1 wt protein.
3. RESULTS AND DISCUSSION
3.1. Subcloning of Dexras1wt Gene in MBP Vector
The quantified plasmid vector Dexras1wt pcDNA3.1 (1009.70 ng/μL) was used to amplify inserted Dexras1wt gene by PCR [Figure 4]. After amplification, the quantified values obtained were 139.55 ng/μL and142.85 ng/μL for Dexras1 and MBP Vector, respectively. The restriction digestion of Vector and gene was confirmed by AGE [Figure 5] and the recombinant colonies were screened and confirmed by PCR of the isolated plasmid [Figure 6]. The cloning of the transformant colony was confirmed by sequencing of the Dexras1 gene in MBP vector, done by Europhins Genomic India Private Limited using the software Applied Biosystem. The confirmed isolated recombinant plasmid was used to transform the culture E. coli BL21 for the expression of the protein.
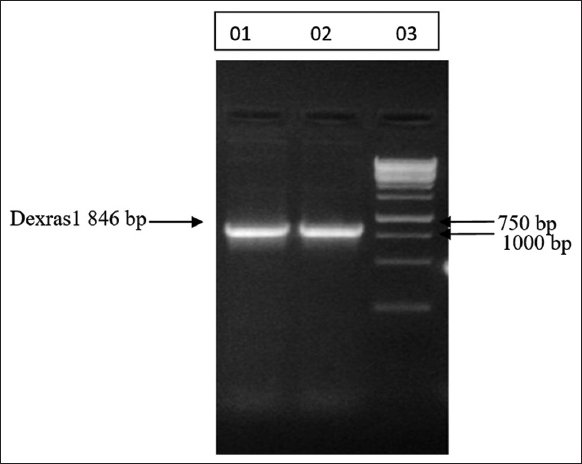 | Figure 4: PCR of Dexras1 wtgene: PCR product of Dexras1 gene. Lane 01 and 02 amplified Dexras1wt gene (846bp) and Lane 03 DNA ladder 1 Kb. [Click here to view] |
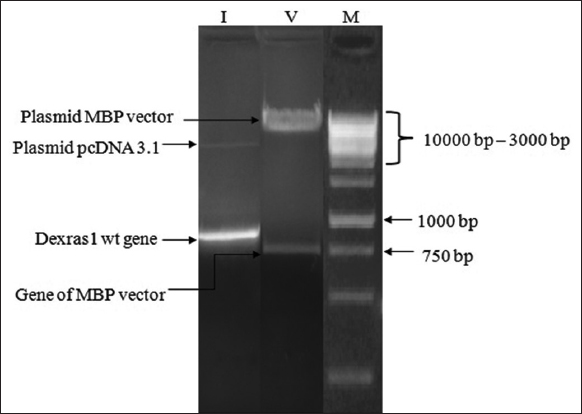 | Figure 5: Agarose gel electrophoresis of restriction digestion of Dexras1wt gene and Vector MBP; lane I (Insert): digested Dexras1wt gene, lane V (Vector) digested MBP vector; lane M (Marker) 1Kb DNA ladder. [Click here to view] |
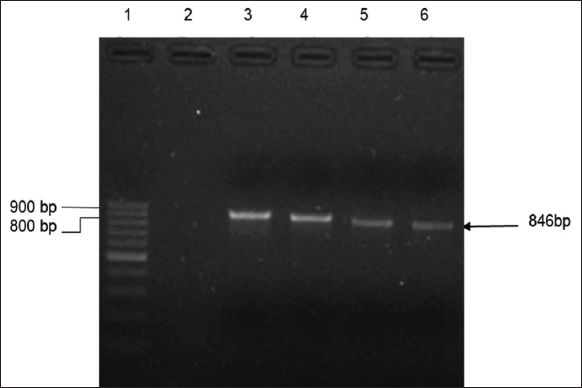 | Figure 6: Dexras1wt gene amplified using isolated vector from assumed clones. Lane 3, 4, 5, and 6 PCR products from the vector of putatively transformed colonies. Lane M: DNA ladder 1Kb. [Click here to view] |
3.2. Expression of Protein
The expression of the protein was confirmed in small-scale culture with 4 h induction with 1 mM IPTG at 37°Con SDS-PAGE [Figure 7] in the presence of Marker (BSA 66 kD, lysozyme 14 kD). For large-scale expression, different expression conditions such as culture growth temp, expression time, and IPTG concentration were analyzed and optimized. The less expression was observed with 1 mM IPTG induction at 20°C for 16–20 h and the expression increased with 0.5 mM IPTG induction at 37°C for 3 h. The expression was highest when the cells are induced with 1 mM IPTG at 37°C for 4 h of incubation time after induction [Figure 8].
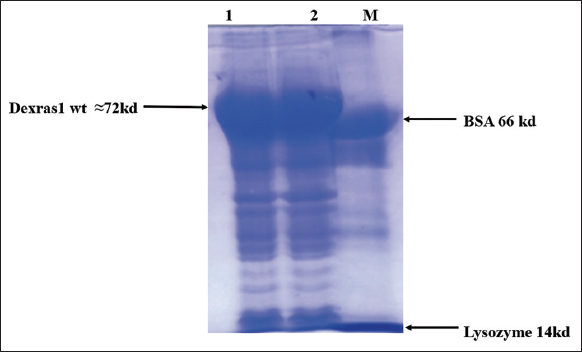 | Figure 7: Expression of protein at 37°C after induction for 4 h. Lane 1 and 2 Dexras1wt gene 31.7kD expressed with MBP 42.5kD (31.7 kD + 42.5 kD ≈74.2 kD fusion protein band). Lane M Marker (BSA 66 kD, lysozyme 14 kD). [Click here to view] |
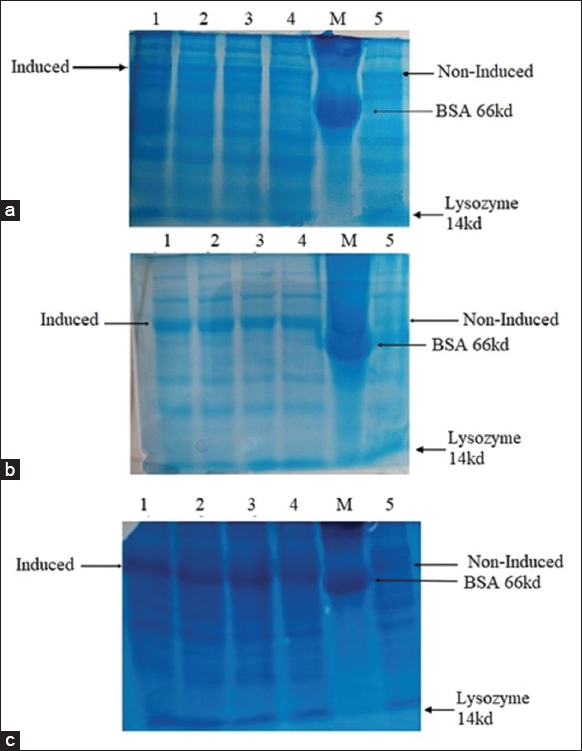 | Figure 8: Expression of the protein in different conditions: a.20°C after induction for 16 h with 1 mM IPTG; b.37°C after induction for 3 h with 0.5 mM IPTG; c. 37°C after induction for 4 h with 1 mM IPTG. In all three (a,b,c) Lane 1, 2, 3, and 4 induced Dexras1wt gene expressed with MBP (31.7kD+42.5 kD≈74.2kD protein band). Lane M: Marker (BSA 66kD). Lane 5 Non-induced Dexras1wt gene. [Click here to view] |
3.3 Purification of Dexras1wt Protein
Protein solubility was determined in both native and denatured forms. Dexras1 wt protein (31.7 kD) ligated to MBP vector (42.5 kD) expressed as a ≈74.2 kD protein and was localized only in cell pellet [Figure 9]. Moreover, the protein was also purified in denaturing conditions and bands of eluted protein were observed on SDS PAGE [Figure 10] which confirms that Dexras1wt protein was expressed in insoluble form and might form inclusion bodies. For a large scale, 1-l culture was induced to express the protein, and Ni-NTA chromatography was used to purify the protein. The bands of eluted protein after renaturation were observed on SDS-PAGE [Figure 11]. The unknown concentration of the eluted protein was approximately 1 μg/μL measured by Bradford assay using the standard graph. The Dexras1wt is involved in signaling of different processes and the structure of Dexras1wt protein showed that it has a CAAX domain and the unique C terminal which shows the prenylation process, by which the protein gets attached to the membrane through its CAAX domain of C terminal and so this membrane binding might be the reason of insoluble form expression of Dexras1 protein.
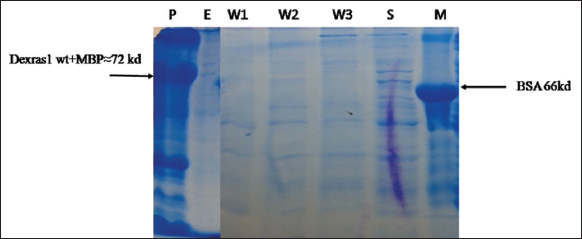 | Figure 9: Expression and protein purification in native form: P: Pellet, E: Elution, W1,W2,W3: wash1, wash2, wash3, S: Supernatant, M: Marker BSA. [Click here to view] |
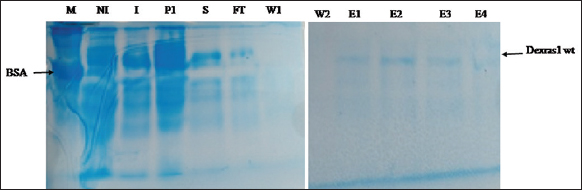 | Figure 10: The expression of protein Dexras1wt with MBP (74.2 kD) in denatured form (small scale). M: Marker BSA 66 kD, NI: Non-induced, I: Induced, P1: Lysed pellet S: Supernatant, FT: Flow through, W1: Wash1, W2: Wash2, E1: Elution1, E2: Elution2, E3: Elution3, E4: Elution4. [Click here to view] |
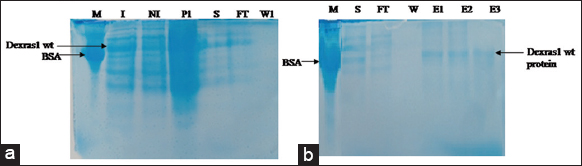 | Figure 11: Protein purification after renaturation (large scale) (a). M: BSA marker 66 kD, I: Induced, NI: Non-induced, P1: Lysed pellet, S: Supernatant, FT: Flow Through, W1: Washing of Protein Dexras1wt, (b) M: BSA marker 66 kD, S: Supernatant, FT: Flow Through, W: Washing, E1, E2, E3: Elution1, Elution 2, Elution 3 of Dexras1wt Protein. [Click here to view] |
3.4. Analyzing Some Structural Determinants of Dexras1wt Protein Using Circular Dichroism
After dialysis and Tev protease treatment, the concentration of the protein was analyzed using a spectrophotometer (BioSpectrometer Basic 230 V/50–60 Hz, Eppendorf) and the highest O.D was observed at 280 nm. The absorbance maxima for Dexras1 protein were observed at 280 nm. The secondary structures of Dexras1wt protein were studied by taking CD spectra from a wavelength of 200–250 nm in a CD spectrometer. The CD spectrum was produced by the curve fitting method using inbuilt Reed’s reference spectra by CD spectrometer. Moreover, the spectrum of molar ellipticity of Dexras1 protein was produced [Figure 12]. On the bases of the CD spectrum, the percentage of a helix, b sheet, turn, and random structures is approximately 12.8%, 34.0%, 2.8%, and 50.4%. The generated spectrum could be used as preliminary data to generate the secondary structure of Dexras1 wt protein in further structural analysis.
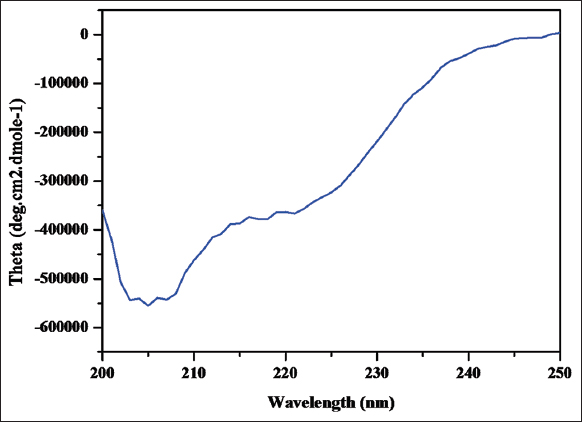 | Figure 12: CD Spectra of Dexras1 wt Protein: A spectra generated by CD Spectrometer(BioSpectrometer Basic 230 V/50–60 Hz, Eppendorf) by Reed’s Reference showing the curve representing helix, turns, and random coils. [Click here to view] |
4. CONCLUSION
Various physiological processes’ signaling is triggered by Dexras1. To know the exact molecular mechanism, structural analysis of Dexras1 is important, but the information about its structure is rather limited. The present study suggests that Dexras1protein is being expressed as insoluble fractions. This is expected because the C-Terminus has a CAAX domain which is a prenylation site and the Dexras1 protein becomes membrane-bound once it is prenylated. Spectra obtained by CD outlines preliminary structural information that suggests the presence of alpha-helix and beta sheets in the entire protein, but it seems like the N terminus might primarily be a non-randomized domain that could have independent functions.
5. ACKNOWLEDGMENT
We are grateful to Dr. Krishna Mohan Poluri, Associate Prof. IIT Roorkee, for his kind support during experiments on sub-cloning expression and purification, and Dr. Brett A. Adams, Dept. of Biology, Utah State University, USA, for providing recombinant vector with complementary Dexras1 gene, Dr. Sheetal Bandhu PDF, IIT Delhi for her kind support and suggestions to use biophysical technique CD.
6. AUTHORS’ CONTRIBUTIONS
The authors confirm contribution to the paper as follows: study conception and design: Ashish Thapliyal, Navin Kumar; data collection, analysis, and interpretation of results Rashmi Verma, Ashish Thapliyal; and draft manuscript preparation: Rashmi Verma. Critical revision of manuscript: Ashsih Thapliyal, Rashmi Verma, Navin Kumar. All authors reviewed the results and approved the final version of the manuscript.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author, [AT], upon reasonable request.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Kemppainen RJ, Behrend EN. Dexamethasone rapidly induces a novel Ras superfamily member-related gene in AtT-20 cells. J Biol Chem 1998;273:3129-31. [CrossRef]
2. Kemppainen RJ, Cox E, Behrend EN, Brogan MD, Ammons JM. Identification of a glucocorticoid response element in the 3'-flanking region of the human Dexras1 gene. Biochim Biophys Acta 2003;1627:85-9. [CrossRef]
3. Tu Y, Wu C. Cloning, expression and characterization of a novel human Ras-related protein that is regulated by glucocorticoid hormone. Biochim Biophys Acta 1999;1489:452-6. [CrossRef]
4. Croix BS, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E,
5. Bernal J, Crespo P. Analysis of Rhes activation state and effector function. Methods Enzymol 2006;407:535-42. [CrossRef]
6. Agretti P, De Marco G, Pinchera A, Vitti P, Bernal J, Tonacchera M. Ras homolog enriched in striatum inhibits the functional activity of wild type thyrotropin, follicle-stimulating hormone, luteinizing hormone receptors and activating thyrotropin receptor mutations by altering their expression in COS-7 cells. J Endocrinol Invest 2007;30:279-84. [CrossRef]
7. Gilman AG. G proteins:Transducers of receptor-generated signals. Annu Rev Biochem 1987;56:615-49. [CrossRef]
8. Lefkowitz RJ. Seven transmembrane receptors:Something old, something new. Acta Physiol (Oxf) 2007;190:9-19. [CrossRef]
9. Cismowski MJ, Takesono A, Ma C, Lizano JS, Xie X, Fuernkranz H,
10. Graham TE, Key TA, Kilpatrick K, Dorin RI. Dexras1/AGS-1, a steroid hormone-induced guanosine triphosphate-binding protein, inhibits 3', 5'-cyclic adenosine monophosphate-stimulated secretion in AtT-20 corticotroph cells. Endocrinology 2001;142:2631-40. [CrossRef]
11. Bourne HR, Sanders DA, McCormick F. The GTPase superfamily:A conserved switch for diverse cell functions. Nature 1990;348:125-32. [CrossRef]
12. Bourne HR, Sanders DA, McCormick F. The GTPase superfamily:Conserved structure and molecular mechanism. Nature 1991;349:117-27. [CrossRef]
13. Stouten PF, Sander C, Wittinghofer A, Valencia A. How does the switch II region of G-domains work?FEBS Lett 1993;320:1-6. [CrossRef]
14. Valencia A, Chardin P, Wittinghofer A, Sander C. The Ras protein family:Evolutionary tree and role of conserved amino acids. Biochemistry 1991;30:4637-48. [CrossRef]
15. Villar KD, Dorin D, Sattler I, Urano J, Poullet P, Robinson N,
16. Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem 1996;271:5289-92. [CrossRef]
17. Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci 2005;118:843-6. [CrossRef]
18. Thapliyal A, Verma R, Kumar N. Small G proteins Dexras1 and RHES and their role in pathophysiological processes. Int J Cell Biol 2014;2014:308535. [CrossRef]
19. Rooney KL, Domar AD. The relationship between stress and infertility. Dialogues Clin Neurosci 2022;20:41-8. [CrossRef]
20. Wang S, Wang C, Wang W, Hao Q, Liu Y. High RASD1 transcript levels at diagnosis predicted poor survival in adult B-cell acute lymphoblastic leukemia patients. Leuk Res 2019;80:26-32. [CrossRef]
21. Zhu LJ, Ni HY, Chen R, Chang L, Shi HJ, Qiu D,
22. Song QQ, Rao Y, Tang GH, Sun ZH, Zhang JS, Huang ZS,
23. Likittrakulwong W, Poolprasert P, Roytrakul S. Morphological trait, molecular genetic evidence and proteomic determination of different chickens (
24. Verma R, Kumar N, Thapliyal A.
25. Zhang Y, Olsen DR, Nguyen KB, Olson PS, Rhodes ET, Mascarenhas D. Expression of eukaryotic proteins in soluble form in
26. Bhat SK, Purushothaman K, Rao AR, Kini KR. Cloning and expression of a GH11 xylanase from
27. Thapliyal A, Bannister RA, Hanks C, BA. The monomeric G proteins AGS1 and Rhes selectively influence Galphai-dependent signaling to modulate N-type (CaV2. 2) calcium channels. Am J Physiol Cell Physiol 2008;295:C1417-26. [CrossRef]
28. Sambrook J, Russel DW. Molecular Cloning, A laboratory Mannual. 3rd ed., Vol. 1. New York, United States:Cold Spring Harbor Press;2001.
29. Green R, Rogers EJ. Transformation of chemically competent
30. Sambrook J, Russel DW. Molecular Cloning, a Laboratory Manual. 4th ed., Vol. 1. New York:Cold Spring Harbor Press;2014.
31. Gupta P, Waheed SM, Bhatnagar R. Expression and purification of the recombinant protective antigen of
32. Dodero VI, Quirolo ZB, Sequeira MA. Biomolecular studies by circular dichroism. Front Biosci (Landmark Ed) 2011;16:61-73. https://doi.org/10.2741/3676 PMid:21196159