1. INTRODUCTION
Cowpea (Vigna unguiculata (L.) Walp.) is a necessary and universally adaptable nutritional legume raised for grain and fodder all over the world. It provides a range of meals as well as revenue and fodder for livestock [1,2]. With a mean yield of 7216.29 tons of dry seeds, the total area of cowpea plants grown in Egypt was estimated to be 1968 hectares [3].
Viral diseases are significantly reducing cowpea yields in Asia, Africa, and Latin America. Worldwide, more than 20 viruses have been reported to naturally infect cowpea, and some are seed-transmitted [4]. Bean common mosaic virus strain blackeye cowpea mosaic (BCMV-BICM) has been found to naturally infect cowpeas and to be transmitted through seed [5-7]. Seed-borne viruses have been disseminated to most cowpea-producing regions of the world through the exchange of seed [8]. In most cases, legume seeds infected by viral pathogens are considered the primary source of infection and the resulting field infection may rise to 100% within 2 months, depending on the abundance and activity of the vector [9].
The (BCMV-BICM, equivalent or another scientific name Blackeye cowpea mosaic virus (BICMV)) [10] and Cowpea aphid-borne mosaic virus (CABMV) are both economically significant and universal seed-borne viruses of cowpea. Analysis of BCMV-BICM coat protein composition [11] and sequencing of the coat protein gene [12] proved the close relationship of the BCMV to what was once considered a separate virus, BCMV-BICM. The identification of strains is supported by serology with strain-specific antibodies and sequence analysis [13,14].
BCMV-BICM, which infects cowpea, is one example of a seed-borne virus that can negatively affect germination, infect seedlings and harm mature plants. To characterize BCMV-BICM infecting cowpea plants, the current study used biological, serological, histological, cytological, and light and electron microscopy approaches. In addition, molecular analyses were used to establish the identification of the viral isolate and to compare the coat protein genes’ nucleotide sequences with some standard isolates recorded in GenBank.
2. MATERIALS AND METHODS
2.1. Isolation and Propagation of Virus Isolate
Leaf samples of naturally infected cowpea (V. unguiculata (L.) Walp.) plants exhibiting virus-like symptoms consisting of various types of mosaic or mottling, vein clearing, vein banding, blistering, necrosis, yellowing, and leaf malformation were obtained from the experimental farm of Giza Agricultural Research Center (ARC), Egypt with zigzag methods as described by Anderson [15] during 2018–2019 growing season.
The BCMV-BICM was maintained in the greenhouse. It was treated for identification in the subsequent experiments. Mechanical inoculation was carried out in the greenhouse to isolate and identify the causal agents as described by Noordam [16]. The inoculum was prepared to inoculate healthy test plants such as Cowpea cv. Kafer El Sheikh and Chenopodium amaranticolor. Inoculated seedlings were kept under observation in an insect-proof greenhouse.
The virus isolate was biologically purified by three consecutive transfers of single local lesions [17] formed on C. amaranticolor. Finally, a single local lesion was propagated in cowpea plants of cv. Kafer El Sheikh, which was then used as a source of the virus isolate.
2.2. Identification of Virus Isolate
2.2.1. Host range and symptomatology
Ten healthy 15-day-old seedlings each of twenty-four plant species and cultivars belonging to four different families were mechanically inoculated as described by Noordam [16], using young, systemically infected leaves of cowpea cv. Kafer El Sheikh for BCMV-BICM.
2.2.2. Modes of transmission
2.2.2.1. Aphid transmission
Apterous Aphids cracivora Koch identified in Plant Protection Res. Institute, ARC were reared on virus- free faba bean cv. Masr1 and starved for 1 h., then given 2 min acquisition access period on symptomatic cowpea leaves infected with BCMV-BICM. Viruliferous individuals were then transferred to ten healthy 7-day-old cowpea cv. Kafer El Sheikh seedlings (5 individuals/plant) for inoculation-feeding periods of 4–6 h., after which they were killed by spraying with insecticide (Actikil 50%). Plants in the control experiment received aphids from asymptomatic ones. Symptoms and the percentage of transmission were recorded for 30 days after inoculation.
2.2.2.2. Seed transmission
An experiment was carried out to study the transmission of the virus isolate through cowpea seeds. Cowpea seeds from the three cultivars tested (Tiba, Kafer El Sheikh, and Dokii 331) were seeded separately in 20-cm-diameter pots (5 seeds/pot) containing sterilized peat-sand-clay (1:1:1) mixture and grown under virus-free glasshouse compartments before inoculation. Two weeks later, the resultant seedlings were mechanically inoculated, either separately with the isolate or with healthy crude sap (the control). Virus-infected plants showing typical symptoms as well as healthy plants were left to mature. Subsamples composed of two hundred seeds, harvested from plants representing different treatments, were sown in clay pots, as mentioned. Grown seedlings were observed for symptom development for 30 days and tested by enzyme-linked immunosorbent assay (ELISA). The percentage of virus seed transmission was estimated in cowpea cultivars tested. Seeds from healthy plants were used as controls and received similar treatments.
2.2.2.3. Virus incidence in seed parts
Another experiment was carried out using a double antibody sandwich ELISA (DAS-ELISA) to assess the virus isolate infected seed parts of cowpea cv. Balady or not. One hundred cowpea seeds raised from infected plants with BCMV-BICM were soaked individually in water for 2 days, avoiding seed-to-seed contamination. The coats were removed manually, whereas individual embryos were aseptically dissected with a razor blade into two cotyledons and embryo axis, then they were washed and triturated separately with a mortar and pestle in seed antigen buffer (0.04 M KH2PO4, 0.46 M Na2HPO4, 0.14 M NaCl, 0.003 M NaN3, 0.003 M KCl, 0.05 [v/v] Tween 20, PH 7.5) at the rate of 1 mL for each coat and cotyledon sample and 0.5 mL for each embryo axis [5]. Each extract was used as an antigen in DAS- ELISA.
2.2.3. Serological diagnosis of the virus isolate
BCMV-BICM was tested by direct-ELISA [18] using obtainable antisera specific to CABMV, BCMV, Bean yellow mosaic virus (BYMV). Results were assessed by measurement of absorbance at 405 nm in a Vinskan ELISA reader. Reading greater than twice the value of healthy controls was considered positive.
2.2.4. Histological and cytological studies
2.2.4.1. Virus-induced inclusions by light microscopy
Epidermal strips obtained from the lower surface of healthy and systemically infected cowpea leaves with BCMV-BICM were treated with 5% Triton X-100 for 10 min [19]. Finally, the treated strips were examined under a light microscope (model Olympus Japan) and photographs were taken with an Olympus C-35AD-4 camera from Japan.VICZ, Germany, after being kept on glass slides.
2.2.4.2. Histological study
Semithin leaf sections were prepared to check the histological and cytological changes induced by BCMV-BICM infection in cowpea leaves. The midrib and small parts of the leaf blades of healthy and infected cowpea leaves were examined histologically. Semithin sections (1 m thick) were prepared using the Leica Ultra Cut UCT ultramicrotome and stained with toluidine blue for 90 s before being examined using the previous light microscope model M-200M at Cairo university research park faculty of agriculture (CURP).
2.2.4.3. Cytological studies
Ultra-thin sections of cowpea leaf cells were investigated to detect the effect of virus infection using the following procedure: the samples were fixed overnight in cold 2.5% glutaraldehyde prepared in 0.1 M potassium phosphate buffer (PH 7.4) and post-fixed in 1% osmium tetroxide (OSO4) in the same buffer for 3 hr. After staining overnight in 1% uranyl acetate, the specimens were dehydrated in the ethanol-acetone series and embedded in Spurr’s medium according to the method described by Rocchetta et al. [20] and adopted by Udayashankar et al. [2] and El-Banna et al. [21]. Ultrathin sections were cut with an ultramicrotome, stained with 5% uranyl acetate for 20 min, and stained with Reynolds’ lead citrate for 10 min. Images were recorded by a JEOL electron microscope (JEM-1400 TE, Japan), as mentioned above.
2.2.5. Particle morphology
To study the morphology of virus particles, a drop of clarified, extracted sap infected with BCMV-BICM as described by Noordam [16], was applied to carbon-reinforced, formvar-coated copper grids for 2 min. A drop of 2% potassium phosphotungstate (a negative stain), pH 6.5, was added. One minute later, the excess stain at the edge of the grid was drained with the aid of filter paper, and the stained grids were then examined in the electron microscope unit of the faculty of agriculture at Cairo University. At the candidate magnification, virus particles were photographed.
2.3. Molecular Detection and Characterization of BCMV-BICM
2.3.1. Reverse transcription-polymerase chain reaction (RT-PCR)
In order to detect BCMV-BICM, total RNA was extracted from collected cowpea cv. Dokii 331 infected and uninfected samples using the Total RNA Mini Kit plant (Geneaid, New Taipei City, Taiwan) following the manufacturer’s instructions and the purified RNA was used as a template for RT-PCR amplifications. Primers of BCMV (F-`3-CCA ATG GTT GAA AAT GCA AAG CC-5`) and (R- `5-CGA CGC GAG ATG CTA ACTG-3) El-kady et al. [22] were used to amplify 404bp of its coat protein gene.
2.3.2. One-step RT-PCR
One-step RT-PCR was performed using Verso TM one-step RT-PCR kit (Thermo Scientific) in a 25 μL reaction mixture containing 3 μL RNA sample, 12.5 μL of one-step PCR master mix (×2),1.5 μL 10 μM of each primer,0.25 μL Verso enzyme mix, 2.5 μL RT Enhancer and 3.75 μL of nuclease-free water. The reaction conditions were as follows: Synthesis of cDNA was done at 50°C for 15 min and denaturation at 95°C for 2 min followed by 35 cycles at 95°C for 1 min, 55°C for 1 min and 72°C for 1 min and final elongation step at 72°C for 10 min for BCMV. PCR products were loaded in 1% agarose gel with 100 bp DNA ladder (BIOMATIK) in ×0.5 TBE buffer, then visualized on 1% UV illumination using the Gel Documentation system (Gel Doc 2000, Bio-Rad, USA). RNA extractions from healthy plants were used as the negative control.
2.3.3. DNA Purification and sequencing
RT-PCR fragments were cut from the gel and purified using the Gel/PCR DNA Fragments Extraction Kit (Geneaid, New Taipei City, Taiwan) following the manufacturer’s instructions. Purified DNA was used for direct sequencing at the Macrogen Company (South Korea). Sequencing results for both viruses were analyzed using DNAMAN Sequence Analysis Software and compared with the isolate of BCMV available in GenBank.
3. RESULTS AND DISCUSSION
3.1. Isolation and Propagation of the Virus Isolate
From naturally infected cowpea plants grown in the experimental farm of Giza ARC, Egypt, one distinct virus isolate representing one different host and morphological virus particles were obtained. Infected plants exhibited the characteristic symptoms of BCMV-BICM, consisting of mosaic or mottling, green vein banding, vein chlorosis, vein yellowing, yellow spots, blistering and distortion of leaves. The virus isolate was distinguished by its susceptibility to infect the indicator test seedlings. The virus was identified based on host range, symptomatology, modes of transmission, serological typing, histological and cytological studies, as well as particle morphology. The identity of the virus isolate was further supported by molecular studies. The isolated virus after identification (as shown later) is BCMV-BICM (synonym, BICMV).
3.2. Identification of BCMV-BICM
3.2.1. Host range and symptomatology
The reactions of twenty-four plant species and cultivars belonging to four different families to virus infection are summarized in Table 1. Among twenty-four plant species and cultivars belonging to four families, it has a restricted host range and infects only eight species [Figure 1] and cultivars belonging to the Chenopodiaceae and Fabaceae. The virus caused local lesions in inoculated C. amaranticolor and Chenopodium quinoa. BCMV-BICM produced yellow local lesions on these hosts. Additionally, cowpea plants infected with BCMV-BICM developed chlorotic local lesions followed by vein clearing, vein banding, and mosaic or mottling. On the other hand, it did not infect pea and faba beans; it produced systemic symptoms only on bean cv. Contender. These BCMV-BICM results appear to be consistent with Hao et al. [23], UdayaShankar et al. [24] and Purcifull et al. [25]. It has been postulated that symptoms produced are dependent on the particular virus, the strain involved, the species and age of the plant, the time of the year, and environmental conditions [7].
Table 1: Reaction of selected host range mechanically inoculated with Bean common mosaic virus strain blackeye cowpea mosaic.
| Family species cultivar | English name | Reaction | |
|---|---|---|---|
| L | S | ||
| Chenopodiaceae | |||
| Ch. amaranticolor Cost and Reyn | Ghost foot | YLL | - |
| Ch. quinoa Wild | YLL | - | |
| Cucurbitaceae | |||
| C. sativus L. cv. Mayadeen | Cucumber | - | - |
| C. melo L. cv. Balady | Melon | - | - |
| C. pepo L. cv. Balady | Squash | - | - |
| Fabaceae | |||
| G. max L. cv. Giza 21 | Soybean | - | - |
| P. vulgaris L. cv. Giza 6 | Green bean | - | - |
| cv. Nebraska | - | - | |
| cv. Contender | - | M or Mo+VB | |
| cv. Mont calm | - | - | |
| cv. Bronco | - | - | |
| cv. Paulista | - | - | |
| cv. Karnak | - | - | |
| P. sativum L. cv. Master B | Pea | - | - |
| cv. Snow wind | - | - | |
| V. faba L. cv. Masr1 | Broadbean | - | - |
| cv. Nobaria | - | - | |
| V. unguiculata (L.) Walp cv. Kareem | Cowpea | CLL | VB+M or Mo |
| cv.Qaha1 | CLL | VB+M or Mo | |
| cv.Dokii331 | CLL | VB+M or Mo | |
| cv. Kafer El-Shiekhe1 | CLL | VB+M or Mo | |
| cv. Balady | CLL | VB+M or Mo | |
| Solanaceae | |||
| N. tabacum L. cv. White Burley | Tobacco | - | - |
| L. esculentum Mill cv. Nada | Tomato | - | - |
LL: Local lesions, S: Systemic infection, CLL: Chlorotic local lesions, NLL: Necrotic local lesions, VB: Vein banding, -: No symptoms, M: Mosaic, Mo: Mottle, YLL: Yellow local lesions, Ch. quinoa: Chenopodium quinoa, Ch. amaranticolor: Chenopodium amaranticolor, C. sativus: Cucumis sativus, C. melo: Cucumis melo, C. pepo: Cucurbita pepo, G. max: Glycine max, P. vulgaris: Phaseolus vulgaris, P. sativum: Pisum sativum, V. faba: Vicia faba, V. unguiculata: Vigna unguiculata, N. tabacum: Nicotiana tabacum, L. esculentum: Lycopersicon esculentum
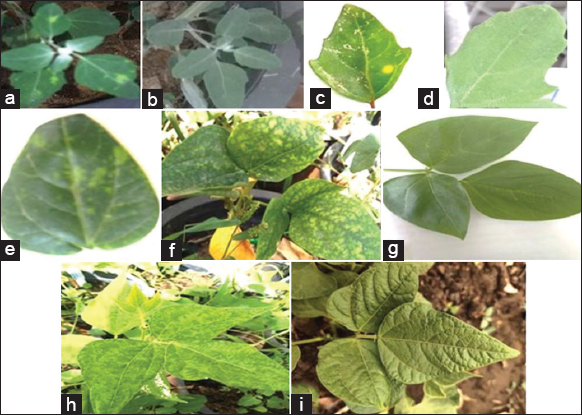 | Figure 1: Plants infected with BCMV-BICM: (a) yellow local lesion produced on either Chenopodium amaranticolor (b) healthy control or Chenopodium quinoa (c and d) Healthy plant. (e) chlorotic local lesions exhibited on cowpea cv. Kafer El Sheikh (cotyledons leaf) followed by systemic mottling and vein banding on trifoliate (f and g) Healthy one. (h) systemic mosaic and vein banding on bean cv. Contender and (i) Healthy plant. [Click here to view] |
3.2.2. Modes of transmission
3.2.2.1. Aphid transmission
Since aphids are important in virus transmission, A. craccivora was used to study the transmission of the virus isolated. The percentage of BCMV-BICM transmission was 40% [Table 2]. This is in agreement with Purcifull et al. [25], Wall et al. [26], and Dijkstra et al. [27]. The difference in the capability of the insect that transmits the virus may reflect a difference in the concentration of the virus in the source plant.
Table 2: Transmission of Bean common mosaic virus strain blackeye cowpea mosaic by Aphis craccivora Koch.
| Test plant | Virus isolate | Number of infected plants/number of inoculated plants | Infection (%) |
|---|---|---|---|
| Cowpea cv. Kafer El-Sheikh | BCMV-BICM | 4/10 | 40 |
BCMV-BICM: Bean common mosaic virus strain blackeye cowpea mosaic.
3.2.2.2. Seed transmission
Seed transmission is an extremely efficient method of introducing viruses into a crop at an early stage, resulting in randomized foci of infection throughout the plants. Subsequently, when some other method of transmission can spread the virus within a crop, In this work, trials have succeeded in transmitting the virus isolate through the seed of three cowpea cultivars. The percentage of seed transmission varied according to the virus isolate and cowpea cultivar. For BCMV-BICM, the percentage ranged from 5 to 8% [Table 3]. These results are in agreement with Hao et al. [23], UdayaShankar et al. [24], and Bashir et al. [28]. Diversity in viral seed-transmission rates would be a consequence of interactions involving host genotype, age of plant infection, viral isolate characteristics, plant vigor and nutrition, temperature, and possible virus-pathotype interaction [22,29,30].
Table 3: Seed transmission of Bean common mosaic virus strain blackeye cowpea mosaic by different cowpea cultivars.
| Cowpea cultivars | BCMV-BICM | |
|---|---|---|
| Ratio | Seed transmission (%) | |
| Tiba | 15/200 | 7.5 |
| Kafer EL-Sheikh | 10/200 | 5 |
| Dokii 331 | 16/200 | 8 |
BCMV-BICM: Bean common mosaic virus strain blackeye cowpea mosaic.
3.2.2.2.1. Virus incidence in seed parts
Results in Table 4 showed that BCMV-BICM infected the coat, cotyledons, and embryo axis of the tested cowpea cv. Blady [Figure 2]. The incidence of positive BCMV-BICM tests in these parts was recorded at 16, 3 and 3%, respectively. These results agree with the previous results obtained by Gillaspie et al. [5], who worked on BCMV-BICM. On the other hand, Sekar et al. [31] mentioned that infectious BCMV-BICM was present in cotyledons and embryo axis in mature seeds of two infected cowpea cultivars, whereas the virus was absent from the seed coats.
Table 4: Incidence of enzyme-linked immunosorbent assay-detectable bean common mosaic virus-blackeye cowpea mosaic in seed parts of infected cowpea tested cultivar.
| Seed parts | Number of infected/number of tested | Infection (%) |
|---|---|---|
| Coat | 16/100 | 16 |
| Cotyledons | 3/100 | 3 |
| Embryo axis | 3/100 | 3 |
 | Figure 2: (a) Seed parts (coat, cotyledon and embryo) of infected cowpea cv. Balady, (b) germinated seeds. [Click here to view] |
3.2.3. Serological diagnosis
Serological tests provide rapid and convenient methods for the identification and definition of plant viruses [30-32]. Positive serological reactions obtained with the virus isolate using direct ELISA indicated the identity of the virus. In the absence of a specific antiserum against BCMV-BICM, we used an antiserum specific to BCMV. It was found that our BCMV-BICM isolate reacted with BCMV antiserum. BCMV-BICM is serologically related to numerous other potyviruses [33], a circumstance that is common within the group. Lana et al. [10] reported that serological tests showed a close, but dissimilar relationship between BCMV and BCMV-BICM. In the description of BCMV-BICM (BICMV) [25], the fact that BCMV is not recorded among the viruses that cause similar symptoms in cowpea indicated that, when found in cowpea, BCMV is classified as BICMV [11].
3.2.4. Histological and cytological studies
3.2.4.1. Virus-induced inclusions by light microscopy
In the present study, amorphous cytoplasmic inclusion bodies induced by BCMV-BICM were observed in infected epidermal strips of cowpea cv. Dokii 331, 14–21 days after inoculation using light microscopy [Figure 3a], compared to healthy epidermal strips [Figure 3b]. Similar results for BCMV-BICM were obtained by Purcifull and Gonsalves [25], Allam et al. [34] and Badr et al. [35].
 | Figure 3: (a) Light microscopy of epidermal strips of cowpea cv. Dokii 331 leaf infected by BCMV-BICM,14 days post-inoculation showing amorphous inclusion bodies (AIB). (b) Healthy epidermal cells. N-Neculos. (X=350). [Click here to view] |
3.2.4.2. Histological study
Clear histological differences were identified between infected BCMV-BICM and healthy plants [Figure 4a and b] at this study point. These changes are mostly limited to the lowering of epidermal thickness, either in the upper or lower epidermal layers. Furthermore, these findings revealed a reduction in the midrib zone, palisade, and spongy tissues, leaf blade thickness, and the length and width of both protoxylem and metaxylem vessels. These findings are consistent with Badr et al. [35] on the BCMV and El-Abhar et al. [36] on the Alfalfa mosaic virus.
 | Figure 4: Infected tissues and healthy tissue (a), light microscopy of semi thin section of BCMV-BICM (b) UE: upper epidermis, LE: Lower epidermis, PM: Palisade mesophyll, SM: Spongy mesophyll, AS: Air space, VB: Vascular bandle (X: Xylem, Ph: Phloem), Co: Collenchyma, S: Stomata. (X=400), BS: bundle sheath cells [Click here to view] |
3.2.4.3. Cytological studies
In Figure 5, BCMV-BICM-infected plants showed alterations in host cells, either in cytoplasmic components or cell organelles, in ultrathin sections. The cell wall, chloroplasts, mitochondria, nucleus, and virus-related inclusion materials are among the abnormalities. The mesophyll tissue of healthy cowpeas was rich in chloroplasts with normal parenchyma cell organization [Figure 5a] and healthy Phloem tissue [Figure 5j], but in BCMV-BICM-infected cultivars, the cell wall was changed and became wrinkled, and the parenchyma cells appeared destructed and arranged irregularly [Figure 5e]. The findings are consistent with Khalil et al. [37], El Banna et al. [38] and Rizk et al. [39]. The cell wall was thicker than it should have been [Figure 5f]. In infected cells, the chloroplasts become spherical and clump together with irregular rows of grana [Figure 5c], whereas the latter becomes disordered and degenerate [Figure 5g]. These findings coincide with Badr et al. [35]. The presence of cytoplasmic cylindrical inclusions such as pinwheels, scrolls, and laminated aggregates is visible [Figure 5b and c]. In epidermal cells and trichomes, the cytoplasmic inclusion bodies were predominant in the parenchyma cells adjacent to the epidermis. Infected cells had typical pinwheels, scrolls [Figure 5d and h], laminated clumps, and endoplasmic reticulum in their cytoplasm. Pinwheel inclusions in the cytoplasm of infected cells are produced by all potyviruses that have been investigated so far by Dougherty and Carrington [40] and Bello et al. [41]. Pinwheels are constructed at the plasmalemma and subsequently moved into the cytoplasm, according to Lawson et al. [42] and Alexandre et al. [43]. The most useful instrument for investigating plant cell ultrastructure as well as direct virus particle detection and its influence on cell organelles [Figure 5i], which are strongly related to the kind of plant-host response to viral infection, this is in agreement with Otulak et al. [44] and Rizk et al. [39], is transmission electron microscopy.
 | Figure 5: (a-j) Ultrastructure of cowpea leaf tissues infected with BCMV-BICM compared with healthy ones. CW: Cell Wall, Ch: Chloroplasts, S: Stroma, G: Grana, Nu: Nucleus, NM: Nuclear Membrane, C: Cytoplasm, CM: Cytoplasm Membrane, PW: Pinwheels, PX: Peroxisomes, M: Mitochondria, Sc: Scrolls, VP: Virus Particles, Va: Vacuoles. [Click here to view] |
3.2.4.4. Particle morphology
For examination of the virus particles in crude extracts or purified preparations, a negative-staining procedure is now used extensively [45,46]. In the present investigation, examination of a clarified infected extract with BCMV-BICM negatively stained with 2% phosphotungstic acid using an electron microscope revealed filamentous viral particles 720–750 nm in length [Figure 6], typical of the Potyvirus group [23,41,47,48].
 | Figure 6: Filamentous flexuous virus particles of BCMV-BICM ranged from 720 to 750 nm in length (Magnification X 50.000). [Click here to view] |
3.3. Molecular Detection and Characterization of BCMV-BICM
3.3.1. Reverse transcription-polymerase chain reaction (RT-PCR)
The BCMV-BICM isolate was further identified by RT-PCR amplification using specific primers for its coat protein genes. The BCMV-BICM isolate was amplified by RT-PCR from fresh cowpea-infected plants, but no products were amplified from RNA extracted from asymptomatic leaf samples when subjected to agarose electrophoresis [Figure 7]. These results for BCMV, cowpea isolate were noted by El-kady et al. [22] and Dada et al. [49].
 | Figure 7: Agarose gel electrophoresis of RT-PCR amplified products. M: 100 bp DNA ladder (BIOMATIK); L 1, 2, 3, 4: Infected leaf samples with BCMV; L5: 1: Healthy sample. [Click here to view] |
3.3.2. DNA purification and sequence analysis
The amplified products for the virus isolate were gel purified and sequenced using DNAMAN Sequence Analysis Software, and the data were evaluated for a consensus sequence and submitted to the GenBank database for BCMV-BICM (OM 891556 seq.). The phylogenetic relationships [Figure 8] were determined. Blast analysis of the coat protein gene of the BCMV cowpea isolates revealed the highest (98.9%) nucleotide identity with BCMV-BICM isolates from Vietnam (DQ925423.1), followed by 98.4% identity with India (MH795801), and 98.1% identity with Vietnam (DQ925421), compared with lower (96.5%) identity with China (AJ312437.1) and India (AJ312438.1) isolates [Table 5]. The close relationship of BCMV-BICM to a previously unknown virus, BICMV demonstrated by Higgins et al. [12], McKern et al. [13] and UdayaShankar et al. [24].
 | Figure 8: The phylogenetic relationships tree among the multiple sequence alignments of the Egyption isolate of BCMV-BICM and other isolates referenced in GenBank. [Click here to view] |
Table 5: Comparison of the nucleotide sequences of the bean common mosaic virus strain blackeye cowpea mosaic coat protein gene isolated in the present study with the corresponding sequences of other isolates available in the Genbank.
| Isolate name | Accession Number | Country | Original Host Plant | Identity% |
|---|---|---|---|---|
| BCMV-BlCVN/BB2-6 | DQ925423.1.seq | Vietnam | Black bean (P. vulgaris) | 98.9 |
| DH-C | MH795801.1.seq | India | V.unguiculata | 98.4 |
| BCMV-BlC-VN/RB2 | DQ925421.1.seq | Vietnam | Red bean (P. vulgaris) | 98.1 |
| BCMV-BlC-VN/RB1 | DQ925420.1.seq | Vietnam | Red bean (P. vulgaris) | 97.8 |
| BCMV-BlC-VN/BB1 | DQ925417.1.seq | Vietnam | Black bean (P. vulgaris) | 97.8 |
| BICMV | AY575773.1.seq | Taiwan | Not available | 97.8 |
| BICMV | AF395678.1.seq | China | Not available | 97.8 |
| Mysore-BCMV strain blackeye cowpea mosaic | HQ864644.1.seq | India | V. mungo cv. LBG-623 | 97.8 |
| BCMV Hempur strain blackeye cowpea mosaic | HQ864645.1.seq | Moscow | V. mungo cv. PU-35 | 97.8 |
| BICMV | Y17823.1.seq | Florida | Cowpea | 97.5 |
| Y strain: “blackeye cowpea mosaic” | AJ312438.1.seq | China | Cowpea | 96.5 |
| R strain: “blackeye cowpea mosaic” | AJ312437.1.seq | China | Cowpea | 96.5 |
| BCMV-BlC-VN/YB1 | DQ925424.1.seq | Vietnam | Yard long bean (V. unguiculata) | 96.2 |
| BCMV-MB | KC832502.1.seq | China | M. bean | 96.2 |
| BCMV-MS1 | EU761198.1.seq | Australia | M. atropurpureum | 95.9 |
| BCMV Northern Western Australia 1 | AH015028.2.seq | Australia | Not available | 95.6 |
| BCMV NY15 | AF083559.1.seq | USA | Not available | 95.4 |
| BCMV-NL4 | L21766.1.seq | Not available | Not available | 95.1 |
| BCMV-PR1 | L21767.1.seq | Puerto Rico | Not available | 95.1 |
| Mexican BCMV | L11890.1.seq | Mexican | Not available | 92.1 |
P. vulgaris: Phaseolus vulgaris, M. atropurpureum: Macroptilium atropurpureum, M. bean: Mung bean, V. unguiculata: Vigna unguiculata, V. mungo: Vigna mungo, BICMV: Blackeye cowpea mosaic virus, BCMV: Bean common mosaic virus, BCMV-BICM: Bean common mosaic virus strain blackeye cowpea mosaic.
4. CONCLUSION
Characterization of BCMV-BICM was achieved by traditional identification methods and by RT-PCR. Since the virus isolates are transmitted through cowpea seeds and spread naturally by aphids, which can cause potentially high yield losses, it is necessary to produce virus-free seeds and establish certification regulations for legumes to protect growers from infected stocks. This is the first report of a BCMV-BICM isolate found spontaneously in Egyptian cowpea plants.
5. ACKNOWLEDGMENT
The authors are thankful to the Plant Pathology Research Institute, Agricultural Research Center, Giza, Egypt, for providing, Serological and Molecular Laboratory, and greenhouse facility.
6. AUTHORS’ CONTRIBUTIONS
All authors contributed to conception and design, data collection, analysis, and interpretation, authoring and editing the article for intellectual content, agreeing to submit to the journal and approving the final version, and agreeing to be accountable for all elements of the work.
7. CONFLICTS OF INTEREST
There are no conflicts of interest that the authors can disclose with regard to this article.
8. FUNDING
There is no funding to report.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
The data that support the findings of this study are openly available in [repository name] at [URL], reference number [reference number].
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Popelka JC, Terryn N, Higgins TJ. Gene technology for grain legumes:Can it contribute to the food challenge in developing countries?Plant Sci 2004;167:195-206. [CrossRef]
2. Udayashankar AC, Nayaka SC, Kumar HB, Mortensen CN, Shetty HS, Prakash HS. Establishing inoculum threshold levels for bean common mosaic virus strain blackeye cowpea mosaic infection in cowpea seed. Afr J Biotechnol 2010;9:8958-69.
3. FAOSTAT Statistical Databases. Food and Agriculture Organization of the United Nations;2021. Available from:https://www.fao.org/faostat/en/#data/QC
4. Mali VR, Thottappilly G. Virus diseases of cowpea in the tropics. Rev Trop Plant Pathol 1986;34:421-522.
5. Gillaspie AG Jr., Hopkins MS, Pinnow DL. Relationship of cowpea seed-part infection and seed transmission of blackeye cowpea mosaic Potyvirus in cowpea. Plant Dis 1993;77:875-7. [CrossRef]
6. Abdullahi I, Ikotun T, Winter S, Thottappilly G, Atiri GI. Investigation on seed transmission of Cucumber mosaic virus in cowpea. Afr Crop Sci J 2001;9:677-84. [CrossRef]
7. Aliyu TH, Balogun OS, Kumar L. Survey of the symptoms and viruses associated with cowpea (Vigna unguiculata (L).) in the Agroecological zones of Kwara state, Nigeria. Ethiop J Environ Stud Manag 2012;5:613-9. [CrossRef]
8. Hampton RO, Thottappilly G, Rossel HW. Viral diseases of cowpea and their control by resistance-conferring genes. Advances in Cowpea Research. 1997:159.
9. Bos L, Makkouk KM. Plant viruses as pests of quarantine significance in seed. In:Quarantine For Seed Fao. Vol. 119. New Delhi:Daya Publishing House;1997. 133.
10. Lana AF, Lohuis H, Bos L, Dijkstra J. Relationships among strains of bean common mosaic virus and blackeye cowpea mosaic virus-members of the Potyvirus group. Ann Appl Biol 1988;113:493-505. [CrossRef]
11. McKern NM, Shukla DD, Barnett OW, Vetten HJ, Dijkstra J, Whittaker LW, et al. Coat protein properties suggest that azuki bean mosaic virus, blackeyecowpea mosaic virus, peanut stripe virus, and three isolates from soybean are all strains of the same Potyvirus. Intervirology 1992a;33:121-34. [CrossRef]
12. Higgins CM, Cassidy BG, Teycheney PY, Wongkaew S, Dietzgen RG. Sequences of the coat protein gene of five peanut stripe virus (PStV) strains from Thailand and their evolutionary relationship with other bean common mosaic virus sequences. Arch Virol 1998;143:1655-67. [CrossRef]
13. McKern NM, Mink GI, Barnett OW, Mishra A, Whittaker LA, Silbernagel MJ, et al. Isolates of bean common mosaic virus comprising two distinct potyviruses. Phytopathology 1992b;82:923-9. [CrossRef]
14. Khan JA, Lohuis D, Goldbach R, Dijkstra J. Sequence data to settle the taxonomic position of bean common mosaic virus and blackeye cowpea mosaic virus isolates. J Gen Virol 1993;74:2243-9. [CrossRef]
15. Anderson LC. Collecting and preparing plant specimens and producing an herbarium. J Dep Biol Sci 1999;20:295-300.
16. Noordam D. Identification of plant Viruses. Methods and Experiments. Wageningen, Netherlands:Centre for Agricultural Publishing and Documentation;1973.
17. Kuhn CW. Separation of cowpea virus mixtures. Phytopathology 1964;54:739-40.
18. Clark MF, Adams AN. Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol 1977;34:475-83. [CrossRef]
19. Christie RG, Edward JR. Light microscopic techniques. Plant Dis 1986;70:273-9. [CrossRef]
20. Rocchetta I, Leonardi PI, Filho GM, de Molina MD, Conforti V. Ultrastructure and X-ray microanalysis of Euglena gracilis (Euglenophyta) under chromium stress. Phycologia 2007;46:300-6. [CrossRef]
21. El-Banna OH, Mikhail M, El-Attar A, Aljamali AA. Molecular and electron microscope evidence for an association of phytoplasma with Sesame phyllody in Egypt. Egypt J Phytopathol 2013;41:1-4. [CrossRef]
22. El-Kady MA, Badr AB, El-Attar AK, Waziri HM, Saker KE. Characterization and molecular studies of bean common mosaic virus isolated from bean plants in Egypt. Egypt J Virol 2014;11:124-35.
23. Hao NB, Albrechtsen SE, Nicolaisen M. Detection and identification of the blackeye cowpea mosaic strain of bean common mosaic virus in seeds of Vigna unguiculata sspp. from North Vietnam. Aust Plant Pathol 2003;32:505-9. [CrossRef]
24. Udayashankar AC, Nayaka CS, Kumar HB, Shetty HS, Prakash HS. Detection and identification of the blackeye cowpea mosaic strain of bean common mosaic virus in seeds of cowpea from southern India. Phytoparasitica 2009;37:283-93. [CrossRef]
25. Purcifull D, Gonsalves D. Blackeye cowpea mosaic virus. In:CMI/AAB Descriptions of Plant Viruses. Wellesbourne, UK:Association of Applied Biology;1985. 305.
26. Wall GC, Kimmons CA, Wiecko AT, Richardson J. Blackeye cowpea mosaic virus (BICMV) in yard-long bean the Mariana islands. Micronesica 1996;29:101-1. [CrossRef]
27. Dijkstra J, Bos L, Bouwmeester HJ, Hadiastono T, Lohuis H. Identification of blackeye cowpea mosaic virus from germplasm of yard-long bean and from soybean, and the relationships between blackeye cowpea mosaic virus and cowpea aphid-borne mosaic virus. Netherlands J Plant Pathol 1987;93:115-33. [CrossRef]
28. Bashir M, Hampton RO. Detection and identification of seed-borne viruses from cowpea (Vigna unguiculata (L.) Walp.) germplasm. Plant Pathol 1996;45:54-8. [CrossRef]
29. Hampton RO. Incidence of the lentil strain of pea seedborne mosaic virus as a contaminant of Lens culinaris germ plasm. Phytopathology 1982;72:695-8. [CrossRef]
30. Kohnen PD, Johansen IE, Hampton RO. Characterization and molecular detection of the P4 pathotype of pea seedborne mosaic potyvirus. Phytopathology 1995;85:789-93. [CrossRef]
31. Sekar R, Sulochana CB. Seed transmission of blackeye cowpea mosaic virus in two cowpea varieties. Curr Sci 1988;57:37-8.
32. Matthews RE. Fundamentals of Plant Virology. United States:Academic Press;2012.
33. Lima JA, Purcifull DE, Hiebert E. Purification, partial characterization, and serology of blackeye cowpea mosaic virus. Phytopathology 1979;69:1252-8. [CrossRef]
34. Allam EK, El-Kady MA, Eid SA, Abdel-Salam AM, Kishtah AA. Purification of Bean yellow mosaic virus. Egypt J Phytopathol 1986;18:59-64.
35. Badr AB, El-Kady MA, Saker KE. Anatomical and cytological changes in bean leaf cells infected with bean common mosaic virus. Egypt J Virol 2014;11:150-8.
36. El-Abhar MA, Elkady MA, Ghanem KM, Bosila HA. Identification, characterization and ultrastructure aspects of Alfalfa mosaic virus infecting potato (Solanum tuberosum L.). J Virol Sci 2018;3:68-77.
37. Khalil RR, Bassiouny FM, El Dougdoug KA, Abo-Elmaty S, Yousef MS. A dramatic physiological and anatomical changes of tomato plants infecting with Tomato yellow leaf curl germinivirus. Int J Agric Technol 2014;10:1213-29.
38. El Banna OH, Hassan AA, Hamed AH. Biological, cytopathological and molecular studies of Potato virus Y isolated from pepper grown under greenhouse conditions in Egypt. Int J Sci Eng Res 2015;6:1281-9.
39. Rizk MN, Ketta HA, Shabana YM. Ultrastructure of cytopathological responses in Solanum tuberosum cv. Spounta infected with necrotic strain of Potato virus Y. Arch Phytopathol Plant Protect 2021;54:1102-20. [CrossRef]
40. Dougherty WG, Carrington JC. Expression and function of potyviral gene products. Ann Rev Phytopathol 1988;26:123-43. [CrossRef]
41. Bello VH, Lorenzi HJ, Rezende JA, Kitajima EW. Bean common mosaic virus and Cucumber mosaic virus naturally infecting Aristolochia spp. plants in Brazil. J Phytopathol 2023;171:57-61. [CrossRef]
42. Lawson RH, Hearon SS, Smith FF. Development of pinwheel inclusions associated with sweet potato russet crack virus. Virology 1971;46:453-63. [CrossRef]
43. Alexandre MA, Duarte LM, Chaves AL, Kitajima EW. Potyvirus infecting ornamental plants grown in the Neotropical region. In:Virus Diseases of Ornamental Plants:Characterization, Identification, Diagnosis and Management. Germany:Springer;2021. 23-59. [CrossRef]
44. Otulak K, Kozie?E, Garbaczewska G. Seeing is believing. The use of light, fluorescent and transmission electron microscopy in the observation of pathological changes during different plant virus interactions. In:Mendez-Vilas A, editor. Microscopy:Advances in Scientific Research and Education. 6th ed. Badajoz, Spain, Formatex Research Center;2014. 367-76.
45. Matthews RE, Hull R. Matthews'Plant Virology. Houston, Texas:Gulf Professional Publishing;2002.
46. Li Q, Zhang B, Hu J, Zhang L, Ji P, Dong J. Complete genome sequence of polygonatum mosaic-associated virus 1, a novel member of the genus Potyvirus in China. Arch Virol 2023;168:42. [CrossRef]
47. Murphy JF, Barnett OW, Witcher W. A Blackeye cowpea mosaic-virus strain from South-Carolina. Phytopathology 1984;74:631.
48. Udayashankar AC, Nayaka CS, Archana B, Nayak U, Niranjana SR, Prakash HS. Strobilurins seed treatment enhances resistance of common bean against bean common mosaic virus. J Phytopathol 2012;160:710-6. [CrossRef]
49. Dada AO, Oresanya A, Akinyosoye ST, Arogundade O. The first report of bean common mosaic virus (BCMV) infection of African yam bean (Sphenostylis stenocarpa) in Nigeria. Mol Biol Rep 2022;49:10133-6. [CrossRef]