1. INTRODUCTION
Purple digitalis or foxglove (Digitalis purpurea, family; Plantaginaceae) is a biennial herbaceous plant native to the Balkans, East Caucasus, West Europe, Turkey, Japan, and India [1,2]. It is also cultivated in several Mediterranean countries, the Americas, and some European countries in commercial quantities [3]; its active component, digoxin, was approved by the FDA in the late 1990s as a treatment for heart failure and atrial fibrillation [4]. Moreover, recent studies indicated future applications in oncology and viral infections [5-8].
In view of the medical importance of D. purpurea, researchers have made many attempts to produce digitalis species containing high levels of glycosides through traditional cultivation. However, the offspring was not stable, and their traits restoration required long-term programs that were cumbersome and expensive [2], especially since the propagation of the plant by traditional methods is linked to a low germination rate and is fraught with risks and is considered an economically unfeasible method [2,9], which makes germination-promoting biotechnologies and tissue culture technologies, an economic necessity to meet the increasing human demand for medicines extracted from this plant.
With the advancement of genetics and technologies used in molecular biology, many researches have focused on biotechnological methods adopted in the study of laboratory-bred plants and comparison of their production of effective biological materials in wild species and the resulting cell strains, and these technologies proved their effectiveness in determining genetic patterns [10,11]. The previous findings showed different concentrations of cardiac glycosides in wild, cultivated, and bred species of digitalis plants [12]. In addition, the effects of seasonal variations on the production as well as the contents of cardiac glycosides, such as digoxin and panaxoside C, in tissue culture have been investigated and were attributed to several factors such as nutrient media components, growth regulators, and culture period [13].
Genetic variations accumulate as a result of adaptation to different environments, and these variations can be advantageous among genotypes [14], especially in the plant genetic improvement programs. In tissue culture, the successful application of biotechnologies in plant propagation in vitro depends on maintaining the genetic stability of the genotype [15]; the main obstacle in cell culture is maintaining this genetic constancy [16,17] where genetic variation between embryonic patterns can be considered a factor affecting the capacity of renewal during cell culture [18]. Somaclonal variability is a common problem in plant tissue cultures, especially those that are kept for long periods [17,19]. The variations that appear in plant tissue cultures depend on several factors, including explant type, culture conditions, medium components, types and concentrations of growth regulators, and the culture age [19,20]. The later age of the tissue culture is a specific factor that strongly influences the effect of variation in cell cultures as a result of the increased incidence of chromosome mutations with the increasing age of the cell culture [17,20]. Somaclonal variances occur at the epigenetic or genetic level and they appear clearly in laboratory cultures and result either from pre-existing autosomal variations within plant extracts or during micropropagation [21]. These autosomal variations include changes in the number and structure of chromosomes, changes in nucleotide sequences, number of gene copies, activation of transposons, and modifications in methylation pattern DNA [17,19,20]. It can be said that autosomal genetic variations are a group of variations in the morphological, biochemical, physiological, and genetic characteristics of plants, and biotechniques based on molecular markers have a great ability to detect these discrepancies more accurately compared to other techniques [15].
Molecular markers are more suitable for analyzing genetic diversity than morphological and biochemical traits because they are not affected by the environment [22]. Among the different molecular markers, the inter-simple sequence repeat (ISSR) technique is highly reliable due to the length of the primers and the high fusion temperature [23]. The researchers use the ISSR technique because it is simple and effective in terms of the stability of results when repeating the experiment, its low cost, and its ability to give high rates of polymorphism [24,25]. ISSR technology has many benefits, such as combining most of the advantages and features of amplified fragment length polymorphism (AFLP) and simple sequence repeats (SSRs), providing a higher reproducibility when compared with random amplified polymorphic DNA (RAPD), determining the polymorphism rate when compared with restriction fragment length polymorphism, and being more cost effective when compared with AFLP [24,26,27]; large number of polymorphic markers are needed to reliably measure genetic relationships and genetic diversity [24].
In cell culture, especially strains with high productivity of biologically active materials, the researcher’s goal is to maintain the genetic stability of these strains to maintain the desired productivity of the active constituents. This paper focused on evaluating the effect of nutrient media on genetic stability in sub-cultures of inducible callus strains selected from tissue culture of D. purpurea. This is the first study that investigated the genetic stability of D. purpurea callus.
2. MATERIALS AND METHODS
2.1. The Source of the Studied Cell Lines
Four cell lines (DPM1, DPM2, DPM3, and DPM4) of D. purpurea callus were obtained after establishing the vegetative cultures of the studied plant and inducing the callus [28]. The study was performed in biotechnology laboratories, National Commission for Biotechnology, and in the plant tissue culture laboratories, Department of Plant Biology, Damascus University, Damascus, Syria. This study was conducted from September 2016 to December 2021. The best cell lines in nutrient media enriched with different concentrations of plant growth regulators were selected [29]. These selected cell lines were regrown on the same media till sub-culture 80 [Table 1]. Four sub-cultures (sub-culture 10, sub-culture 20, sub-culture 40, and sub-culture 80) for each cell line were used to study the genetic stability index in the studied nutritional media. A seed sample of D. purpurea was imported from Hortus Company, Blenheim, New Zealand (batch No. Y481), and the in vitro grown plant resulting from the seeds was used as two controls.
Table 1: The studied cell lines for Digitalis purpurea callus.
| Cell Line | Medium | Concentration PGRS |
|---|---|---|
| DPM1 | MS | (2 mg 2.4–D+1 mgBA) L-1 |
| DPM2 | B5 | (2 mg 2.4–D+1 mgBA) L-1 |
| DPM3 | LS | (2 mg NAA+2 mgBA) L-1 |
| DPM4 | 5C01 | (2 mg NAA+1 mgKin) L-1 |
2.2. DNA Isolation
DNA was isolated from 200 mg of callus for each studied sample by cetyltrimethylammonium bromide as previously described [30] with adopting some modifications according to the recommendations of Sa’ et al. [31], with the addition of 2 μl of RNAase at the end of the reaction to eliminate any amount of RNA that may be produced during the extraction process to ensure high yield and higher purity of DNA.
2.3. Quantitative and Qualitative Determination of DNA
The amount of DNA in the samples was determined by reading optical densities of the samples at wavelengths 260/280 nm using (Hitachi UV Spectrophotometer) for three sample replicates, and the concentration of DNA was calculated from the following equation [32]:
DNA Concentration (g/l) = OD260 × expansion factor × 50
Where: OD260 represents the optical density of DNA absorption in micrograms at the wavelength of 260 nm, the DNA samples were extended to obtain a concentration of 50 ng. μl-1 and DNA were qualitatively determined on an agarose gel of 0.8%.
2.4. ISSR-Polymerase Chain Reaction (PCR) Reaction
The DNA was amplified by PCR according to the protocol of Williams et al. [33], with adopting some modifications to have a final reaction volume of 25 μl consisting of (2 μl of DNA, 12.5 μl of Green Master Mix, 1.25 μl of primer, and 9.25 μl of nuclease-free water). The PCR reaction was performed using the Kapa2G Fast Ready-Mix PCR kit according to the manufacturer’s instructions and according to the conditions shown in Table 2.
Table 2: Thermal program for the PCR.
| Stage | Temperature | Time (sec) | Cycles number |
|---|---|---|---|
| DNA strand break | 95°C | 300 | 1 |
| Denaturation | 95°C | 30 | 40 |
| Annealing | 38–58°C | 60 | |
| Extension | 72°C | 60 | |
| Final stage | 72°C | 600 | 1 |
| Indefinite | 4°C | ∞ | - |
A 3μl of SYBER DNA Stain (11×) was added to each reaction as an alternative to the other dyes, and then, 12 ml of the total reaction volume was injected into a 2% agarose gel in 1x TBE solution in the presence of DNA Ladder (1 Kb and 100 bp) in each reaction [34]. The bands were separated by electrophoresis for 2 h with an electric current of100 volts, and the gel was photographed with an Image Analyzer Eagle Eye II Teratogen. These procedures were repeated twice for each sample. ISSR technology was applied to study the genetic stability of the studied cell lines within the nutrient media. Table 3 shows the nucleotide sequence of the used primers and the degree of their coalescence.
Table 3: Primers, nucleotide sequence, thermal, and polymorphism properties.
| PR | Sequence | TM | T-B | P-MS | P-B5 | P-LS | P-5C01 |
|---|---|---|---|---|---|---|---|
| 1 | ACA CAC ACA CAC ACA CCG | 56 | 10 | 80% | 80% | 70% | 0 |
| 2 | CAC CAC CAC CAC CAC | 50 | 0 | 0 | 0 | 0 | 0 |
| 3 | GTG GTG GTG GTG GTG | 50 | 11 | 45.4% | 27.2 | 54.5% | 0 |
| 4 | CAC ACA CAC ACA G | 40 | 16 | 81.2% | 81.2% | 75% | 0 |
| C3 | CTC TCT CTC TCT CTC TGC | 51.5 | 13 | 69.2% | 84.6% | 38.5% | 38.5% |
| D4 | CAC ACA CAC ACA AC | 42.0 | 11 | 54.5% | 63.6% | 63.6% | 27% |
| E5 | CAC ACA CAC ACA GT | 42.0 | 8 | 100% | 62.5% | 62.5% | 12.5% |
| F6 | CAC ACA CAC ACA AG | 42.0 | 11 | 81.8% | 63.6 | 72.7% | 27% |
| G7 | CAC ACA CAC ACA GG | 44.0 | 12 | 66.6% | 33.3% | 75% | 0 |
| H8 | GAG AGA GAG AGA GG | 44.0 | 11 | 72.7% | 54.5% | 45.5% | 0 |
| I9 | GTG TGT GTG TGT GG | 44.0 | 8 | 0 | 75% | 62.5% | 0 |
| J10 | GAG AGA GAG AGA CC | 44.0 | 16 | 81.2% | 68.7% | 75% | 6% |
| K11 | GTG TGT GTG TGT CC | 44.0 | 11 | 81.8% | 63.6% | 81.8% | 9% |
| M13 | GAG GAG GAG GC | 38.0 | 11 | 90.9% | 72.7% | 36.3 | 0 |
| P15 | GTG GTG GTG GC | 38.0 | 11 | 90.9% | 72.7% | 81.8% | 27% |
| Q16 | GTG TGT GTG TGT GTG TC | 52.3 | 13 | 84.6% | 61.5% | 61.5% | 15.3% |
| R17 | ACA CAC ACA CAC ACA CG | 54.3 | 18 | 38.8% | 72.2% | 55.5% | 0 |
| S18 | AGG AGG AGG AGG AGG AGG | 57.2 | 18 | 55.5% | 55.5% | 55.5% | 0 |
| T19 | GAG AGA GAG AGA GAG AGA T | 50.0 | 12 | 83.3% | 66.6% | 50% | 0 |
| U20 | GAC AGA CAG ACA GAC A | 47.9 | 11 | 72.7% | 54.5% | 81.8% | 9% |
| 232 | 69.8% | 64.2% | 62.9% | 8% |
2.5. Statistical Analysis
The ISSR-PCR results were analyzed with the Total Lab 1 (Ultra-Lum Inc., Claremont-California) program, where the size of the DNA bands was determined. The appearance of the bands was encoded with the symbol 1 and its absence with the symbol 0. The matrix of percent disagreement values (PVD) was calculated based on the Jaccard similarity coefficient. The matrix was used in the cluster analysis by applying the unweighted pair group method with arithmetic averaging, and a genetic kinship tree was drawn using the power marker program.
3. RESULTS
The results of DNA extraction experiments from the selected cell lines of D. purpurea callus and the in vitro grown plants showed that the DNA purity at A260/A280 ranged between 1.98 and 1.94, except for the seed sample, where the DNA purity was 1.74. On the other hand, the average total DNA concentration per 1g of the sample ranged between 1609 μg/μl and 2845 μg/μl, while the average total DNA production per 1 g of seeds was 965.3 μg/μl. This was due to the fact that the callus samples and the micropropagation in vitro sample were fresh and extracted at the peak stages of cell division 21 days, in addition to the fact that the growth and germination conditions of the callus samples and the in vitro grown plant were in optimal and accurate conditions.
3.1. Polymorphism of the Cell Lines from D. Purpurea Callus
The genetic study included the use of 20 primers, 19 of which produced 232 bands (figures of 20 primers have been attached as submental materials) with an average of 12.8 bands for each primer. All bands were polymorphic, where the polymorphism percentage reached 69.8% in MS, 64.2% in B5, 62.9% in LS, and 8% in 5C01. The primer S18 and primer (R17) exhibited the highest number of bands, 18 bands each [Figure 1a and b]. In contrast, primer I9 and primer (E5) showed the lowest number of bands, with an average of eight bands each. The primers (1, 3, D4, E5, G7, H8, J10, M13, Q16, R17, S18, and U20) were unique by giving 25 unique bands, whereas the primers (E5, H8, and J10) excelled with three unique bands for each [Figure 1c-e]. The discrepancy in bands number could be attributed to several factors including the type of the primer, the nucleotide sequence of the primer, the fusion temperature during amplification, and the transcription point at which the primer finds its complementary sequence on the DNA template in the sample.
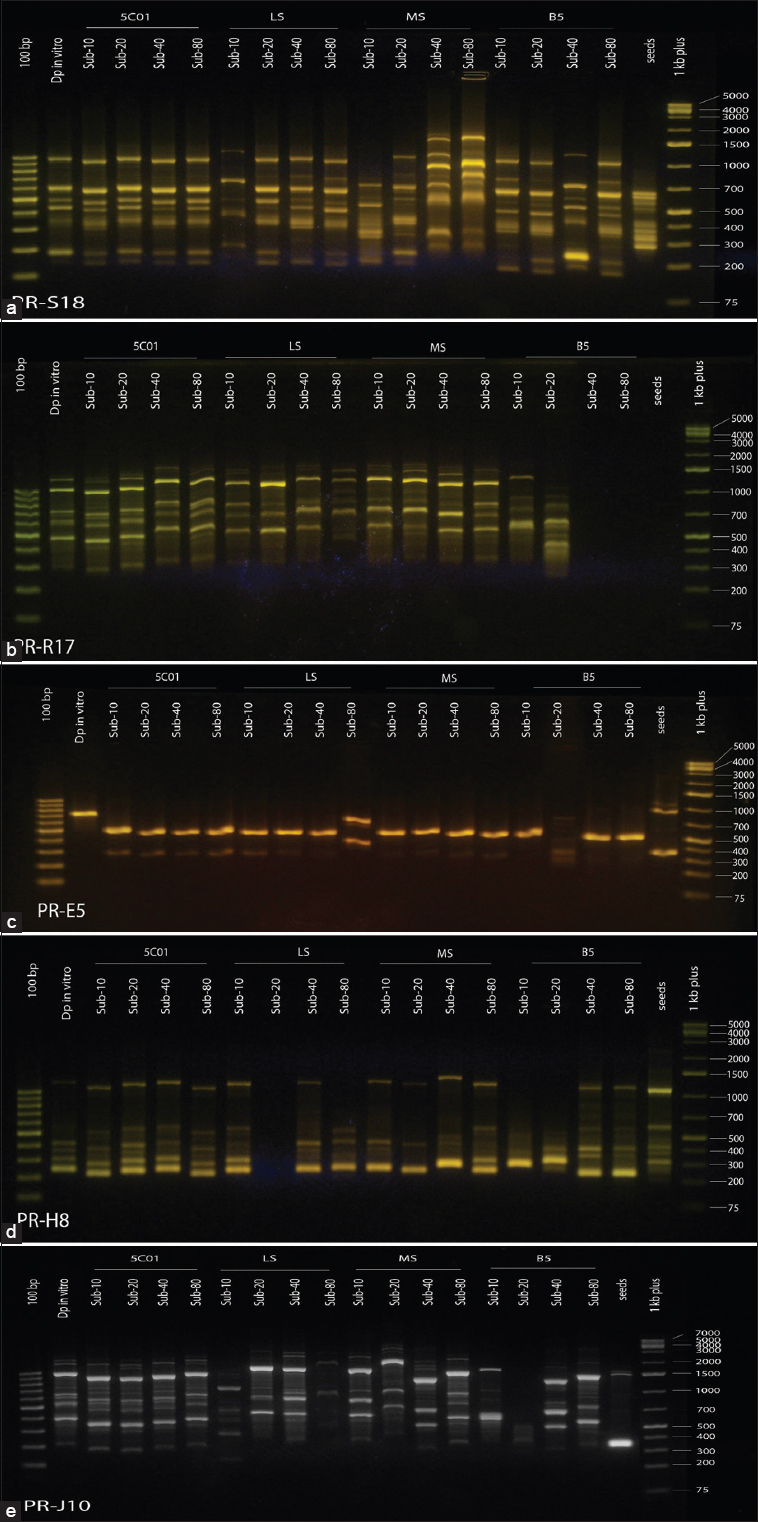 | Figure 1: Migration images of ISSR-PCR products of cell lines on agarose gel (a: primer S18; b: primer R17; C: primer E5; d: primer H8; e: primer J10) [Click here to view] |
3.2. Genetic Stability in Cell Lines
When applying the matrix of PVD for cell strains in the nutrient media [Table 4], the results showed that the nutrient medium 5C01 was more stable for the callus, where the value of PVD ranged between 0.0714 and 0.1231 during 5 years indicating a very low genetic diversity ratio. On the other hand, the coefficient of variance in the MS medium ranged between 0.5034 and 0.6178 in the callus subcultures, and the B5 and the LS media showed genetic instability, and the proportions of genetic variation in the callus subcultures ranged between 0.3805–0.7801 and 0.4580–0.7934, respectively. These findings confirmed the existence of genetic variation in the MS, LS, and B5 media. This comparison indicated the possibility of using the 5C01 medium in the development of the callus for long periods when the goal is to maintain the cell line in the stable and steady production of secondary compounds. Moreover, the results showed the presence of somaclonal variations that affected the level of genetic stability in the strains studied in the subcultures in the MS, LS, and B5 media, which made them suitable media in the production of the improved and productive strains of crops and in the production of new species that may be suitable for studying some of the stress-resistant properties or selecting some strains to improve their productivity of secondary compounds.
Table 4: Matrix of PVD of the studied cell lines of Digitalis purpurea.
| Micro- Pr | Sub5C- 10 | Sub5C- 20 | Sub5C- 40 | Sub5C- 80 | SubLS- 10 | SubLS- 20 | SubLS- 40 | SubLS- 80 | SubMS- 10 | SubMS- 20 | SubMS- 40 | SubMS- 80 | SubB5- 10 | SubB5- 20 | SubB5- 40 | SubB5- 80 | Seeds | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Micro- Pr | 0 | |||||||||||||||||
| Sub5C- 10 | 0.5168 | 0 | ||||||||||||||||
| Sub5C- 20 | 0.5163 | 0.0800 | 0 | |||||||||||||||
| Sub5C- 40 | 0.5067 | 0.0887 | 0.0714 | 0 | ||||||||||||||
| Sub5C- 80 | 0.5097 | 0.1231 | 0.0916 | 0.0853 | 0 | |||||||||||||
| SubLS- 10 | 0.6327 | 0.4030 | 0.3704 | 0.3926 | 0.3885 | 0 | ||||||||||||
| SubLS- 20 | 0.5685 | 0.5101 | 0.4698 | 0.4797 | 0.4636 | 0.4580 | 0 | |||||||||||
| SubLS- 40 | 0.5646 | 0.4552 | 0.4247 | 0.4452 | 0.4082 | 0.5217 | 0.4179 | 0 | ||||||||||
| SubLS- 80 | 0.8140 | 0.7424 | 0.7445 | 0.7293 | 0.7338 | 0.7934 | 0.7680 | 0.7907 | 0 | |||||||||
| SubMS- 10 | 0.4965 | 0.4595 | 0.4503 | 0.4392 | 0.4133 | 0.5248 | 0.5306 | 0.4648 | 0.7786 | 0 | ||||||||
| SubMS- 20 | 0.6081 | 0.6226 | 0.6111 | 0.6038 | 0.5776 | 0.6554 | 0.5423 | 0.6159 | 0.8016 | 0.5705 | 0 | |||||||
| SubMS- 40 | 0.5962 | 0.5032 | 0.4841 | 0.4839 | 0.4684 | 0.5278 | 0.5621 | 0.5395 | 0.8286 | 0.5513 | 0.6178 | 0 | ||||||
| SubMS- 80 | 0.5986 | 0.4577 | 0.4589 | 0.4690 | 0.4422 | 0.5038 | 0.5625 | 0.5385 | 0.7920 | 0.5510 | 0.6027 | 0.5034 | 0 | |||||
| SubB5- 10 | 0.6929 | 0.6483 | 0.6443 | 0.6554 | 0.6447 | 0.7059 | 0.6370 | 0.6015 | 0.8468 | 0.6232 | 0.6391 | 0.6803 | 0.6593 | 0 | ||||
| SubB5- 20 | 0.7482 | 0.7778 | 0.7862 | 0.7662 | 0.7688 | 0.8322 | 0.7500 | 0.7174 | 0.8558 | 0.7517 | 0.7536 | 0.7329 | 0.7445 | 0.6930 | 0 | |||
| SubB5- 40 | 0.6028 | 0.6000 | 0.5974 | 0.5800 | 0.5897 | 0.6620 | 0.7059 | 0.6014 | 0.8455 | 0.6027 | 0.6735 | 0.6133 | 0.6458 | 0.7801 | 0.7364 | 0 | ||
| SubB5- 80 | 0.6338 | 0.6200 | 0.6169 | 0.6093 | 0.6090 | 0.6831 | 0.6644 | 0.5929 | 0.8607 | 0.5944 | 0.6939 | 0.6601 | 0.6383 | 0.7099 | 0.7302 | 0.3805 | 0 | |
| Seeds | 0.7267 | 0.7407 | 0.7195 | 0.7301 | 0.7337 | 0.7483 | 0.7613 | 0.7152 | 0.8760 | 0.7389 | 0.7230 | 0.7607 | 0.7647 | 0.7704 | 0.8106 | 0.6567 | 0.6385 | 0 |
When comparing the micropropagation sample, which was a source of explants used in callus induction, and the cellular varieties studied in the media, the coefficient of variation values ranged 0.4965–0.8140. Furthermore, the coefficient of variation values between the seed sample and the studied callus samples ranged 0.6385–0.8760. This may return to the emergence or disappearance of some genes associated with some differentiation characteristics, structures, or physiological processes associated with light and the development of plant structures.
3.3. Cluster Analysis of Cell Lines of D. Purpurea Callus, Seeds, and Plants Growing In Vitro (Micropropagation)
The results of the cluster analysis in Figure 2 showed the separation of cell lines based on the degrees of similarity, where the studied samples were separated into seven groups “clusters,” a set of sub-cultures cell lines of callus (sub 5C-10; 5C-20; 5C-40; 5C-80; LS-10; MS10; LS-40; LS20; MS-40; and MS-80) gathered with the in vitro grown plant into one group and a number of sub-groups. In addition, the cell lines growing in the 5C01 medium were separated into one sub-group, the cell lines (Sub B5-80 and B5-40) separated into one group, and the cell lines (MS-20; B5-10; B5-20; and LS-80) were separated into four groups, each strain in a separate group, while the seed samples were separated in another separate group.
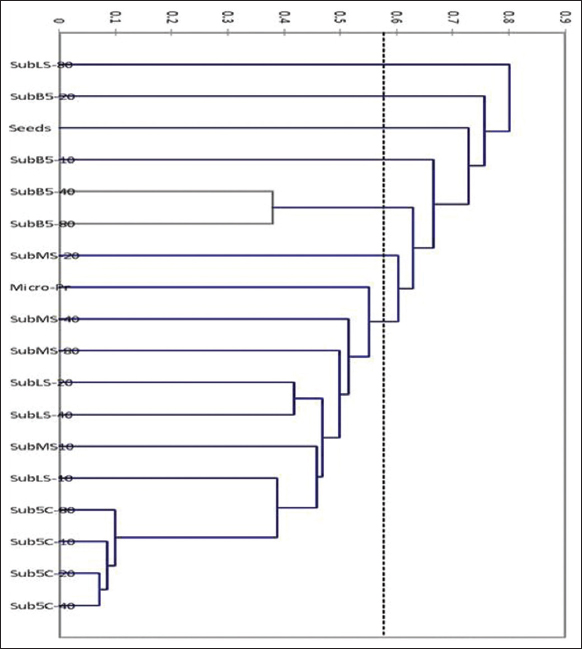 | Figure 2: Cluster analysis of the studied cell lines, seeds and plants growing in vitro [Click here to view] |
The results of the distances between the clusters showed that the distance between the in vitro grown plant, the explants source, and the cellular strains was an average point of variation and similarity in both respects and that the clear cases of genetic variation occurred, especially in the B5 medium, were due to the fact that this medium enhanced the level of somaclonal variations and may be a suitable medium for genetic mutations. In the LS medium, the strain SubLS-80 showed a single group, and it was the furthest in terms of distance and indicated that the LS medium may be stable to a specific level in sub-cultures. In the strain SubLS-40, the genetic stability was acceptable, changed very much, and deviated toward an independent group path in the SubLS-80.
A few sub-cultures (Sub-Culture 20) in B5 and MS media showed a temporary deviation to form single cell lines and were genetically differentiated. This deviation returned to the normal course in the subsequent sub-cultures. This may be attributed to temporary somaclonal variations that may arise in callus cultures, and these were revising variations, or to the emergence and disappearance of a few genes as a result of adaptation in the nutrient medium, or to the difference in the mitotic stages at the level of the callus, where the divisions in the callus were not synchronized.
4. DISCUSSION
Genetic diversity studies are gaining importance in tissue culture according to the type and course of the study. Genetic diversity is important in genetic improvement programs of crops or the production of plants resistant to stresses and plants free of viruses. However, genetic instability is a problem in programs for producing secondary compounds from tissue cultures for medicinal plants.
The callus induction is an initial step in the path of secondary components synthesis. The stability and permanence of callus growth are the most important indicators since the ability to maintain callus production varies with different species, where the callus can lose its properties after several sub-cultures in the species Rosmarinus officinalis and Rosa canina [35,36] while Lavandula angustifolia maintained its ability after the 20th sub-culture in the presence of regular transfer every 3–4 weeks [37].
The importance of studying genetic diversity among berry cultivars in selecting varieties with higher yields in fruits was previously reported [22]. It has been found that genetic variation was positive between genotypes and is useful for selecting appropriate groups for genomic mapping [14], and these genetic variations accumulated as a result of adaptation to different nutrient environments in artificial media, biotechnology methods that adopt bacteria to transfer the desirable traits in corps such as rice [38], thus stimulating physical, genetic variations as a result of adaptation, or non-adaptation to the components of the nutrient medium [10]. These findings support our results that showed different values of genetic stability in the studied media containing different types of plant growth regulators.
The DNA purity in our study ranged between 1.98 and 1.94. These results lay within a normal percentage of pure DNA preparations (the normal range is between 1.8 and 2). Our results outperformed the results of Mir et al. [39] and matched with the results of Tshilenge et al. [40] on coffee species Coffea robusta. The high DNA purity in our study could be attributed to the modifications made to the adopted extraction protocol resulting in efficient purification of the DNA from proteins and RNA, which go online with previous explanations [31]. The high degree of purity of the isolated DNA in our study was manifested by clear, separate, unbroken, and well-defined bands of DNA in the migration images on the gel [Figure 1]. This DNA purity supported the quality of the subsequent PCR reactions, which also had been previously reported [39].
When samples of Crocus sativus L. were analyzed using 8 ISSR primers, four of them produced plurality bands while the rest of the primers did not show any suitable amplified bands [39]. In our study, 20 primers were used, 19 of them produced bands with polymorphism and one primer did not show any bands. The large number of primers used in our study was consistent with the recommendations of Sunar et al. [24] emphasized the need to use a large number of multiple nucleotide sequence markers to measure genetic relationships and genetic diversity in a reliable way. The discrepancy in the genetic stability of callus cell cultures in nutrient media was attributed to the component’s variance of the nutrient medium, the difference in the electrolytes quality of the minor and major elements that make up the nutrient media, the difference in the quality and concentrations of enhanced plant growth regulators, and the effect of these regulators on the levels of somaclonal Variations. Kunakh [41] suggested that changes in DNA methylation, amplification of special sequences from it or additional synthesis, changes in genetic stock or the number of chromosomes, changes in mitotic phases, and the accompanying abnormalities were factors that affect the genetic variations in callus cultures.
Our study excelled in the average number of bands (12.2 bands) for each primer in the overall interactions compared with the study of Liu et al. [42] which produced bands with an average of 5.9 bands for each ISSR primer with 88.28% polymorphism. The molecular weight of the bands in our study ranged between 75 and 3500 bp while it ranged between 200 and 2000 bp in Lycium species or cultivars in the same study [42].
In studying the genetic variance of Lycium varieties [42], the genetic distances between different cultivars or species had a Jacquard coefficient index of 0.36–0.98 and generally converged with the results of our study in MS, LS, and B5 media but differed in the 5C01 medium, which showed clear genetic stability.
Genetic variation in the soft fruit cultures was observed in the fifth sub-cultures, and the maximum variation was 28% [43]. The variation could reach high levels, especially when the cultural conditions were favorable for this phenomenon. For example, the analysis using RAPD for plants resulting from tissue culture of pineapple for 4 years showed a degree of variance of 95.9% [44]. Negi and Sexena noticed no genetic variation until the sub-culture 33 of Bambusa balcooa using ISSR-PCR technique [45]. Myersandsimon reported genetic instability in callus cultures of Allium sativum that were sustained for long periods of time on medium supplemented with 2,4-D [46]. Simoes et al. [47] noticed a gradual decrease in embryonic efficiency when re-implanting for the 2nd time (second sub-culture) on the same medium, due to the effect of using 2.4-D for a long time. The embryonic efficiency of sweet potato callus lines decreased during continuous exposure to 2.4-D, and this was explained by the decrease in the gene expression associated with the aging of culture when using 2.4-D [48]. Changes in the cellular efficiency of embryo formation were observed as well in the presence of 2.4-D in sugarcane callus cultures [49], where the use of 2.4-D led to rapid cell divisions in the early stages, followed by a difference in the growth of the formed callus mass and a decrease in the rate of cell division [50]. Atrophy of some callus cells occurred as a result of ethylene release in large quantities when 2.4-D was used for long periods [51].
ISSR primers were used by several studies to assess the genetic fidelity of in vitro bred Stevia rebaudiana (Bert) plants [52-55] and to assess the genetic similarity between different groups and treatments in S. rebaudiana [56,57]. The RAPD and ISSR primers for wild species and tissue culture-derived plants of Oryza sativa did not show morphological pluralism, according to Krishnan et al. [17]. The SSR method was employed in identification of stable quantitative trait loci for fiber quality traits in Gossypium hirsutism [58]. In our study, the cytokinins were the most prominent factor for inducing genetic variations through polymorphism. The difference in cytokinin type in the 5C01 medium enhanced with Kin compared to the MS, LS, and B5 media enhanced with BA was an essential indicator of genetic stability in synergy with some other related to the quality of the nutrient medium components and the mineral electrolytes. These findings would open the way for the possibility of benefiting from the genetic stability assessment study in evaluating the chemical constituents of cardiac glycosides of the cell lines of D. purpurea callus, and working to induce and increase production in the selected cell lines.
5. CONCLUSIONS
From the results of the present study, it could be concluded that the elimination of proteins and RNA during the DNA extraction protocol enhanced the purity of DNA and the extraction yield of DNA. Furthermore, the somaclonal variations that occurred at the level of sub-cultures were often temporary and varied according to the nutrient medium and the plant growth regulators that promote it. Importantly, the genetic variation was affected by the mineral electrolytes concentration in the nutrient media of the sub-cultures of D. purpurea callus. Similarly, the type and concentration of cytokinins are important factors in the genetic variation index in nutrient media enriched with plant growth regulators. It is necessary to conduct genetic variation studies through a wide range of primers to investigate the somaclonal variations and morphological polymorphism in callus cultures.
6. ACKNOWLEDGMENTS
Thanks go to Prof. Fahed Albiski, Dr. Shaza Besher, and Eng. Buthina Al-Salama from the Department of Molecular Biology, National Commission for Biotechnology, Damascus –Syria, for their advices and support during achieving this work. We would like to thank two anonymous reviewers and the editor for their helpful comments to improve the manuscript.
7. AUTHORS’ CONTRIBUTIONS
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Mohammed Ahmed AL-Oqab. The first draft of the manuscript was written by Mohammed Ahmed AL-Oqab, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
8. FUNDING
The authors did not receive support from any organization for the submitted work.
9. CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
10. ETHICAL APPROVAL
This research did not involve experiments with human or animal participants.
11. DATA AVAILABILITY
The data presented in this study are available on request from the corresponding author.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Olmstead RG. Whatever happened to the
2. Verma SK, Das AK, Cingoz GS, Gurel E.
3. Willis AJ, Memmott J, Forrester RI. Is there evidence for the post-invasion evolution of increased size among invasive plant species?Ecol Lett 2000;3:275-83. [CrossRef]
4. Chen S, Khusial T, Patel D, Singh S, Yakubova T, Wang A,
5. Platz EA, Yegnasubramanian S, Liu JO, Chong CR, Shim JS, Kenfield SA,
6. Durlacher CT, Chow K, Chen XW, He ZX, Zhang X, Yang T,
7. Lindblom L, Ekman S. Genetic variation and population differentiation in the lichen-forming ascomycete
8. Cho J, Lee YJ, Kim JH, Kim SI, Kim SS, Choi BS,
9. Probert R, Adams J, Coneybeer J, Crawford A, Hay F. Seed quality for conservation is critically affected by pre-storage factors. Aust J Bot 2007;55:326-35. [CrossRef]
10. Sabit H, Abdel-Ghany S, Al-Dhafar Z, Said OA, Al-Saeed Ja, Alfehaid Ya,
11. Ishizaki K, Nishihama R, Yamato KT, Kohchi T. Molecular genetic tools and techniques for
12. Bhusare BP, John CK, Bhatt VP, Nikam TD.
13. Gurel E, Yucesan B, Aglic E, Gurel S, Verma SK, Sokmen M,
14. Souframanien J, Gopalakrishna T. A comparative analysis of genetic diversity in blackgram genotypes using RAPD and ISSR markers. Theor Appl Genet 2004;109:1687-93. [CrossRef]
15. Wójcik D, Trzewik A, Kucharska D. Field performance and genetic stability of micropropagated gooseberry plants
16. Rout GR, Mohapatra A, Jain SM. Tissue culture of ornamental pot plant:A critical review on present scenario and future prospects. Biotechnol Adv 2006;24:531-60. [CrossRef]
17. Krishnan SR, Priya AM, Ramesh M. Rapid regeneration and ploidy stability of 'cv IR36'indica rice (
18. Park SY, Cho HM, Moon HK, Kim YW, Paek KY. Genotypic variation and aging effects on the embryogenic capability of
19. Bairu MW, Aremu AO, van Staden J. Somaclonal variation in plants:Causes and detection methods. Plant Growth Regul 2011;63:147-73. [CrossRef]
20. Rani V, Raina SN. Genetic fidelity of organized meristem-derived micropropagated plants:A critical reappraisal.
21. Deepthi VP. Somaclonal variation in micro propagated bananas. Adv Plant Agric Res 2018;8:624-7.
22. Abdelaziz SM, Medraoui L, Alami M, Pakhrou O, Makkaoui M, Boukhary AO,
23. Ramzan M, Sarwar S, Kauser N, Saba R, Hussain I, Shah AA,
24. Sunar S, Korkmaz M, Sigmaz B, Agar G. Determination of genetic relationships among
25. Omondi EO, Debener T, Linde M, Abukutsa-Onyango M, Dinssa FF, Winkelmann T. Molecular markers for genetic diversity studies in African leafy vegetables. Adv Biosci Biotechnol 2016;7:188-97. [CrossRef]
26. Yang D, Ma P, Liang X, Liang Z, Zhang M, Shen S,
27. Culley TM, Sbita SJ, Wick A. Population genetic effects of urban habitat fragmentation in the perennial herb
28. AL-Oqab M, Zaid S, Al-Ammouri Y. Effect of different concentration of plant growth regulators on the germination and micropropagation of
29. Al-Oqab M, Zaid S, Al-Ammouri Y. Effect of the basic media enhanced with plant growth regulators on the inducting and growing of callus from
30. Doyle JJ. Isolation of plant DNA from fresh tissue. Focus 1990;12:19-5.
31. SáO, Pereira JA, Baptista P. Optimization of DNA extraction for RAPD and ISSR analysis of
32. Marietta MA, Yoon PS, Iyengar R, Leaf CD, Wishnok JS. Molecular Cloning. A Laboratory Manual. Vol. 27. United States:Cold Spring Harbor Laboratory, Cold Spring Harbor:Telford Press;1988.
33. Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 1990;18:6531-5. [CrossRef]
34. Sambrook J, Maccallum P, Russell D. Molecular Cloning:A Laboratory Manual, 2nd ed. Cold Spring Harbor, NY:Cold Spring Harbor Laboratory Press;1989.
35. Prisecaru M, Ghiorghi??G, Iliescu C, Petrescu D. Studiul Organogenezei ?G, Iliescu C, Petrescu D. Studiul Organogenezei
36. Ghiorghi??G, Nicu??D, Maftei DE, Maftei ID. Preliminary data of the
37. Ghiorghi?? G, Maftei DE, Nicu?? D. Some aspects concerning the
38. Hasan M, Rong-Ji D, Bano A, Hassan SG, Iqbal J, Awan U,
39. Mir MA, Mansoor S, Sugapriya M, Alyemeni MN, Wijaya L, Ahmad P. Deciphering genetic diversity analysis of saffron (
40. Tshilenge P, Nkongolo KK, Mehes M, Kalonji A. Genetic variation in
41. Kunakh VA. Evolution of cell populations
42. Liu X, Du J, Khan A, Cheng J, Wei C, Mei Z,
43. Khan S. Variations Arising Through Tissue Culture in Soft Fruits. Scotland, UK:The University of Edinburgh;1997.
44. Roostika I, Khumaida N, Ardie SW. RAPD analysis to detect somaclonal variation of pineapple
45. Negi D, Saxena S. Ascertaining clonal fidelity of tissue culture raised plants of
46. Myers JM, Simon PW. Regeneration of garlic callus as affected by clonal variation, plant growth regulators and culture conditions over time. Plant Cell Rep 1999;19:32-6. [CrossRef]
47. Simões C, Albarello N, Callado CH, De Castro TC, Mansur E. Somatic embryogenesis and pant regeneration from callus cultures of
48. Padmanabhan K, Cantliffe DJ, Koch KE. Auxin-regulated gene expression and embryogenic competence in callus cultures of sweetpotato,
49. Guiderdoni E, Demarly Y. Histology of somatic embryogenesis in cultured leaf segments of sugarcane plantlets. Plant Cell Tissue Organ Cult 1988;14:71-88. [CrossRef]
50. Corchete MP, Sanchez JM, Cacho M, Moran M, Fernandez-Tarrago J. Cardenolide content in suspension cell cultures derived from root and leaf callus of
51. Trigiano RN, Gray DJ. Plant Tissue Culture Concepts and Laboratory Exercises. 2nd ed. England, UK:Routledge;2018.
52. Lata H, Chandra S, Techen N, ElSohly M, Khan IA. Determination of genetic stability of micropropagated plants of
53. Soliman HI, Metwali EM, Almaghrabi OA. Micropropagation of
54. Singh P, Dwivedi P, Atri N.
55. Lata H, Chandra S, Techen N, Wang YH, Khan IA. Molecular analysis of genetic fidelity in micropropagated plants of
56. Sharma N, Kaur R, Era V. Potential of RAPD and ISSR markers for assessing genetic diversity among
57. Othman HS, Osman M, Zainuddin Z. Genetic variabilities of
58. Jamshed M, Jia F, Gong J, Palanga KK, Shi Y, Li J,