1. INTRODUCTION
Colon cancer (CC) is the second- and third-most common cancer in women and men, respectively. The pathology of CC is largely influenced by hereditary [1]. The steady accumulation of genetic and epigenetic alterations that activate oncogenes and inactivate tumor suppressor genes is a characteristic of cancer. These modifications lead to malignant transformation of colon cells through repeated cycles of clonal proliferation that select for cells with aggressive behavior [2,3]. According to one prevalent hypothesis, a common therapy holds that a stem cell or a cell that resembles a stem cell that is found close to the base of the colon crypts is responsible for the majority of colon cancers [4]. Mutations may occur in oncogenes, tumor suppressor genes, and DNA repair system genes [5].
Despite starling evidence in cancer biology over the past few decades, treatment for the vast majority of patients remains a pipe dream. This unsatisfactory situation, which can lead to chemotherapy failure and a fatal prognosis for patients, is largely caused by the development of drug resistance. Drug resistance is a critical problem for not just well-known cytotoxic medications but also to more recent finest molecules and treatment antibodies that specifically target tumor cell locations [6]. Consequently, the race to discover and develop new medications is still on. Drug resistance is often complex and multiple pathways may contribute to tumor insensitivity to drugs [7].
Scopoletin (Sc) is found in a variety of medicinal plants, including species of the genus Scopolia [8]. It is known to be extremely harmful to tumor cells [9]. It exhibits anti-inflammatory and antioxidant properties and activates programmed cell death and autophagy [10]. It also has antidepressant, anti-thyroid, anti-hyperglycemic, hypouricemic, neuroprotective, and endocrine activities [11,12]. Recent studies on Sc have shown that some Sc derivatives possess potent antitumor properties [13].
Network pharmacology explores the complex connections between factors, diseases, and targets by combining systems biology, pharmacology, and computer analysis technologies. It helps us analyze the mechanism of action of Sc in the treatment of diseases by analyzing components and disease-related targets and providing biological processes and pathways through which Sc may play a role. Through an integrated system of bioinformatic analysis, the current experimental study aims to predict the possible role of Sc in the prevention and control of colon cancer. Data were obtained from Gene-disease associations (DisGeNET) and Comparative Toxicogenomics Database [CTD], which were subsequently analyzed to obtain intersection genes and assessed to diagnose major genes associated with colon cancer. The process map of the experiment is shown in Figure 1.
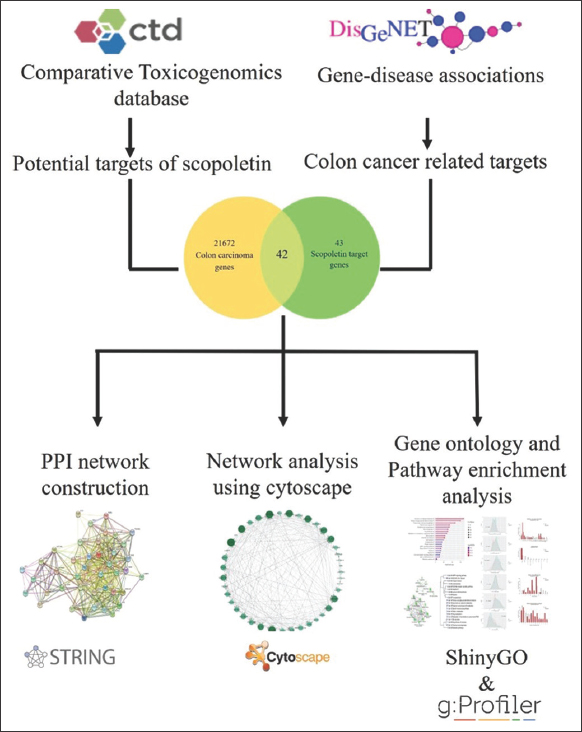 | Figure 1: Systematic representation of the experimental study. [Click here to view] |
2. MATERIALS AND METHODS
2.1. Source and Search Strategy for Data Collection
CTD (http://ctdbase.org/) was used to find Sc’s target genes by inputting “Sc” into the search box [14]. Some other Sc-target genes were selected from previous literature studies [15-22]. The terms “Colon carcinoma” and “colon cancer” were used in a database called DisGeNET (http://www.disgenet.org/). The above database serves to find the potential target genes for colon cancer and chronic inflammation.
2.2. Protein-Protein Interaction (PPI) Analysis
To create the PPI study, the intersection of genes was obtained by first mapping Sc-targeted genes to colon carcinoma genes. Crossover genes have been used as potential targets of Sc in the treatment of colon cancer. Using specified criteria, PPI assessment was carried out. The following are the entries to be done in the database, Enter “official gene symbol” into the “Multiple proteins” field, with organism “Homo sapiens” with a medium confidence score of above 0.4 in the string database [https://string-db.org/] [23].
2.3. Network Analysis
All of the PPI data were processed through the Cytoscape (https://cytoscape.org/) [24]. A network analyzer tool was employed for performing multicomponent and multi-target effects for Scopoletin. It analyzes biological pathways and determines network topology metrics such as network dimension, count of typical neighbor node, and number of connected node pairs. In addition, it calculates the distribution of more complex network parameters such as shortest path length, average clustering coefficient, and node degree.
2.4. Identification of Hub Genes
Hub nodes are those with higher degree, closeness, and betweenness scores than the mean, which are determined. The Maximal Clique Centrality (MCC) algorithm has been shown to be a successful tool for identifying hub nodes in a coexpression network. Using CytoHubba and Centiscape 2.2, a Cytoscape plugin, the MCC of each node was determined. The top 10 MCC values are used to classify genes as hub genes [25].
2.5. Identification and Validation of Clusters
Subclusters from densely connected clusters and functional units in PPI networks were identified using the Molecular Complex Detection (MCODE) in plugin accessible in the Cytoscape [26]. MCODE scores >5, degree cutoff = 2, max depth = 100, and k-score = 5 were the selection criteria.
2.6. Gene ontology (GO) Analysis of the Hub Genes
ShinyGOv0.6 was used to determine GO of potential targets (http://bioinformatics.sdstate.edu/go/). All the intersection genes were inputted in the search box and the best matching species was given as “Homo sapiens” with P-value (FDR) cutoff of 0.05 [27].
2.7. Pathway Enrichment Analysis of the Hub Genes
To identify and collect the networks connected to target genes, gProfiler was used for pathway enrichment analysis (https://biit.cs.ut.ee/gprofiler/gost) [28]. Put all the intersection genes in the gene box input list with a threshold value of 0.05, mark the ordered query, organism – Homo sapiens, and run the query.
3. RESULTS
3.1. Data Acquisition and PPI Network
A total of 2833 colon cancer genes were generated from the DisGeNET database [Figure 2a]. A total of 51 Sc-targeted genes were selected. Out of which, 28 genes (ACACA, ACOX1, ACSL1, ADIPOQ, ADIPOR2, CIDEA, FASN, GPT, IL10, IL6, LIPE, MYD88, PNPLA2, SREBF1, TICAM1, TLR4, TNF, UGT1A1, UGT1A10, UGT1A3, UGT1A6, UGT1A7, UGT1A8, UGT1A9, UGT2B15, and XPC) are collected from CTD database, and the remaining 23 genes (BCL2, BCL2L1, BCL2L7, CALB2, CAS3, CDH17, EGFR, GPA33, HMOX1, NOS2, KEAP1, LAMP3, MAPK1LC3A, MKI67, MUC2, NFE2L2, TP53, PTGS2, SIRT1, SQSTM1, STAT1, TLR2, and VEGF) were selected from previous literature. Fifty intersection genes were obtained by mapping Sc-targeted genes to colon cancer genes. As shown in Figure 2a, all 50 intersection genes were used as prospective Sc targets against colon cancer. The STRING software was used to examine the physical and functional relationships between the genes and 50 intersecting genes were performed to construct the PPI network. The findings revealed that the network has 49 nodes and 551 edges. Figure 2b shows an average node degree of 22.5, an average local clustering coefficient of 0.776, and a predicted number of 204 edges, with a medium confidence of 0.4 for its interaction score.
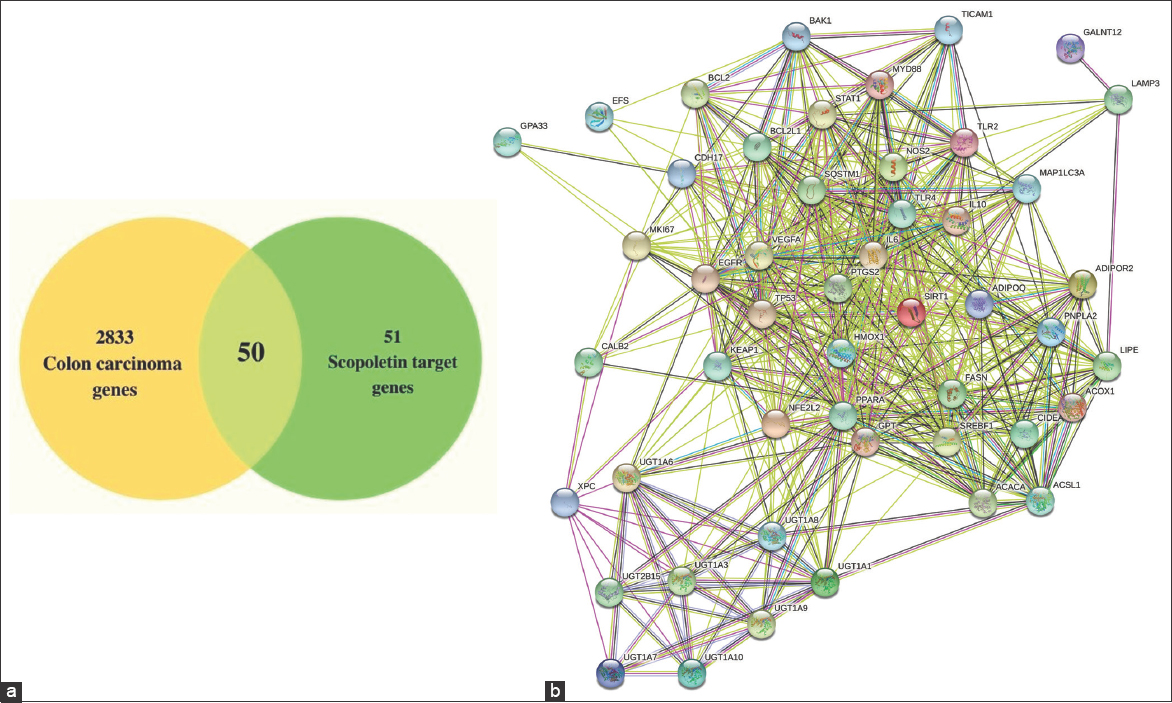 | Figure 2: (a) Venn diagram depicting Scopoletins (Sc) interaction targets with colon cancer (b) Protein–protein interactions intersection target network for Sc-treated colon cancer (medium confidence score - 0.4). [Click here to view] |
3.2. Network Analysis and Localization of Hub Genes
According to the analysis, the tightly packed network contains 49 nodes and 551 edges. Other statistics include network diameter = 4, network radius = 2, characteristic path length = 1.618, clustering coefficient = 0.756, network density = 0. 469, network heterogeneity = 0.459, and network centralization = 0359. Figure 3 shows the top-ranked genes along with other parameters.
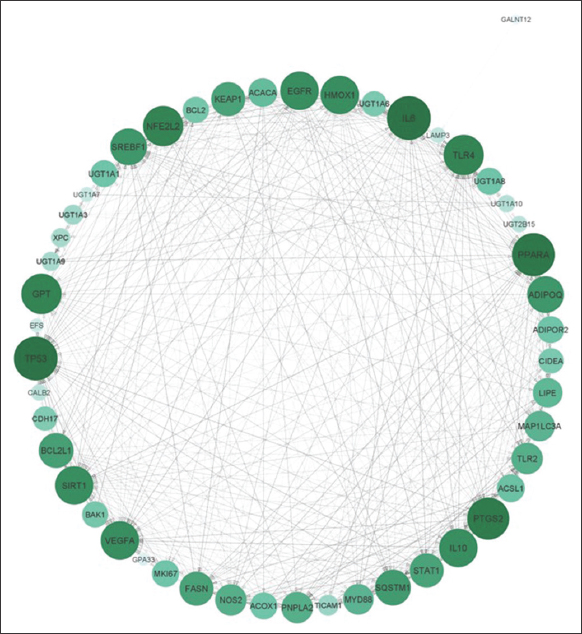 | Figure 3: Protein–protein interactions network analysis using Cytoscape. The software called Cytoscape is used to retrieve the nodes and edges from the STRING software and display them. Gene names are represented as nodes in various hues of green, while interactions are represented as grey edges. The size and color of the circles represent the node degree value, while the thickness of the border represents the total score between two nodes. Degree increases with node size. [Click here to view] |
3.3. Identification of Hub Genes
The IL6, PTGS2, TP53, SIRT1, PPARA, HMOX1, NFE2L2, SQSTM1, ADIPOQ, GPT, SREBF1, IL10, TLR4, VEGF, EGFR, BCL2L1, KEAP1, STAT1, MYD88, and TLR2 are the 20 top genes, indicating that their important roles in the network are described in the Supplementary File (S1). Higher the node size indicates higher the degree-ranked targets called the hub genes. The hub gene, as determined by the MCC scores, is shown in Table 1 as the gene that is connected to the key module.
Table 1: Genes are ranked depending on MCC scores.
| Rank | Name | MCC Score |
|---|---|---|
| 1 | IL6 | 5.12E+18 |
| 2 | PTGS2 | 5.12E+18 |
| 3 | TP53 | 5.12E+18 |
| 4 | SIRT1 | 5.12E+18 |
| 5 | PPARA | 5.12E+18 |
| 6 | HMOX1 | 5.12E+18 |
| 7 | NFE2L2 | 5.12E+18 |
| 8 | SQSTM1 | 5.12E+18 |
| 9 | ADIPOQ | 5.12E+18 |
| 10 | GPT | 5.12E+18 |
| 11 | SREBF1 | 5.12E+18 |
| 12 | IL10 | 5.12E+18 |
| 13 | TLR4 | 5.12E+18 |
| 14 | VEGFA | 5.12E+18 |
| 15 | EGFR | 5.12E+18 |
| 16 | BCL2L1 | 5.12E+18 |
| 17 | KEAP1 | 5.11E+18 |
| 18 | STAT1 | 5.11E+18 |
| 19 | MYD88 | 4.87E+18 |
| 20 | TLR2 | 4.87E+18 |
| 21 | MAP1LC3A | 2.56E+18 |
| 22 | NOS2 | 2.55E+18 |
| 23 | FASN | 2.50E+17 |
| 24 | PNPLA2 | 6.83E+15 |
| 25 | LIPE | 4.09E+14 |
| 26 | ACACA | 9.42E+13 |
| 27 | ACOX1 | 2.75E+13 |
| 28 | BCL2 | 2.09E+13 |
| 29 | ADIPOR2 | 9.15E+12 |
| 30 | ACSL1 | 6.54E+12 |
| 31 | CIDEA | 2.62E+12 |
| 32 | MKI67 | 1.31E+12 |
| 33 | BAK1 | 9.44E+10 |
| 34 | CDH17 | 8.72E+10 |
| 35 | UGT1A8 | 1.44E+09 |
| 36 | UGT1A1 | 9.62E+08 |
| 37 | UGT1A6 | 4.79E+08 |
| 38 | TICAM1 | 4722480 |
| 39 | UGT1A3 | 17280 |
| 40 | UGT1A9 | 12240 |
| 41 | UGT1A10 | 6480 |
| 42 | XPC | 5790 |
| 43 | UGT2B15 | 5760 |
| 44 | UGT1A7 | 720 |
| 45 | CALB2 | 248 |
| 46 | LAMP3 | 145 |
| 47 | EFS | 48 |
| 48 | GPA33 | 4 |
| 49 | GALNT12 | 1 |
MCC: Maximal clique centrality.
3.4. Identification and Validation of Clusters
MCODE addon was utilized to investigate PPI network’s functional module, and six corresponding relevant modules were identified [Figure 4a]. Nuclear factor erythroid-related factor-2 (Nrf2) signaling pathway (Module 1), inflammatory pathway (Module 2), autophagy pathway (Module 3), apoptosis pathway (Module 4), cell proliferation signaling (Module 5), and insulin-sensitizing pathway (Module 6) are the tightly related modules. MCODE clustering recognizes heavily connected clusters from the entire network. MCODE clustering identifies highly connected clusters from across the network (MN). These clusters are visually depicted in Figure 4b-e. The detailed MCODE clusters link area is shown in tabular form [Table 2].
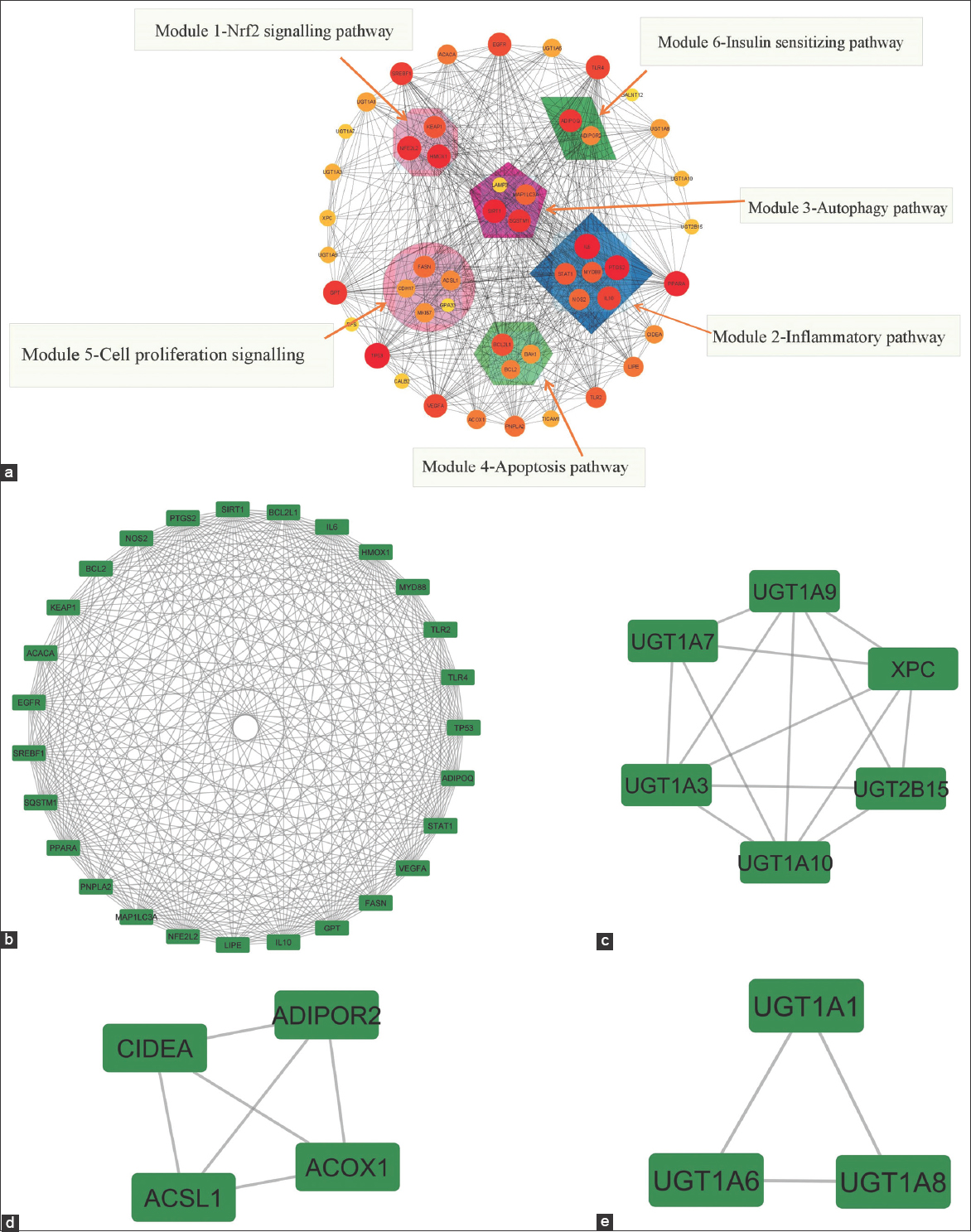 | Figure 4: (a) Interaction targets for module analysis. (Diameter of the circle-node degree; thickness of the edges-combined two node scores). With the use of MCODE plugin within cytoscape, four subnetworks from the protein-protein interactions analysis with a cluster score over three were determined. The margins are gray, whereas the nodes are emphasized in green. (b) MN Cluster-1, (c) MN Cluster-2, (d) MN Cluster-3, and (e) MN Cluster-4. [Click here to view] |
Table 2: The most interlinked regions were clustered from intersecting gene targets using MCODE.
| Cluster | Score | Number of nodes | Number of edges | Node IDs |
|---|---|---|---|---|
| 1 | 24.538 | 27 | 319 | STAT1, EGFR, PPARA, TP53, KEAP1, ACACA, NFE2L2, LIPE, GPT, HMOX1, SQSTM1, SIRT1, VEGFA, BCL2L1, TLR4, MYD88, PTGS2, BCL2, TLR2, SREBF1, PNPLA2, MAP1LC3A, IL10, FASN, ADIPOQ, NOS2, IL6 |
| 2 | 5.6 | 6 | 14 | UGT1A7, UGT1A10, UGT1A3, UGT2B15, XPC, UGT1A9 |
| 3 | 4 | 4 | 6 | ACSL1, CIDEA, ADIPOR2, ACOX1 |
| 4 | 3 | 3 | 3 | UGT1A1, UGT1A8, UGT1A6 |
3.5. GO Analysis by Hub Genes
The GO was used to pinpoint the distinctive biological traits. GO is divided into three functional groups: Biological process (BP) [Figure 5], molecular function (MF) [Figure 6], and cellular component (CC) [Figure 7]. With a false discovery rate (FDR) cutoff size of 0.05, the top five GO-enriched pathways were examined. Flavone metabolic process (GO: 0051552), flavonoid glucuronidation (GO: 0052696), xenobiotic glucuronidation (GO: 0052697), flavonoid metabolic process (GO: 0009812), and cellular glucuronidation (GO: 0052695) are all enriched in the BP analysis. The MF analysis indicated that the genes are involved in retinyl-palmitate esterase activity (GO: 0050253), lipopolysaccharide immune receptor activity (GO: 0001875), BH3 domain binding (GO: 0051434), retinoic acid binding (GO: 0001972), and glucuronosyltransferase activity (GO: 0015020). CC genes are enriched in lipid droplet (GO: 0005811), organelle outer membrane (GO: 0031968), outer membrane (GO: 0019867), nuclear membrane (GO: 0031965), and nuclear envelope (GO: 0005643). All the genes involved in the pathways were listed in the Supplementary files (S3, S4, and S5).
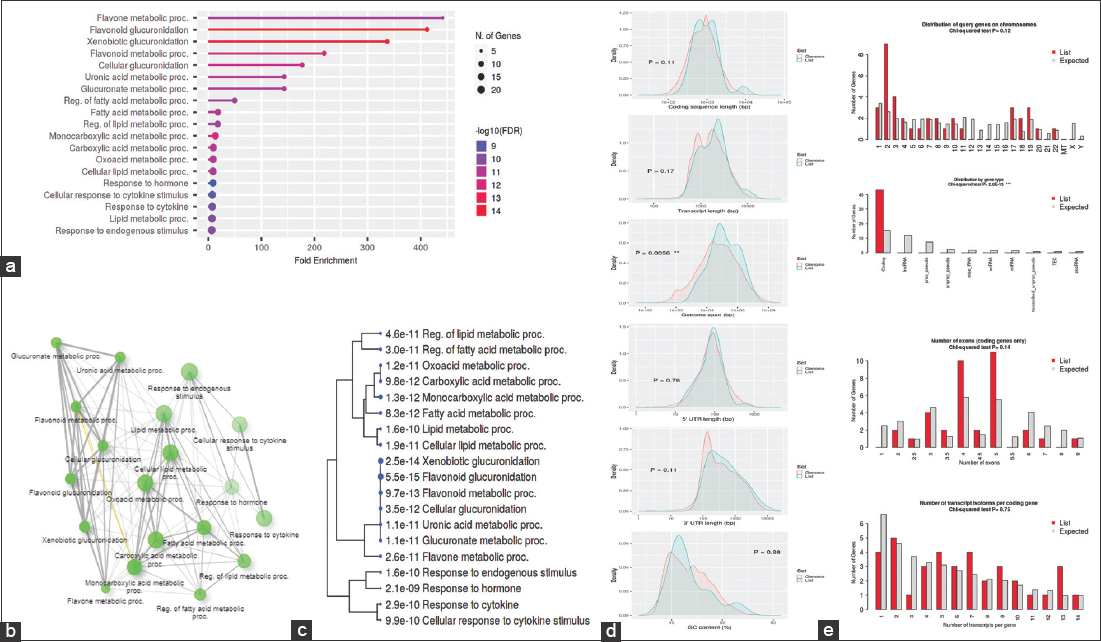 | Figure 5: Gene ontology analysis of biological process: (a) Fold Enrichment graph – Fold enrichment is defined as the percentage of genes that belong to a pathway divided by the percentage of genes that belong to that pathway It shows how much a particular pathway’s genes are overrepresented (b) Interactive plot – The diagram depicts the connections between enriched routes. Two pathways are associated if they share 20% or more genes of their genes. Gene sets with darker nodes are considerably enriched, larger nodes imply greater gene sets, wider edges show gene overlap (c) Tree plot – An illustration of the relationship between the pathways shown in the enrichment tab using a hierarchical clustering tree. Pathways with a large number of cgenes in common are grouped together, a larger points mean higher P-values (d) Student’s t-test plot – Gene features are compared to those of other genes in the genome, and a query is run to examine the gene list’s unique characteristics compared to those of other genes (e) Chi-squared test plot – It works similarly to student’s t-tests. [Click here to view] |
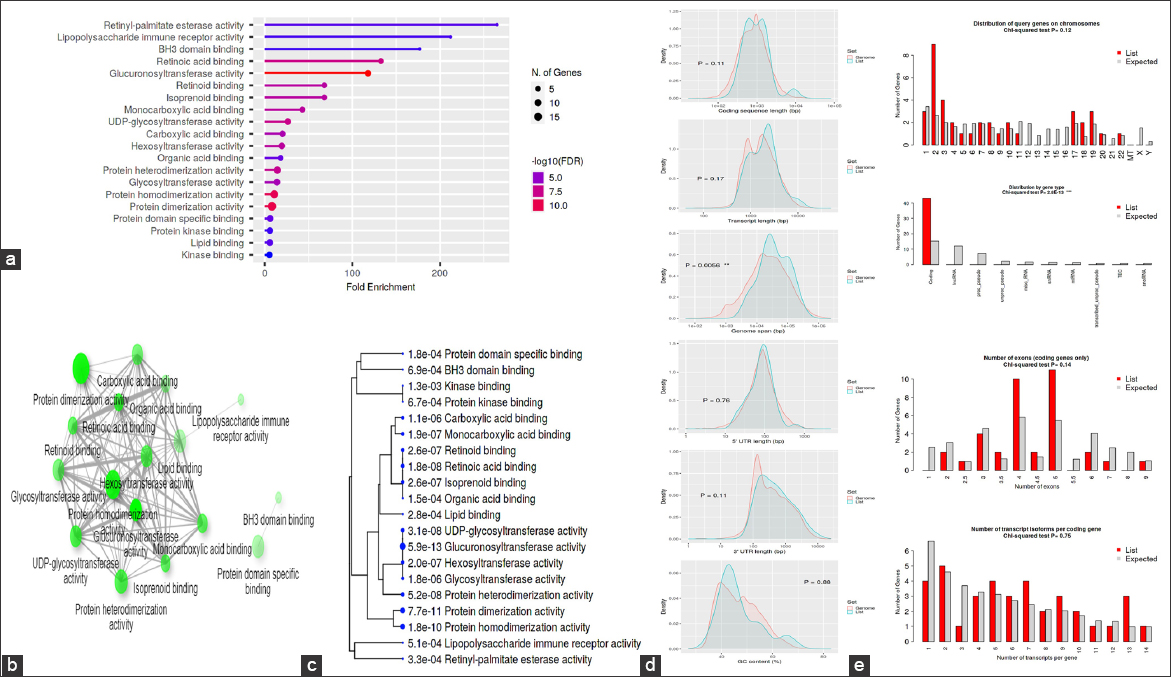 | Figure 6: Gene ontology analysis of molecular function: (a) Fold Enrichment graph, (b) Interactive plot, (c) Tree plot, (d) Student’s t-test plot, and (e) Chi-squared test plot. [Click here to view] |
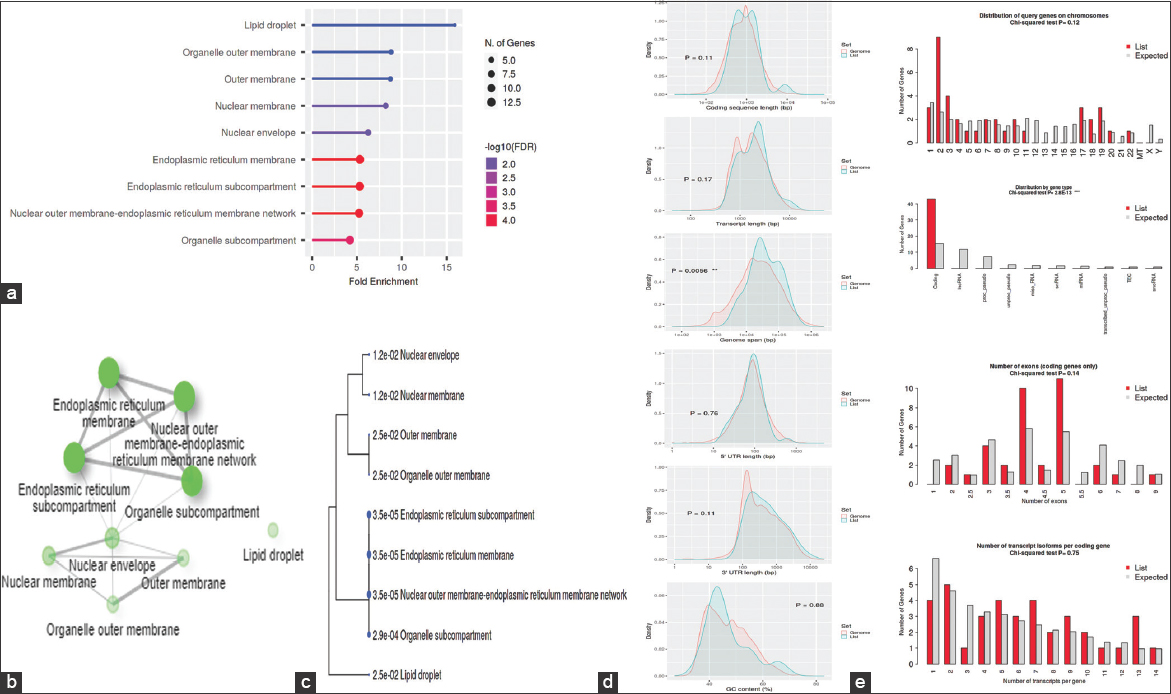 | Figure 7: Gene ontology analysis of colon cancer: (a) Fold Enrichment graph, (b) Interactive plot, (c) Tree plot, (d) Student’s t-test plot, and (e) Chi-squared test plot. [Click here to view] |
3.6. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
To determine whether genes are activated or repressed in different classes of networks, gene regulation data are linked to KEGG domains. Figure 8 shows the 20 leading enhanced networks with an FDR threshold value of 0.05. They are highly clustered in several signaling pathways that are ascorbate and aldarate metabolism, pentose and glucuronate interconversions, porphyrin and chlorophyll metabolism, steroid hormone biosynthesis, retinol metabolism, drug metabolism, metabolism of xenobiotics by cytochrome P450, bile secretion, alcoholic liver disease, leishmaniasis, PD-L1 expression and PD-1 checkpoint pathway in cancer, toxoplasmosis, chemical carcinogenesis, Chagas disease, AMPK signaling pathway, biosynthesis of cofactors, measles, hepatitis B, lipid and atherosclerosis, and metabolic pathways.
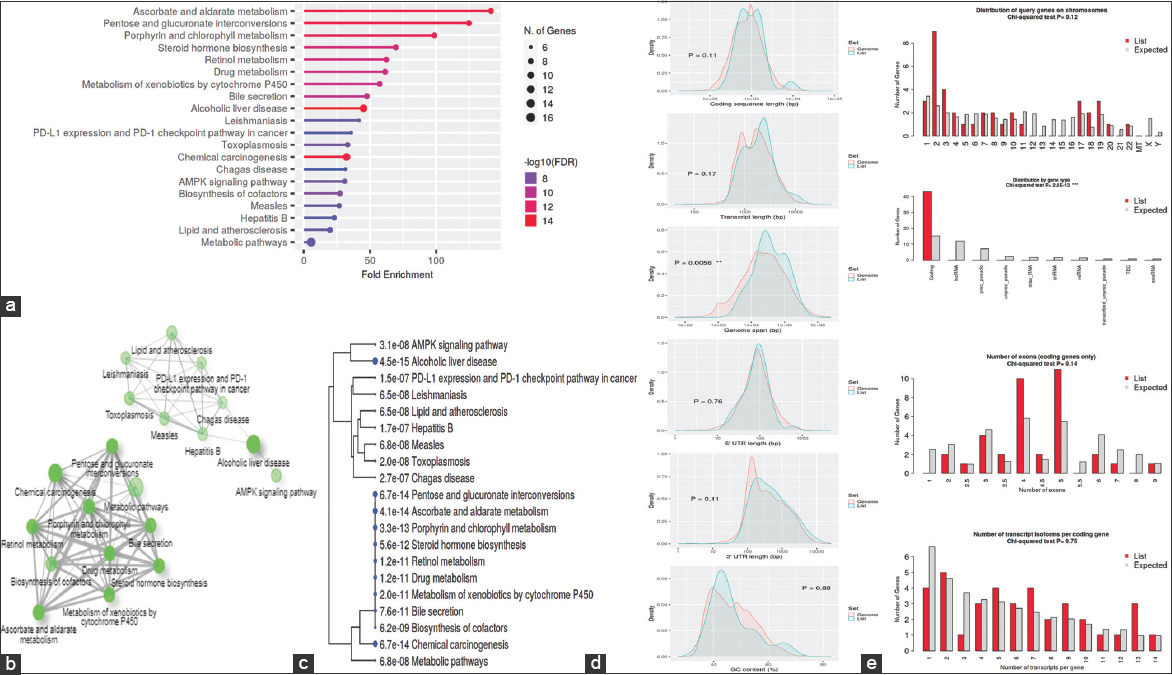 | Figure 8: Kyoto encyclopedia of genes and genomes enrichment analysis: (a) Fold Enrichment graph, (b) Interactive plot, (c) Tree plot, (d) Student’s t-test plot, and (e) Chi-squared test plot. [Click here to view] |
3.7. Pathway Enrichment Analysis
g: Profiler was used for the bioinformatics analysis to allow users to visually view the enrichment information of these potential genes. g: Profiler significantly increased the number of words in the GO, KEGG, REAC, and WP databases. There are 421 pathways in BP, 32 pathways in MF, 14 pathways in CC, 44 pathways in KEGG, 22 pathways in REAC, and 53 pathways in WP which are depicted in Figure 9.
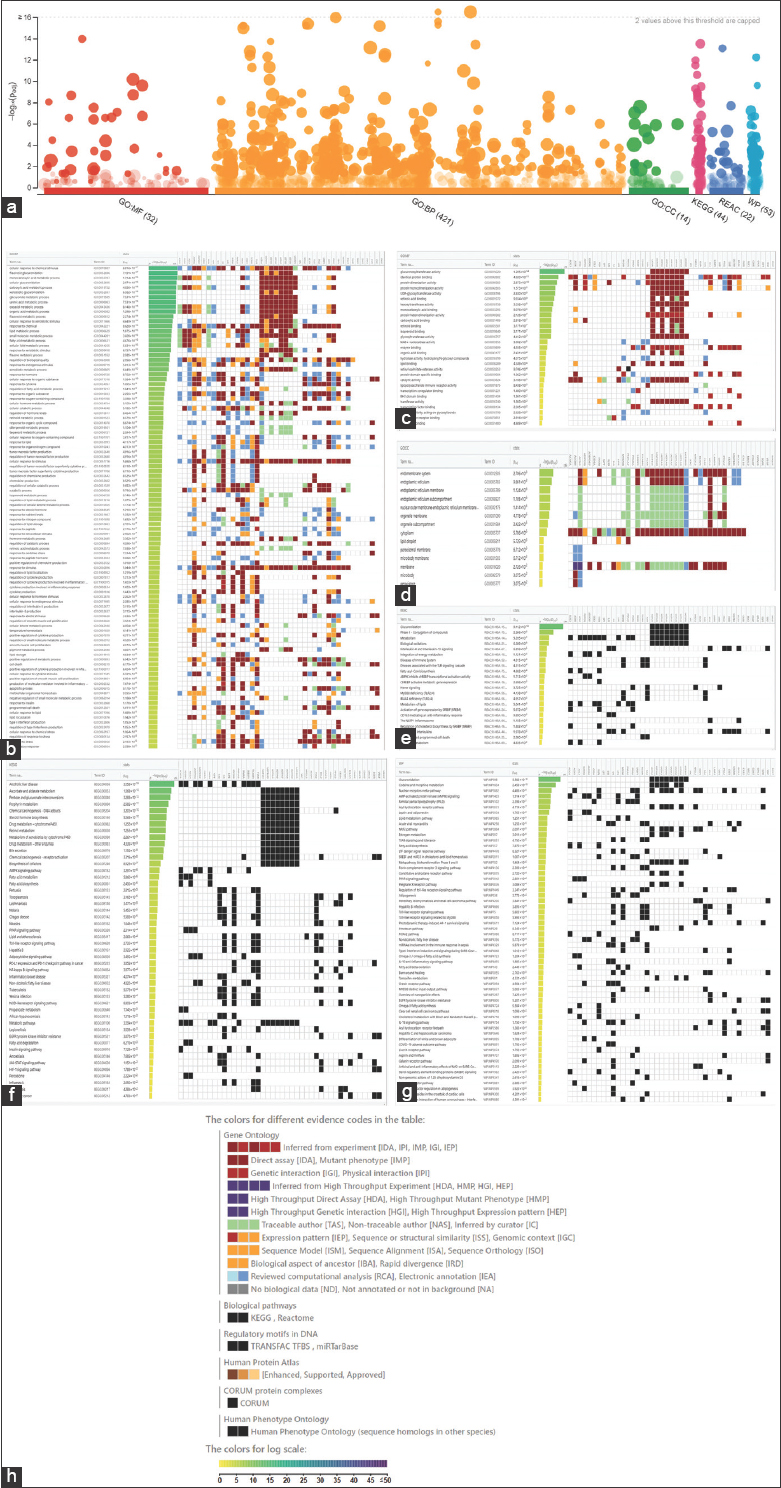 | Figure 9: (a) Results of computational studies employing intersection genes using gProfiler- Significantly enriched terms by gene ontology, Kyoto encyclopedia of genes and genomes, Reactome (REAC), and Wikipathways (WP) databases. Pathway enrichment analysis: (b) Biological process, (c) Molecular function, (d) Colon cancer, (e) REAC, (f) Kyoto encyclopedia of genes and genomes, (g) WP, and (h) Graphical Illustration. [Click here to view] |
4. DISCUSSION
A total of 50 Sc target genes were generated from the CTD and DisGeNET databases. The PPI network was generated with the STRING resource and viewed with the Cytoscape. This PPI network enables us to understand the GO (BP, MF, and CC) and KEGG networks. In BP, flavone metabolic process, flavonoid glucuronidation, xenobiotic glucuronidation, flavonoid metabolic process, and cellular glucuronidation are all enriched. GO MF analysis involved retinyl-palmitate esterase activity, lipopolysaccharide immune receptor activity, BH3 domain binding, retinoic acid binding, and glucuronosyltransferase activity. CC-exposed genes are enriched in lipid droplet, organelle outer membrane, outer membrane, nuclear membrane, and nuclear envelope. Five-enhanced networks identified using the KEGG database. Ascorbate and aldarate metabolism, pentose and glucuronate interconversions, porphyrin and chlorophyll metabolism, chemical carcinogenesis, and steroid hormone biosynthesis all have highly clustered genes.
Using MCODE plug-in of Cytoscape, four main network clusters were analyzed, and six functional modules were obtained. Modules 1–6 include the Nrf2 signaling pathway, the inflammatory pathway, the apoptosis pathway, the autophagy pathway, cell proliferation signaling and insulin-sensitizing pathway. Functional module 1 is Nrf2 signaling pathway, which includes three genes (NFE2L2, KEAP1, HMOX1) and is associated with oxidative stress. (Nrf2, Gene name: NFE2L2), a crucial oxidative stress gene encoding factor that controls a number of chemoprotective and antioxidant genes is being considered as a potential CC therapeutic target. By binding to antioxidant response elements on target promoter regions, Nrf2 acts as a master regulator of genes encoding antioxidant enzymes, proteins, and detoxifying enzymes [29,30]. Thus, Nrf2 pathway contributes to Sc’s protection against oxidative stress-induced damage in colon cancer.
The second module is the inflammatory route includes six genes (IL6, IL10, PTGS2, NOS2, MYD88, and STAT1), where nitric oxide (NO) and prostaglandin (PG) E2 generated by NO synthase type 2 (NOS2) and cyclooxygenase type 2 (COX2) are inflammatory mediators. NOS2 and COX2 are complex, and many of the stimuli that enhance NOS2 expression (e.g., microbial component and cytokines), also increase COX2 expression. NO and PGE2 inhibit the synthesis of PGE2 and NO, respectively, by mechanisms that are still unknown, including transcriptional and posttranscriptional effects. Inflammation can be reduced by NOS2- and COX2- specific inhibitors. Sc thus has inhibitory actions on NOS2 and PTGS2 along with IL-10 and IL-6 [31].
Autophagy is the third functional module includes four genes (LAMP3, MAP1LC3A, SIRT1, and SQSTM1) and apoptosis is the fourth functional module including three genes (BCL2, BCL2L1, and BAK1), where BCL2L1/BCL-XL, and BCL2 itself, binding to the BH3 domains of pro-apoptotic BH3 family members, such as BCL2L11/BIM, to prevent them from activating apoptosis through BAX and BAK1. Overexpression of the BCL2 gene has a significant effect on the lipidation of LC3B or the formation of autolysosomes, thereby enabling autophagy [32].
The fifth module, cell proliferation signaling, includes five genes: CDH17, FASN, ACSL1, MKI67, and GPA33. The CDH17 gene regulates integrin activation and signaling, resulting in the activation of focal adhesion kinase, Ras, extracellular signal-regulated kinase, Jun N-terminal kinase, and cyclin D1 leading to CC progression [21]. Fatty acid synthase (FASN) is a multifunctional enzyme catalyzing the fatty acid synthesis from acetyl-CoA, malonyl-CoA and nicotinamide adenine dinucleotide phosphate (NADPH) as a cofactor, a crucial enzyme for long-chain fatty acids generation. In CC, FASN depends on enzymatic activity for existence and proliferation [33]. Overexpression of ACSL1 causes a rise in epithelial to mesenchymal transition (EMT) markers N-cadherin and Slug as well as increases proliferation [34]. MKI67 is linked to intrinsic cell population proliferative stimulation in malignant cancer cells [35]. GPA33 is a membrane antigen that is highly expressed (95%) in colorectal cancer. It makes for a great target for immunotoxins that attack colon cancer cells [36].
The sixth pathway, which includes two genes, is the insulin-sensitizing pathway (ADIPOQ and ADIPOR2). Experimental evidence witnessed that mice lacking the ADIPOQ gene showed enhanced colon carcinogenesis. ADIPOR2 is vital in the regulation ADIPOQ’s anticancer effects, which promotes epithelial cell proliferation [37].
As a result, activating the oxidative stress pathway, which includes Nrf2/KEAP1, leads to detoxification, antioxidant benefits, and anti-inflammatory effects. It also induces apoptosis in tumor cells, along with autophagy signaling. A number of genes that increase cell proliferation also play a significant impact in the survival of cancer cells. Hence, potential gene-drug interactions were identified by the above-mentioned functional module genes are examined for molecular docking in a subsequent study to provide a thorough understanding of Sc protection against colon cancer cells.
5. CONCLUSION
This study revealed potential biomarkers associated with colon cancer progression. Furthermore, this study emphasizes the significance of PPI network analysis as a powerful framework for gaining insight into the main hub nodes and identifying the possible biomarkers of colon cancer. Sc can be a promising candidate in promoting colon health by inhibiting inflammation and suppressing oxidative stress through a multi-pathway network.
6. ACKNOWLEDGMENT
The authors would like to thank the management of PSG College of Arts and Science for providing financial assistance in the form of PSGCAS Seed Grant Scheme (Ref No. PSGCAS/SGS/2019-2020/Biochem/020) to Dr.K.M.Sakthivel for the present study.
7. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, et al. Colorectal cancer. Nat Rev Dis Primers 2015;1:15065. [CrossRef]
2. Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 2008;135:1079-99. [CrossRef]
3. Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996;87:159-70. [CrossRef]
4. Zeki SS, Graham TA, Wright NA. Stem cells and their implications for colorectal cancer. Nat Rev Gastroenterol Hepatol 2011;8:90-100. [CrossRef]
5. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61:759-67. [CrossRef]
6. Wahl O, Oswald M, Tretzel L, Herres E, Arend J, Efferth T. Inhibition of tumor angiogenesis by antibodies, synthetic small molecules and natural products. Curr Med Chem 2011;18:3136-55. [CrossRef]
7. Efferth T, Grassmann R. Impact of viral oncogenesis on responses to anti-cancer drugs and irradiation. Crit Rev Oncog 2000;11:165-87.
8. Dou Y, Tong B, Wei Z, Li Y, Xia Y, Dai Y. Scopoletin suppresses IL-6 production from fibroblast-like synoviocytes of adjuvant arthritis rats induced by IL-1b stimulation. Int J Immunopharmacol 2013;17:1037-43. [CrossRef]
9. Liu XL, Zhang L, Fu XL, Chen K, Qian BC. Effect of scopoletin on PC3 cell proliferation and apoptosis. Acta Pharmacol Sin 2001;22:929-33.
10. Witaicenis A, Seito LN, da Silveria Chagas A, de Almeida LD Jr., Luchini AC, Rodrigues Orsi P, et al. Antioxidant and intestinal anti-inflammatory effects of plant-derived coumarin derivatives. Phytomedicine 2014;21:240-6. [CrossRef]
11. Ding Z, Dai Y, Wang Z. Hypouricemic action of scopoletin arising from xanthine oxidase inhibition and uricosuric activity. Planta Med 2005;71:183-5. [CrossRef]
12. Sakthivel KM, Vishnupriya S, Dharshini LC, Rasmi RR, Ramesh B. Modulation of multiple cellular signalling pathways as targets for anti-inflammatory and anti-tumorigenesis action of scopoletin. J Pharm Pharmacol 2022;74:147-61. [CrossRef]
13. Bhattacharyya SS, Paul S, De A, Das D, Samadder A, Boujedaini N, et al. Poly (lactide-co-glycolide) acid nanoencapsulation of a synthetic coumarin:Cytotoxicity and bio-distribution in mice, in cancer cell line and interaction with calf thymus DNA as target. Toxicol Appl Pharmacol 2011;253:270-81. [CrossRef]
14. Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, et al. The comparative toxicogenomics database:Update 2017. Nucleic Acids Res 2017;45:D972-8.
15. Shijia W, Hong L, Yongheng B. Nrf2 in cancers:A double-edged sword. Cancer Med 2019;8:2252-67. [CrossRef]
16. Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology 2010;138:2101-14. [CrossRef]
17. Watson AJ. Apoptosis and colorectal cancer. Gut 2004;53:1701-9. [CrossRef]
18. Devenport SN, Shah YM. Functions and implications of autophagy in colon cancer. Cells 2019;8:1349. [CrossRef]
19. Hsu HP, Lai MD, Lee JC, Yen MC, Weng TY, Chen WC, et al. Mucin 2 silencing promotes colon cancer metastasis through interleukin-6 signaling. Sci Rep 2017;7:5823. [CrossRef]
20. Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol Med Rep 2015;11:1566-72. [CrossRef]
21. Bartolome RA, Barderas R, Torres S, Fernandez-Acenero MJ, Mendes M, Garcia-Foncillas J, et al. Cadherin-17 interacts with a2b1 integrin to regulate cell proliferation and adhesion in colorectal cancer cells causing liver metastasis. Oncogene 2014;33:1658-69. [CrossRef]
22. Blum W, Pecze L, Rodriguez JW, Steinauer M, Schwaller B. Regulation of calretinin in malignant mesothelioma is mediated by septin 7 binding to the CALB2 promoter. BMC Cancer 2018;18:475. [CrossRef]
23. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017:Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8.
24. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape:A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [CrossRef]
25. Li CY, Cai JH, Tsai JJ, Wang CC. Identification of hub genes associated with development of head and neck squamous cell carcinoma by integrated bioinformatics analysis. Front Oncol 2020;10:681. [CrossRef]
26. Bandettini WP, Kellman P, Mancini C, Booker OJ, Vasu S, Leung SW, et al. MultiContrast delayed enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance:A clinical validation study. J Cardiovasc Magn Reson 2012;14:83. [CrossRef]
27. Ge SX, Jung D, Yao R. ShinyGO:A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020;36:2628-9. [CrossRef]
28. Reimand J, Isserlin R, Voisin, V, Kucera M, Tannus-Lopes C, Rostamianfar A, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 2019;14:482-517. [CrossRef]
29. Boutten A, Goven D, Artaud-Macari E, Boczkowski J, Bonay M. NRF2 targeting:A promising therapeutic strategy in chronic obstructive pulmonary disease. Trends Mol Med 2011;17:363-71. [CrossRef]
30. Kim J, Cha YN, Surh YJ. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat Res 2010;690:12-23. [CrossRef]
31. Weingberg JB. Nitric oxide synthase 2 and cyclooxygenase 2 interactions in inflammation. Immunol Res 2000;22:319-41. [CrossRef]
32. Lindqvist LM, Vaux DL. BCL2 and related prosurvival proteins require BAK1 and BAX to affect autophagy. Autophagy 2014;10:1474-5. [CrossRef]
33. Lee KH, Lee MS, Cha EY, Sul JY, Lee JS, Kim JS, et al. Inhibitory effect of emodin on fatty acid synthase, colon cancer proliferation and apoptosis. Mol Med Rep 2017;15:2163-73. [CrossRef]
34. Sebastiano MR, Konstantinidou G. Targeting long chain acyl-CoA synthetases for cancer therapy. Int J Mol Sci 2019;20:3624. [CrossRef]
35. Mulyawan IM. Role of Ki67 protein in colorectal cancer. Int J Res Med Sci 2019;7:647. [CrossRef]
36. Carreras-Sangra N, Tome-Amat J, Garcia-Ortega L, Batt CA, Onaderra M, Martinez-del-Pozo A, et al. Production and characterization of a colon cancer-specific immunotoxin based on the fungal ribotoxin a-sarcin. Protein Eng Des Sel 2012;25:425-35. [CrossRef]
37. Ou Y, Chen P, Zhou Z, Li C, Li J, Tajima K, et al. Associations between variants on ADIPOQ and ADIPOR2 with colorectal cancer risk:A Chinese case-control study and updated meta-analysis. BMC Med Genet 2014;15:137. [CrossRef]