ARTICLE HIGHLIGHTS
This research delves into the prevalence and mechanisms of plasmid-mediated antimicrobial resistance (AMR) genes in gut microbiomes, with special emphasis on Prevotella and Bifidobacterium genera. By exploring various screening methods and statistical analysis, this study showcases the resistance genes present in both genera and an in-depth analysis of the mechanisms of these resistance genes and how they can potentially affect public health.
1. INTRODUCTION
Antimicrobial resistance (AMR), is a silent pandemic that the world is currently facing. Antimicrobial drugs are effective tools against bacterial infections that could put the health of both people and animals in danger. Microorganisms gain resistance through various methods like mutation [1], Horizontal gene transfer [2], Efflux pump [3], selective pressure and improper or over use of antibiotics [4]. Its prevalence reduces the effectiveness of medications and, thus, the ability to treat illnesses successfully. AMR is the greatest present threat to human as well as animal health [5]. Day by day bacteria is gaining more resistance to a variety of antibiotics. AMR is a characteristic that may develop as a result of mutations or maybe horizontally transmitted by bacteria that have already developed resistance to antibiotics [6]. The improper use of antibacterial drugs or improper sanitation leads to increased resistance in the gut microbiota of animals. As a result, the ruminant digestive system has AMR genes that might serve as a reservoir for the development and spread of AMR. Gut bacteria lead to the spread of these AMR genes from animals to humans [5,7]. This can cause the microorganisms already present in our gut to acquire these AMR genes from the probiotics through horizontal gene transfer. Gut microbiomes vary greatly between people and populations. Multiple studies have indicated that Bifidobacterium and Prevotella are among the most often identified genera in Asian populations’ gut microbiomes [8,9].
Bifidobacterium is a genus of Gram-positive bacteria belonging to the family Bifidobacteriaceae. It is predominantly found in the human stomach and intestine [10,11]. Establishing Bifidobacterium in the human gut is associated with health benefits, including immune development, neuromodulation, pathogen inhibition, and modulation of the intestinal microbiota composition [12]. Bifidobacteria were first isolated from the faeces of breastfed infants in the year 1899 by Gomes et al. Bifidobacteria are isolated from a wide range of ecological niches like the mouth, sewage and also from insect and mammalian gut, and more recently from water kefir [13].
Prevotella is a genus of bacteria that belongs to the family Prevotellaceae. It is a Gram-negative, anaerobic organism that is commonly found in the human oral cavity, gastrointestinal tract, and female reproductive tract [14]. Prevotella has been linked to various diseases, including periodontitis, endodontic infections, and bacteremia (infection of the blood) [15]. It has also been related to the evolution of different cancer types, like colon and breast cancer. Prevotella is abundant in the eastern population of the world, due to their plant-rich diet. The association between a plant-rich diet and the presence of Prevotella is a beneficial microbe [16].
So this study will mainly focus on finding the prevalence of AMR genes in plasmids of Prevotella and Bifidobacterium and a statistical analysis will be conducted to compare the plasmid size and GC content.
2. MATERIALS AND METHODS
2.1. Plasmid Selection
The plasmids for both Prevotella and Bifidobacterium were collected from the plasmid database PLSDB [17] and from NCBI [18]. The term “Bifidobacterium” was used to search plasmids of Bifidobacterium and the term “Prevotella” was used to search plasmids of the Prevotella genus in the search browser given. 61 plasmid sequences were collected for Bifidobacterium [Table A]; they included Bifidobacterium longum DJO10A and B. longum subsp. infantis 157F, Bifidobacterium choerinum strain FMB-1 and other strains of Bifidobacterium. The size of the plasmids ranged from 0.0026 Mb to 0.198 Mb and their GC content ranged between 55% and 68%.
For Prevotella, a total of 32 plasmid sequences were collected [Table B]; they included different strains of Prevotella like Prevotella scopos JCM 17725, Prevotella nigrescens, and Prevotella copri DSM 18205 and others. The size of the plasmid ranged between 0.004 Mb and 1 Mb and their GC content ranged between 32% and 46%.
2.2. Phylogenetic Tree Construction with Plasmids
The phylonium tool was used to determine the evolutionary distances between genomes that are closely related. Compared to alignment-based methods of phylogeny reconstruction, it is faster and more accurate in comparison to other methods that are alignment-free. Using these evolutionary distances, a phylogenetic tree was constructed [19].
2.3. Screening Plasmids for AMR Genes
The collected plasmids were subjected to screening to check the prevalence of AMR genes using the ResFinderFG 2.1 web server [20]. The database contains fifteen antibiotic drug classes and all of them were used for screening. Resfinder will identify the presence of AMR genes in a given sequence by contrasting as well as locating genes that confer antibiotic resistance. They included various drug classes, including beta-lactam (BL), fluoroquinolone, macrolide (ML)-lincosamide (LS)-streptogramin B (SGB), tetracycline (TC), and more.
Bifidobacterium, is a gram-positive bacteria so drugs exclusively used to cure gram-negative bacteria do not have any significant negative effect on them, in the same way, the gram-positive drug does not have any significant negative effect on Prevotella as it is a Gram-negative bacteria. So, the gram-negative and gram-positive drug classes are used as a control respectively.
The sequences were uploaded to the web server, and the AMR gene criteria were set to detect the presence of resistance genes to all drug classes available on the server. The percentage identity was determined at 90%, and the percentage of perfect alignment was set to 100%. The nucleotide number that is similar to the gene providing the best-matching resistance in the database and the respective sequence in the plasmid were compared to determine the percentage identity. By default, a percentage above 60% in nucleotide sequence overlap with the resistant gene was considered to be a hit. The plasmid that contained the most AMR genes in the database was evaluated using a minimum identity setting of 30 and a maximum identity value of 100%.
2.4. Potential Microbial Resistance Index
Potential multiple antibiotic resistance also known as the p-MAR index was computed for the plasmids screened for resistance, from the aforementioned results. Previously published methods were used to calculator the index [21,22]. The p-MAR index is defined as the ratio between genes that show antibiotic resistance and genes that show antibiotic susceptibility.
2.5. Verification of the Acquired AMR Genes
The results obtained from ResFinderFG 2.1 database are verified using another database called KmerResistance 2.2 [23,24]. The database was set to resistant genes, while the identity threshold is set to 70%, involving a depth correction threshold at 10%. The results outputted were recorded and cross checked with the outputs given by the ResFinderFG database. KmerResistance 2.2 also shows the template sequence and query coverage of the uploaded sequence in its output.
2.6. Determining Antibiotic Resistance Determinants (ARDs)
The plasmid that confers resistance genes was collected and they were screened to find the presence of ARDs using another software/database called ResfinderFG 1.0 [20].Using functional metagenomic ARDs, ResFinderFG identifies resistant phenotypes through the usage of a functional genomic database. Default parameters were used. Identity percentage was set to 98% and the minimum query length set to 60%. “Assembled contigs/genomes” was the read type preferred, and these sequences were cross checked for the 13 determinant families in the database [25,26].
3. RESULTS AND DISCUSSION
3.1. Acquired AMR Genes
To determine the prevalence of AMR genes in both Bifidobacterium and Prevotella in the ResFinder database. ResFinder finds the presence of AMR genes in the genome using the available known genes procured in the database. The query sequence uploaded by the user is aligned or compared using an alignment program called BLAST. The output received is recorded for further use.
Totally 61 plasmid sequences of Bifidobacterium were collected and screened for anti-microbial drug resistance. Out of which 9 plasmids confer resistance genes to the selected drug classes [Table C]. Bifidobacterium strains resisted drug classes like SGB, LS, ML and TC. The genes responsible for the resistance are erm(X), tet(W), and lnu(C). Among them, the most frquent gene was the erm(X), and is seen 4 times out of 10 while the tet(W) resistance gene, was seen 5 times on 10 observations. The erythromycin ribosome methylase (erm) resistance gene is an rRNA methyltransferase that projects the ribosome from inactivation due to antibiotic binding and is widely found in Bifidobacterium species and Corynebacterium species and is responsible for antibiotic resistance for SGB, LS and ML [29,30]. Traditional MLs such as erythromycin and azithromycin have limited clinical utility due to the increased spread and broad antibiotic resistance spectrum of erm [31]. The tet(W) gene codes for a protection protein that binds to the ribosome and modifies the ribosomal conformation that prevents TC from binding, enabling protein synthesis to continue. Of all TC resistance gene classes, this is the most common [32].
For Prevotella, 32 plasmids were collected and screened for AMR genes, out of which 5 genes confer resistance genes [Table D]. Strains of Prevotella were resistant to two main drug classes: BL and TC. The genes responsible for the resistance are cfxA6, cfxA3, cfxA4, cfxA5 and tet(Q). The cfxA6, cfxA3, cfxA4, and cfxA5 belong to class A beta-lactamase, where they catalyse the opening and hydrolysis of the BL ring. Some Prevotella strains exhibit resistance to penicillin by expressing the beta-lactamase genes, and they may have gained resistance to this drug class as penicillin is a systemic antibiotic prescribed to patients with dentoalveolar infections, where it involves anaerobic bacteria residing in the oral cavity like Prevotella and Fusobacterium [33,34]. Tet(Q) is a TC-resistant determinant that confers resistance by a ribosome protection mechanism.
3.2. Verification of the Acquired AMR genes
The data collected from the ResFinder software is once again verified using the database called KmerResistance 2.2. The output of KmerResistance 2.2 gives a detailed view of template coverage, Query coverage (query coverage is the length of the matching query sequence divided by the template length), Q_value (Q_value tests whether the current template is a significant hit) and P_value. The obtained outputs, shown in Tables 1 and 2, are compared with the output from ResFinder and no significant difference was noted. The verified results were noted, and they were used in the next step.
Table 1: Resistance to different classes of antimicrobial drugs and the p-MAR index of the 9 Bifidobacterium plasmids listed showing the 4 drug classes.
| S. No. | Organism/Plasmid | SGB | LS | ML | TC | Total | p-MAR |
|---|---|---|---|---|---|---|---|
| 1. | B. breve BR3 chromosome | 3 | 2 | 1 | 6 | 0.4 | |
| 2. | B. choerinum FMB-1 | 3 | 2 | 1 | 6 | 0.4 | |
| 3. | B. longum BG7 | 3 | 3 | 0.2 | |||
| 4. | B. longum I2-2-3 | 3 | 2 | 1 | 6 | 0.4 | |
| 5. | B. longum K2-21-4 | 3 | 2 | 1 | 6 | 0.4 | |
| 6. | B. longum NBRC 114370 | 3 | 3 | 0.2 | |||
| 7. | B. longum DJO10A | 3 | 3 | 0.2 | |||
| 8. | B. pullorum CACC 514 | 2 | 2 | 0.13 | |||
| 9. | B. longum NBRC 114494 | 2 | 2 | 0.13 |
SGB: Streptogramin B, LS: Lincosamide, ML: Macrolide, TC: Tetracycline, p-MAR: Potential multiple antibiotic resistance, B. breve: Bifidobacterium breve, B. choerinum: Bifidobacterium choerinum, B. longum: Bifidobacterium longum, B. pullorum: Bifidobacterium pullorum
Table 2: Resistance to different classes of antimicrobial drugs and the p-MAR index of the 5 Prevotella plasmids listed showing the 2 drug classes.
| S. No. | Organism/Plasmid | TC | BL | Total | p-MAR |
|---|---|---|---|---|---|
| 1. | P. copri YF2 | 1 | 1 | 0.06 | |
| 2. | P. copri DSM 18205 FDAARGOS_1573 | 3 | 3 | 0.2 | |
| 3. | P. melaninogenica GAI 07411 | 3 | 3 | 0.2 | |
| 4. | P. melaninogenica F0299 | 3 | 3 | 0.3 | |
| 5. | P. scopos JCM 17725 W2052 | 1 | 1 | 0.06 |
BL: Beta-lactam, TC: Tetracycline, p-MAR: Potential multiple antibiotic resistance. P. copri: Prevotella copri, P. melaninogenica: Prevotella melaninogenica, P. scopos: Prevotella scopos
3.3. Determining ARDs
To determine the presence of ARDs, the plasmids with the AMR genes were screened in the database ResFinderFG 1.0. ResFinderFG 1.0 contains 13 ARD families in its database they are 16S_rRNA_methyltransferase, aminoglycosides (AGs) acetyl-transferases, AGs nucleotidyltransferase, AGs phospho-transferases, beta-lactamases (all sub-families), chloramphenicol acetyl-transferases, dihydrofolate reductase, efflux pumps, quinolone resistance, spanning tet(A) and variants, spanning tet(M) and variants, TC monooxygenases, homologues of D-Ala-D-x ligases (selected on D-cycloserine).
Three ARD families were detected in Bifidobacterium. The most occurring ARD was dihydrofolate reductase (dfr) followed by spanning tet(M) and variants (tet_protection).
Five ARD families were detected in Prevotella. They are beta-lactamase (all sub-families), dihydrofolate reductase (dfr), homologues of D-Ala-D-x ligases (van_ligase) followed by spanning tet(M) and variants (tet_protection) and chloramphenicol acetyl-transferases (cat).
3.4. Size and GC content of the Bifidobacterium and Preovetlla Plasmids
To establish a correlation between the size and the GC content of the plasmid, PCA was carried out on the 9 plasmids of Bifidobacterium and 5 plasmids of Prevotella that contained the AMR genes. The following summarises the sizes of the plasmids and the GC content, where Bifidobacterium plasmids had a lowest of 0.003 and highest of 0.198 in size (MB) and the mean being 0.067 ± 0.091, and the lowest GC content was roughly 56% and highest 66%, and a mean of 60% ± 3.98. The Prevotella plasmid’s lowest size (MB) was 0.006 and highest of 0.167 in size (MB) and the mean being 0.061 ± 0.076, and the lowest GC content was approximately 37% and a highest of 42%, with a mean of 40% ± 1.96.
For Bifidobacterium, the negative Pearson correlation (r = −0.68, α = 0.95) indicates no linear correlation between the GC content and the plasmid size [Figure 1].
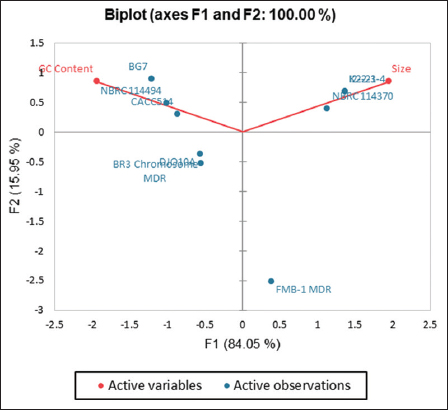 | Figure 1: A principal component analysis (PCA) biplot of variable size (MB) and GC (%) content for 9 plasmids of Bifidobacterium (labelled in blue). Factor 1 and Factor 2 of the PCA show their percentage contribution in brackets. [Click here to view] |
For Prevotella, the positive correlation (r = 0.24, α = 0.95) indicates that there is a linear relationship between the GC content and the plasmid size [Figure 2].
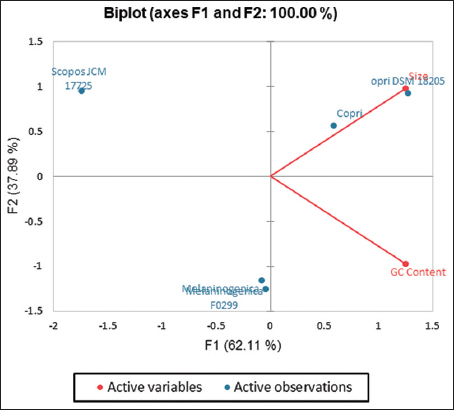 | Figure 2: A principal component analysis (PCA) biplot of variable size (MB) and GC (%) content for five plasmids of Prevotella (labelled in blue). Factor 1 and Factor 2 of the PCA show their percentage contribution in brackets. [Click here to view] |
Recent advancements in the study of GC content and analysis of newer plasmids are important, as they are involved in horizontal gene transfer, which can give more information on natural evolution and the increase of AMR genes [35]. The GC content gives extensive data regarding the evolutionary information and the genomic size of bacteria [36-38]. The range of GC content seen here falls in the average range reported for species, which is 13–75% [39], but higher than the average range reported for bacteria, which is 50.76% [40]. It may be inferred that plasmids with a greater GC content are introduced earlier than plasmids with a relatively lower GC content in comparison, which may be due to environmental differences or the phylogenetic composition [41].
3.5. Statistical Analysis of Sequences
Using phylonium, the sequences were analysed, and the pairwise distances were calculated. Using these distances, a phylogenetic tree was constructed, and standard error estimates were calculated by toggling to the bootstrap method (1000 replicates). Figures 3 and 4 showcase the phylogenetic trees for the Bifidobacterium and Prevotella species [42].
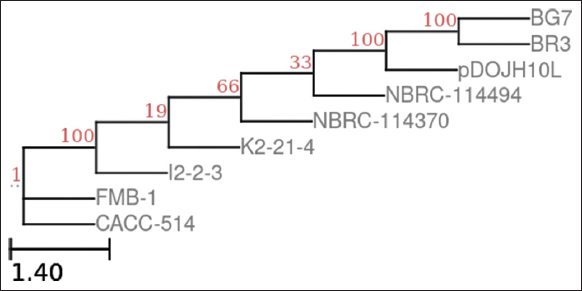 | Figure 3: The phylogeny of 9 Bifidobacterium genomes; This phylogeny was computed using phylonium with bootstrap method (1000 replicates), neighbour-joining and then visualised with ETE [30]. [Click here to view] |
 | Figure 4: The phylogeny of 5 Prevotella genomes; This phylogeny was computed using phylonium with bootstrap method (1000 replicates), neighbour-joining and then visualised with ETE. [Click here to view] |
Various studies have been conducted to prove the presence of AMR genes in gut microbiomes. This study mainly focused on the presence of AMR in Bifidobacterium and Prevotella, and there have also been various studies that prove the presence of antimicrobial genes of other gut microorganisms such as Lactobacillus, Escherichia coli, and Bacteroides fragilis.
Lactobacillus is a genus of bacteria that is commonly found in the human gut and is known for its probiotic properties. Studies have proved the presence of AMR genes in Lactobacillus against antibiotics like TC, erythromycin, and clindamycin [43].
B. fragilis is a common anaerobic bacterium found in gut microbiota. Isolates of B. fragilis from clinical samples contain cfiA(C) genes resistant to carbapenem antibiotics [44].
Advancements in genomic sequencing technology have allowed for the identification and characterization of the antimicrobial genes. Lactobacillus reuteri forms antimicrobial substances such as reuterin and 3-hydroxypropionaldehyde [45]. Anti-microbial genes in E. coli are found to be involved in the synthesis of colicins and other antimicrobial peptides [46].
Understanding the mechanism of antimicrobial genes of different gut microbes can help researchers develop new strategies for promoting the growth of beneficial bacteria and preventing the spread of pathogenic bacteria in the gut.
4. CONCLUSION
Following the in silico analysis of the Bifidobacterium and Prevotella plasmids for acquired AMR genes showed that there was no positive linear correlation between size and GC content for the Bifidobacterium and that there is a positive linear correlation between size and GC content for the Prevotella set that was analysed. The phylogenetic tree between the species of Bifidobacterium and Prevotella shows the change in the composition of AMR genes in the ancestry line. There is not much difference between the composition of resistance genes in Bifidobacterium and Prevotella when compared to the ancestry line, with the AMR genes being confined to 5 classes in Bifidobacterium and 2 classes in Prevotella. Although the presence of these AMR genes has been confirmed, it does not directly translate that it will lead to a strong phenotypic expression. Further research can be conducted to determine the phenotypic expression of the AMR genes in both Bifidobacterium and Prevotella.
5. ACKNOWLEDGMENT
We the authors thank the Biotechnology Department of SRMIST for all its support and encouragement to do this work.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data generated and analysed are included within this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol 2005;3:e176. [CrossRef]
2. Brown-Jaque M, Calero-Cáceres W, Muniesa M. Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 2015;79:1-7. [CrossRef]
3. Li G, Zhang J, Guo Q, Jiang Y, Wei J, Zhao LL, et al. Efflux pump gene expression in multidrug-resistant Mycobacterium tuberculosis clinical isolates. PLoS One 2015;10:e0119013. [CrossRef]
4. Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis 2013;13:1057-98. [CrossRef]
5. Tóth AG, Csabai I, KrikóE, T?zsér D, Maróti G, Patai ÁV, et al. Antimicrobial resistance genes in raw milk for human consumption. Sci Rep 2020;10:7464. [CrossRef]
6. Cameron A, McAllister TA. Antimicrobial usage and resistance in beef production. J Anim Sci Biotechnol 2016;7:68. [CrossRef]
7. Guo H, Pan L, Li L, Lu J, Kwok L, Menghe B, et al. Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J Food Sci 2017;82:724-30. [CrossRef]
8. Nakayama J, Watanabe K, Jiang J, Matsuda K, Chao SH, Haryono P, et al. Diversity in gut bacterial community of school-age children in Asia. Sci Rep 2015;5:8397. [CrossRef]
9. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222-7. [CrossRef]
10. Turroni F, van Sinderen D, Ventura M. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol 2011;149:37-44. [CrossRef]
11. Sgorbati B, Biavati B, Palenzona D. The genus Bifidobacterium. In:The Genera of Lactic Acid Bacteria. Boston, MA:Springer;1995. 279-306. [CrossRef]
12. Turroni F, Duranti S, Milani C, Lugli GA, van Sinderen D, Ventura M. Bifidobacterium bifidum:A key member of the early human gut microbiota. Microorganisms 2019;7:544. [CrossRef]
13. Gomes AM, Malcata FX. Bifidobacterium spp. and Lactobacillus acidophilus:Biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci Technol 1999;10:139-57. [CrossRef]
14. Dayakar MM, Bhat S, Lakshmi KN. Prevotella intermedia-an overview and its role in periodontitis. J Adv Clin Res Insights 2021;8:79-82. [CrossRef]
15. Ley RE. Gut microbiota in 2015:Prevotella in the gut:Choose carefully. Nat Rev Gastroenterol Hepatol 2016;13:69-70. [CrossRef]
16. Shah HN, Chattaway MA, Rajakurana L, Gharbia SE. Prevotella. In:Bergey's Manual of Systematics of Archaea and Bacteria. Hoboken, NJ:John Wiley and Sons;2015. 1-25. [CrossRef]
17. Schmartz GP, Hartung A, Hirsch P, Kern F, Fehlmann T, Müller R, et al. PLSDB:Advancing a comprehensive database of bacterial plasmids. Nucleic Acids Res 2021;50:D273-8. [CrossRef]
18. NCBI. National Center for Biotechnology Information;2019. Available from:https://www.ncbi.nlm.nih.gov [Last accessed on 2022 Dec 14].
19. Klötzl F, Haubold B. Phylonium:Fast estimation of evolutionary distances from large samples of similar genomes. Bioinformatics 2019;36:2040-6. [CrossRef]
20. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 2020;75:3491-500. [CrossRef]
21. Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 1983;46:165-70. [CrossRef]
22. Davis R, Brown PD. Multiple antibiotic resistance index, fitness and virulence potential in respiratory Pseudomonas aeruginosa from Jamaica. J Med Microbiol 2016;65:261-71. [CrossRef]
23. Clausen PT, Zankari E, Aarestrup FM, Lund O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J Antimicrob Chemother 2016;71:2484-8. [CrossRef]
24. Clausen PT, Aarestrup FM, Lund O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 2018;19:307. [CrossRef]
25. Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder:A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother 2017;72:2764-8. [CrossRef]
26. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+:Architecture and applications. BMC Bioinformatics 2009;10:421. [CrossRef]
27. XLSTAT Statistical Software for Excel. XLSTAT, Your Data Analysis Solution;2019. Available from:https://www.xlstat.com/en. [Last accessed on 2023 Jan 18].
28. Vidal NP, Manful CF, Pham TH, Stewart P, Keough D, Thomas R. The use of XLSTAT in conducting principal component analysis (PCA) when evaluating the relationships between sensory and quality attributes in grilled foods. MethodsX 2020;7:100835. [CrossRef]
29. Martínez N, Luque R, Milani C, Ventura M, Bañuelos O, Margolles A. A gene Homologous to rRNA methylase genes confers erythromycin and clindamycin resistance in Bifidobacterium breve. Appl Environ Microbiol 2018;84. [CrossRef]
30. Li B, Chen D, Lin F, Wu C, Cao L, Chen H, et al. Genomic island-mediated horizontal transfer of the erythromycin resistance gene erm(X) among Bifidobacteria. Appl Environ Microbiol 2022;88:e0041022. [CrossRef]
31. Aires J, Doucet-Populaire F, Butel MJ. Tetracycline resistance mediated by tet(W), tet(M), and tet(O) genes of Bifidobacterium isolates from humans. Appl Environ Microbiol 2007;73:2751-4. [CrossRef]
32. Flo?rez AB, Ammor MS, A?lvarez-Marti?n P, Margolles A, Mayo B. Molecular analysis of tet(W) gene-mediated tetracycline resistance in dominant intestinal Bifidobacterium species from healthy humans. Appl Environ Microbiol 2006;72:7377-9. [CrossRef]
33. Iwahara K, Kuriyama T, Shimura S, Williams DW, Yanagisawa M, Nakagawa K, et al. Detection of cfxA and cfxA2, the beta-lactamase genes of Prevotella spp., in clinical samples from dentoalveolar infection by real-time PCR. J Clin Microbiol 2006;44:172-6. [CrossRef]
34. Wade WG, Spratt DA, Dymock D, Weightman AJ. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin Infect Dis 1997;25 suppl 2:S235-6. [CrossRef]
35. Sørensen SJ, Bailey M, Hansen LH, Kroer N, Wuertz S. Studying plasmid horizontal transfer in situ:A critical review. Nat Rev Microbiol 2005;3:700-10. [CrossRef]
36. Romiguier J, Roux C. Analytical biases associated with GC-content in molecular evolution. Front Genet 2017;8:16. [CrossRef]
37. Musto H, Naya H, Zavala A, Romero H, Alvarez-Valín F, Bernardi G. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem Biophys Res Commun 2006;347:1-3. [CrossRef]
38. Agashe D, Shankar N. The evolution of bacterial DNA base composition. J Exp Zool B Mol Dev Evol 2014;322:517-28. [CrossRef]
39. Li XQ, Du D. Variation, evolution, and correlation analysis of C+G content and genome or chromosome size in different kingdoms and phyla. PLoS One 2014;9:e88339. [CrossRef]
40. Shintani M, Sanchez ZK, Kimbara K. Genomics of microbial plasmids:Classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol 2015;6:242. [CrossRef]
41. Reichenberger ER, Rosen G, Hershberg U, Hershberg R. Prokaryotic nucleotide composition is shaped by both phylogeny and the environment. Genome Biol Evol 2015;7:1380-9. [CrossRef]
42. Huerta-Cepas J, Serra F, Bork P. ETE 3:Reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 2016;33:1635-8. [CrossRef]
43. Campedelli I, Mathur H, Salvetti E, Clarke S, Rea MC, Torriani S, et al. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl Environ Microbiol 2019;85. [CrossRef]
44. Bush K, Bradford PA. Epidemiology of β-lactamase-producing pathogens. Clin Microbiol Rev 2020;33:e00047-19. [CrossRef]
45. O'Toole PW, Marchesi JR, Hill C. Next-generation probiotics:The spectrum from probiotics to live biotherapeutics. Nat Microbiol 2017;2:17057. [CrossRef]
46. Min H, Baek K, Lee A, Seok YJ, Choi Y. Genomic characterization of four Escherichia coli strains isolated from oral lichen planus biopsies. J Oral Microbiol 2021;13:1905958. [CrossRef]
Table A: List of Bifidobacterium strains that were screened for antimicrobial resistance.
| Serial number | Organism name | Strain |
|---|---|---|
| 1. | B. breve | BIF195 |
| 2. | B. longum subsp. infantis | 157F |
| 3. | B. longum | DJO10A |
| 4. | B. longum subsp. longum | KACC 91563 |
| 5. | B. longum | NCC2705 |
| 6. | B. breve | BR3 |
| 7. | B. longum | BG7 |
| 8. | Bifidobacterium | FMB-1 |
| 9. | B. longum | ICIS-505 |
| 10. | B. pullorum subsp. gallinarum | CACC-514 |
| 11. | B. longum | NBRC 114370 |
| 12. | B. longum | K2-21-4 |
| 13. | B. longum | ICIS-500 |
| 14. | B. longum | I2-2-3 |
| 15. | B. pseudocatenulatum | YIT11953 |
| 16. | B. longum subsp. longum | NBRC 114494 |
| 17. | B. longum | AGR2137 |
| 18. | B. catenulatum | DSM16992 |
| 19. | B. longum | E18 |
| 20. | B. longum | BORI |
| 21. | B. longum | NCC2705 |
| 22. | B. longum | BBMN68 |
| 23. | B. longum | CECT 7347 |
| 24. | B. longum | GT15 |
| 25. | B. longum | F8 |
| 26. | B. catenulatum subsp. kashiwanohense | JCM 15439 |
| 27. | B. catenulatum subsp. kashiwanohense | DSM21854 |
| 28. | B. breve | 12L |
| 29. | B. longum | 216816 |
| 30. | Bifidobacterium spp. | A24 |
| 31. | B. longum subsp. infantis | 157F |
| 32. | B. choerinum | DSM20434 |
| 33. | B. bifidum | BGN4 |
| 34. | B. bifidum | IPLA20015 |
| 35. | B. breve | DPC6330 |
| 36. | B. breve | JCM7017 |
| 37. | B. pseudocatenulatum | D2CA |
| 38. | B. pseudolongum subsp. globosum | DSM20092 |
| 39. | B. breve | 31L |
| 40. | B. asteroids | DSM20089 |
| 41. | B. asteroids | PRL2011 |
| 42. | B. longum | 2L |
| 43. | B. longum subsp. longum | KACC-9153 |
| 44. | B. pullorum subsp. gallinarum | DSM20607 |
| 45. | B. breve | HPH 0326 |
| 46. | B. longum subsp. longum | 72B |
| 47. | B. longum subsp. longum | EK-13 |
| 48. | B. breve | S27 |
| 49. | B. longum subsp. longum | 1-6B |
| 50. | B. breve | MCC0121 |
| 51. | B. longum subsp. longum | 2-2B |
| 52. | B. longum subsp. infantis | EK3 |
B. longum: Bifidobacterium longum, B. breve: Bifidobacterium breve, B. choerinum: Bifidobacterium choerinum, B. pullorum: Bifidobacterium pullorum, B. pseudocatenulatum: Bifidobacterium pseudocatenulatum, B. catenulatum: Bifidobacterium catenulatum, B. bifidum: Bifidobacterium bifidum, B. pseudolongum: Bifidobacterium pseudolongum, B. asteroids: Bifidobacterium asteroids.
Table B: List of Prevotella strains that were screened for antimicrobial resistance.
| Serial number | Organism name | Strain |
|---|---|---|
| 1. | P. copri | YF2 |
| 2. | P. copri DSM 18205 | FDAARGOS_1573 |
| 3. | P. corporis | OB21 FMU 4 |
| 4. | P. dentalis | DSM 3688 |
| 5. | P. denticola | F0115 |
| 6. | P. melaninogenica | GAI 07411 |
| 7. | P. melaninogenica | F0299 |
| 8. | P. melaninogenica | F0300 |
| 9. | P. nigrescens | FDAARGOS_1486 |
| 10. | P. nigrescens | F0109 |
| 11. | P. nigrescens | F0103 |
| 12. | P. scopos JCM 17725 | W2052 |
| 13. | Prevotella spp. | E2-28 |
| 14. | Prevotella spp. oral taxon 299 str. | F0039 |
| 15. | Prevotella spp. oral taxon 313 | F0648 |
| 16. | P. veroralis | F0319 |
| 17. | P. melaninogenica | ADL-403 |
| 18. | P. melaninogenica | D18 |
| 19. | P. intermedia | JCM1150 |
| 20. | P. ruminicola | KHP1 |
| 21. | P. dentalis | DSM 3688 |
| 22. | P. dentalis | JCM 13448 |
| 23. | P. nigrescens | CC14M |
| 24. | P. nigrescens | F010 |
| 25. | P. denticola | F0289 |
| 26. | P. veroralis | F0319 |
| 27. | P. melaninogenica | D18 |
| 28. | P. melaninogenica | DNF00666 |
| 29. | P. nigrescens | F0103 |
| 30. | P. scopos | JCM 17725 |
P. melaninogenica: Prevotella melaninogenica, P. copri: Prevotella copri, P. scopos: Prevotella scopos, P. corporis: Prevotella corporis, P. dentalis: Prevotella dentalis, P. denticola: Prevotella denticola, P. nigrescens: Prevotella nigrescens, P. veroralis: Prevotella veroralis, P. intermedia: Prevotella intermedia, P. ruminicola: Prevotella ruminicola.
Table C: List of Bifidobacterium strains that are resistant to listed antibiotics along with their resistance genes.
| Name of the organism and strain name | AMR | Resistance gene | |
|---|---|---|---|
| Drug | Class | ||
| B. breve (BR3) | Virginiamycin s | Streptogramin b | ermX |
| Clindamycin | Lincosamide | ||
| Quinupristin | Streptogramin b | ||
| Erythromycin | Macrolide | ||
| Pristinamycin ia | Streptogramin b | ||
| Lincomycin | Lincosamide | ||
| B. choerinum (FMB-1) | Virginiamycin s | Streptogramin b | ermX lnu (C) |
| Quinupristin | Streptogramin b | ||
| Lincomycin | Lincosamide | ||
| Clindamycin | Lincosamide | ||
| Erythromycin | Macrolide | ||
| Pristinamycin ia | Streptogramin b | ||
| B. longum (BG7) | Tetracycline | Tetracycline | tet (W) |
| Doxycycline | Tetracycline | ||
| Minocycline | Tetracycline | ||
| B. longum (I2-2-3) | Quinupristin | Streptogramin b | erm (X) |
| Pristinamycin ia | Streptogramin b | ||
| Erythromycin | Macrolide | ||
| Lincomycin | Lincosamide | ||
| Clindamycin | Lincosamide | ||
| Virginiamycin s | Streptogramin b | ||
| B. longum (K2-21-4) | Virginamycin | Streptogramin b | erm (X) |
| Clindamycin | Lincosamide | ||
| Quinupristin | Streptogramin b | ||
| Erythromycin | macrolide | ||
| Lincomycin | Lincosamide | ||
| Pristinamycin ia | Streptogramin b | ||
| B. longum (NBRC 114370) | Doxycycline | Tetracycline | tet (W) |
| Tetracycline | tetracycline | ||
| Minocycline | Tetracycline | ||
| B. longum subsp. longum (DJO10A) | Doxycycline | Tetracycline | tet (W) |
| Tetracycline | Tetracycline | ||
| Minocycline | Tetracycline | ||
| B. pullorum subsp. gallinarum (CACC 514) | Doxycycline | Tetracycline | tet (W) |
| Minocycline | Tetracycline | ||
| B. longum subsp. longum (NBRC 114494) | Doxycycline | Tetracycline | tet (W) |
| Minocycline | Tetracycline | ||
B. longum: Bifidobacterium longum, B. breve: Bifidobacterium breve, B. choerinum: Bifidobacterium choerinum, B. pullorum: Bifidobacterium pullorum, AMR: Antimicrobial resistance.
Table D: List of Prevotella strains that are resistant to listed antibiotics along with their resistance genes.
| Name of the organism and strain name | AMR | Resistance gene | |
|---|---|---|---|
| Drug | Class | ||
| P. copri (YF2) | Unknown beta lactam | Beta lactam | cfxA4 cfxA6 |
| P. copri DSM 18205 (FDAARGOS_1573) | Doxyciline | Tetracycline | tet (Q) |
| Tetracycline | Tetracycline | ||
| Minocycline | Tetracycline | ||
| P. melaninogenica (GAI 07411) | Ampicillin | Beta lactam | cfxA |
| Cefoxitin | Beta lactam | cfxA3 cfxA4 | |
| Unknown beta-lactam | Beta lactam | ||
| Cephamycin | Beta lactam | cfxA5 | |
| P. melaninogenica (F0299) | Doxycycline | Tetracycline | tet (Q) |
| Tetracycline | Tetracycline | ||
| Minocycline | Tetracycline | ||
| P. scopos JCM 17725 (W2052) | Ampicillin | Beta lactam | cfxA cfxA3 cfxA4 cfxA5 |
P. melaninogenica: Prevotella melaninogenica, P. copri: Prevotella copri, P. scopos: Prevotella scopos, AMR: Antimicrobial resistance.