1. INTRODUCTION
Coal mining has a series of repercussions on land resources and enormous pressure on the ecosystem. Extensive coal mining processes such as stripping, excavation, transportation, and dumping destroy the nutritional and functional properties of soil by disrupting soil texture, organic matter, minerals, and microbial population. Moreover, the rehabilitation of mined soil may result in increased spatial heterogeneity of soils [1,2]. At present, the reclamation process by vegetative cover is considered the most desirable, environmentally friendly, and sustainable technique to overcome the hazardous and ecological problems caused due to coal mining activities. According to Dubey et al. [3], in most mined regions of India, genuine and sincere reclamation actions based on scientific findings have not been performed. The initiative revegetation of the mining sites of country is not attaining the anticipated outcomes due to a high mortality rate and delayed growth of planted species. The coal mine tailing is a very hostile substrate for the growth and development of plant and soil microorganisms due to inhospitable physicochemical conditions such as high salinity, high concentrations of heavy metals, lower water retention due to coarse fraction prevalence, and deficiencies in soil organic matter. Therefore, improvement of physicochemical properties of mine tailing is very crucial before rehabilitation process [4].
The existing method of reclaiming the mined area is capital and energy-intensive. The conventional method consists of backfilling the area level in a proper direction to avoid any water-logging or water run-off situation; laying an additional top soil layer; application of mulches and/or adhesive chemical stabilizers to the surface soil that helps to prevent erosion and encourage the formation of vegetative cover; application of soil amendments such as lime or gypsum to neutralize the acidity or alkalinity, respectively, and amendments of fertilizer according to the deficiency of macro and micronutrients periodically to sustain the plant growth [5]. First two steps necessitate extensive machinery, which increases the cost of the reclamation process while amendments of fertilizer is not practicable or requires continuous fertigation on monthly basis. However, when fertigation is stopped or fertilizers are washed away after initial applications, the soil is still not able to support plant growth on its own. In addition, chemical fertilizers have a low nutrient usage efficiency; thus, they are applied in much larger quantities than are needed. This over-fertigation results in leaching of nitrogen and other fertilizers, thereby causing pollution of surrounding areas that can include ground water contamination and eutrophication of water bodies [6]. As a result, the goal of the coal mined site or backfilled restoration to cultivable land through conventional practice is not achieved in an economically viable and environmentally friendly manner.
As per the conventional practice of backfilled reclamation suggested by the Indian Bureau of Mines, there is a process of laying the topsoil on the backfilled area at a thickness of around 30 cm. If the dressing is done with the topsoil, after some time, due to the seasonal effect, the salt lying under the topsoil layer will gradually come upward and reach to the rhizosphere of the plant, which in turn leads to the salt stress [5]. The rhizobacteria acclimatized to salt stress, when used innovatively, can help the plants to grow successfully in similar salt stress conditions. The use of plant growth-promoting rhizobacteria (PGPR) for reclamation enable barren landfills to become fertile in a short period, boosting soil water holding capacity, and generating topsoil that can support high-quality flora perpetually. Furthermore, site-specific microorganisms have high metabolic versatility, allowing them to acclimate in low nutrition levels as well as in the deleterious chemical and physical conditions of mine tailings. Thus, the acclimatized PGPR could aid in the development of a low-cost and sustainable method for rehabilitating mining regions with an increased output. In these regard, a stronger focus is required on isolation and characterization of PGPR populations acclimatized to harsh conditions, which can provide significant benefits to plants under various stress. In addition, the PGPR populations can also be accelerated by supplementing the organic wastes locally available from the nearby industries such as sugar mills, paper mills, and food processing. This can provide long-term sustainability by increasing the carbon level of the soil periodically, which also reduces the cost of the reclamation process.
The cost-effective and sustainable methods such as enriching the PGPR of coal mine soils and application of acclimated PGPR should be explored in view of the disadvantages of conventional reclamation methods and the urgent need to accelerate the reclamation process. In this regard, the present study was conducted to identify diversity amongst the PGPR populations found in the rhizosphere of plants growing in lignite coal mines. Physicochemical properties were studied to understand the stress tolerance of PGPR. Isolation and phylogenetic analysis was conducted for the culturable PGPR with the intention that a better understanding of PGPR diversity at the coal mines will assist in reclamation process at coal tailing sites.
2. MATERIALS AND METHODS
All the reagents and media used in the study were of analytical grade. Ready to used media of Luria Bertani agar, Pikovskaya’s agar, Aleksandrow agar, and sheep blood agar media plates were acquired from Himedia, India. Jensen’s agar media and zinc-solubilizing media were prepared in laboratory [7]. Jensen’s agar was comprised of (g/L): sucrose, 20.0; Fe2(SO4)3·H2O, 0.1; K2HPO4, 0.5; MgSO4·7H2O, 0.5; NaCl, 0.5; CaCO3, 2.0; Na2MoO4·2H2O, 0.005; agar, 20 and pH, 7.2. Zinc-solubilizing medium was comprised of (g/L): glucose, 10; ZnO, 1.0; (NH4)2SO4, 0.5; KCl, 0.2; yeast extract, 0.5; FeSO4·7H2O, 0.01; MnSO4, 0.01; K2HPO4, 0.5; agar, 20 and pH, 7.2 [7,8].
2.1. Study Area and Sample Collection
Samples were collected from the rhizosphere of the plants from Amod G-19 Extension Lignite Mine. It is an opencast mining sector run by Gujarat Mineral Development Corporation, Ltd [9]. It deals with excavating lignite, silica sand, and ball clay. It is located near Amod village of Jhagadia taluka, Bharuch, Gujarat, India (latitude: 21°42’12” to 21°43’14” and longitude: 73°13’02” to 73°14’12”). The average depth for lignite seam may be up to 150 m and cut off depth for lignite seam is 100 m. The mines cover a total 385 hectare (ha) lease area out of which 301.34 ha is occupied by lignite. Total 168 ha area was excavated till September 2018 with production rate of 1 million ton per annum. The reclamation program was initiated at Amod G-19 Extension Lignite Mine by planting in 17.42 ha area. A total 50 ha area has been proposed to be converted into agricultural land by the end of coal mine life. A cost of Rs. 110/m2 has been estimated in the development of the backfilled area for agricultural uses as per the 2019–2020 report [10].
A total of 37 samples were collected from the different locations of coal mine sites using random sampling method [Figure 1]. Samples were collected by digging the rhizospheric zone and placed in sterile plastic bags. The samples were kept at 4°C until use. The mine soil was assessed for physicochemical properties such as pH, appearance, electrical conductivity (EC), moisture content, organic matter, organic carbon, total nitrogen, total phosphorus, total soluble sulfates, cation exchange capacity, and heavy metals according to agri biochem research lab (ABRL) standard protocol. The ratios of water to soil were 1:2 and 1:4 for measurement of pH and EC, respectively.
 | Figure 1: (a) The sample collection site and (b) map of sampling. [Click here to view] |
2.2. Isolation of PGPR
Isolation was carried out by suspending 10 g of soil sample in 90 ml of sterile normal saline followed by shaking at 150 rpm, 37°C for 1 h. The supernatant was then serially diluted, and 0.1 ml of suspension was spread on selective media for the isolation of PGPR. The differential media, namely, Jensen’s medium, Pikovskaya’s agar, Aleksandrow agar, and zinc-solubilizing medium, were used for the isolation of nitrogen-fixing, phosphate, potash, and zinc-solubilizing rhizobacteria, respectively. The plates were incubated at 28 ± 2°C for 5–6 days. Different types of isolates were then selected based on morphological characteristics and zone of solubilization. Subculturing was performed on the same medium for further purification of bacterial isolates. Putative nitrogen-fixing bacteria were preserved on Jensen agar slants whereas other isolates were preserved on Luria Bertani agar slants.
2.3. Screening of Potent PGPR
The screening of potent phosphate, potash, and zinc-solubilizing rhizobacteria was carried out by determining the zone of solubilization on Pikovskaya’s agar, Aleksandrow agar, and zinc-solubilizing medium, respectively. The isolates were spread onto respective media and incubated for 5–7 days at 28 ± 2°C. The positive isolates were considered by the presence of a halo zone around the bacterial colony. The solubilization index was calculated as given in equation (1) [11]
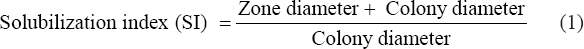 |
2.4. Hemolytic Activity
The pathogenicity test was carried out by culturing the selected PGPR on blood agar plates (HiMedia, Cat#MP1301). The plates were incubated at 28 ± 2°C. Bacterial isolates were selected based on their ability to produce different types of hemolysins test. The formation of distinct hemolytic halo around the colonies indicated β-hemolysis. A greenish and dark color under the colony indicated alpha-hemolysis. In case of gamma-hemolysis or non-hemolysis, the agar media did not change any color or opacity [12].
2.5. Molecular Identification of Bacterial Isolates
2.5.1. DNA isolation, amplification, and sequencing
The bacterial isolates were sent for the 16S ribosomal RNA (rRNA) gene sequencing at Bionivid Technology Pvt. Ltd. Bengaluru. The genomic DNA extraction was carried out using Wizard® Genomic DNA Purification Kit (Cat# TM050). PCR reactions were performed on Veriti® thermal cycler (Applied BioSystems) under the following conditions: 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 90 s, with a final extension at 72°C for 10 min. The PCR reactions were performed in a 20 μl volume comprising DreamTaq Green PCR Master mix (Cat# K1081), 10 pmol of primers 27F and 1492R [13], and 50 ng of genomic DNA. The PCR product was visualized on 2% agarose gel and Purelink™ Quick PCR Purification kit (Cat# K310001) was used to purify the amplified PCR product. Successful PCR purified products were used for 16S rRNA gene sequencing. To confirm the identity of bacterial species, the obtained sequences were compared with the available sequences in the NCBI (National Centre for Biotechnology Information) database using Basic Local Alignment Search Tool. The sequences were identified using a 99% base pair match to the closest accessible reference sequences [14]. DNA barcodes for nucleotide sequences were generated using the Barcode of Life Data System (BOLD).
2.5.2. Phylogenetic analysis
The phylogenetic tree was built using 16S rRNA gene sequencing data of isolated bacterial strains and closely related species obtained from GenBank, NCBI. The nucleotide sequences were aligned with Clustal [15] integrated in MEGA X [16]. Partition Finder was used to analyze aligned data and choose the optimum partitioning scheme [17]. Phylogenetic relationships in RAxML were inferred using Maximum Likelihood (ML) analysis [18]. ML analysis was carried out for 1000 bootstrap replicates using GTR + 1 model.
3. RESULTS AND DISCUSSION
3.1. Physicochemical Characteristics of Coal Mine Spoils
The plant growth at the mined site is associated with the physicochemical properties of mine spoils. Therefore, to carry out the selection of site specific PGPR from these coal mine sites, it becomes important to understand its physicochemical parameters. A total of 37 samples were collected from the different locations of the coal mine site. The pH of the mine spoils was found to be acidic to neutral. The soil contained soluble salts between 38.4 ppm and 7712.0 ppm. The results clearly depicted that the pH and EC of soil affects the presence of PGPR [Table 1]. PGPR were not isolated from the soil samples having the pH below 2.5. Similarly, PGPR were not isolated from the soil samples having EC above 2.6 mS/cm. The soil is to be considered slight saline if the EC values are above 2 mS/cm which may have negative effect on growth of microorganisms [19]. Therefore, PGPR were not isolated from the soil samples with high EC values. Maximum PGPR populations (82%) were isolated from the soil samples having pH range between 6.0 and 7.8 whereas 18% PGPR populations were isolated from the soil samples having pH range between 3.0 and 5.5.
Table 1: Details of sample collection, soil characteristics and isolation of PGPR.
| S. No. | Sample ID | Collection date | Latitude | Longitude | Soil type | Soil color | Soil pH | Soil EC (mS/cm) | Nitrogen fixers | PSB | KSB | ZSB | Total PGPR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PJ294 | 20.02.2021 | 21 42 19.55 N | 73 13 14. 98 E | Sandy | Dark brown | 7.21 | 0.17 | 0 | 1 | 0 | 0 | 1 |
| 2 | PJ295 | 20.02.2021 | 21 42 19.40 N | 73 13 13.94 E | Sandy | Light brown | 4.04 | 0.18 | 0 | 1 | 0 | 0 | 1 |
| 3 | PJ296 | 20.02.2021 | 21 42 19.13 N | 73 13 13.10 E | Sandy | Brown | 5.31 | 0.29 | 2 | 2 | 0 | 1 | 5 |
| 4 | PJ297 | 20.02.2021 | 21 42 19.02 N | 73 13 13.27 E | Sandy | Dark gray | 6.02 | 0.14 | 0 | 2 | 0 | 0 | 2 |
| 5 | PJ298 | 20.02.2021 | 21 42 18.57 N | 73 13 13.37 E | Sandy | Brown | 6.20 | 0.24 | 0 | 1 | 0 | 0 | 1 |
| 6 | PJ299 | 20.02.2021 | 21 42 19.41 N | 73 13 13.06 E | Sandy | Dark brown | 6.84 | 0.20 | 0 | 1 | 0 | 0 | 1 |
| 7 | PJ300 | 20.02.2021 | 21 42 18.77 N | 73 13 12.45 E | Sandy | Brown | 4.05 | 0.46 | 0 | 1 | 0 | 0 | 1 |
| 8 | PJ301 | 20.02.2021 | 21 42 17.23 N | 73 13 12.40 E | Loamy | Black | 2.44 | 2.76 | 0 | 0 | 0 | 0 | 0 |
| 9 | PJ302 | 20.02.2021 | 21 42 14.86 N | 73 13 11.84 E | Chalky | Dark brown | 3.01 | 2.14 | 1 | 2 | 0 | 0 | 3 |
| 10 | PJ303 | 20.02.2021 | 21 42 15.61 N | 73 13 12.13 E | Sandy | Brown | 6.44 | 0.06 | 2 | 0 | 0 | 1 | 3 |
| 11 | PJ304 | 20.02.2021 | 21 42 35.59 N | 73 13 54.28 E | Clay | Red | 6.44 | 0.11 | 2 | 2 | 1 | 0 | 5 |
| 12 | PJ305 | 20.02.2021 | 21 42 34.57 N | 73 13 54.91 E | Sandy | Brown | 6.80 | 0.06 | 2 | 0 | 0 | 0 | 2 |
| 13 | PJ306 | 20.02.2021 | 21 42 33.57 N | 73 13 56.90 E | Sandy | Light brown | 6.48 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| 14 | PJ307 | 20.02.2021 | 21 42 33.67 N | 73 13 56.88 E | Sandy | Light brown | 6.49 | 0.07 | 2 | 0 | 0 | 0 | 2 |
| 15 | PJ308 | 20.02.2021 | 21 42 29.70 N | 73 13 54.02 E | Sandy | Light brown | 6.39 | 0.06 | 2 | 1 | 0 | 0 | 3 |
| 16 | PJ309 | 20.02.2021 | 21 42 29.33 N | 73 13 54.10 E | Sandy | Gray | 7.09 | 0.22 | 3 | 0 | 0 | 0 | 3 |
| 17 | PJ310 | 20.02.2021 | 21 42 18.38 N | 73 13 13.27 E | Sandy | Light yellow | 1.62 | 12.05 | 0 | 0 | 0 | 0 | 0 |
| 18 | PJ311 | 20.02.2021 | 21 42 15.80 N | 73 13 12.41 E | Clay | Black | 5.48 | 0.82 | 0 | 1 | 0 | 1 | 2 |
| 19 | PJ312 | 20.02.2021 | 21 42 26.57 N | 73 13 51.53 E | Sandy | Light brown | 6.33 | 0.10 | 2 | 2 | 0 | 2 | 6 |
| 20 | PJ313 | 20.02.2021 | 21 42 15.64 N | 73 13 48.22 E | Sandy | Brown | 7.37 | 0.99 | 2 | 1 | 0 | 2 | 5 |
| 21 | PJ314 | 20.02.2021 | 21 42 15.77 N | 73 13 48.27 E | Silty | Gray | 7.65 | 0.55 | 1 | 0 | 0 | 2 | 3 |
| 22 | PJ315 | 20.02.2021 | 21 42 13.44 N | 73 13 51.37 E | Sandy | Light brown | 7.56 | 0.11 | 0 | 0 | 0 | 0 | 0 |
| 23 | PJ316 | 20.02.2021 | 21 42 13.32 N | 73 13 52.80 E | Sandy | Light brown | 7.79 | 0.33 | 4 | 0 | 0 | 0 | 4 |
| 24 | PJ317 | 20.02.2021 | 21 42 12.63 N | 73 13 52.85 E | Sandy | Light brown | 7.55 | 0.09 | 1 | 0 | 0 | 0 | 1 |
| 25 | PJ318 | 20.02.2021 | 21 41 11.60 N | 73 13 52.10 E | Loamy | Gray | 7.57 | 0.51 | 2 | 0 | 0 | 0 | 2 |
| 26 | PJ319 | 20.02.2021 | 21 41 9.05 N | 73 13 51.09 E | Sandy | Orange | 7.56 | 0.19 | 3 | 0 | 0 | 1 | 4 |
| 27 | PJ320 | 20.02.2021 | 21 42 7.02 N | 73 13 50.64 E | Sandy | Brown | 7.20 | 0.11 | 1 | 0 | 0 | 0 | 1 |
| 28 | PJ321 | 20.02.2021 | 21 42 4.92 N | 73 13 47.60 E | Sandy | Light brown | 7.79 | 0.26 | 3 | 0 | 0 | 0 | 3 |
| 29 | PJ322 | 20.02.2021 | 21 42 2.63 N | 73 13 46.11 E | Sandy | Light brown | 7.73 | 0.24 | 2 | 0 | 0 | 0 | 2 |
| 30 | PJ323 | 20.02.2021 | 21 42 1.60 N | 73 13 45.22 E | Sandy | Dark brown | 7.62 | 0.83 | 2 | 1 | 0 | 1 | 4 |
| 31 | PJ324 | 20.02.2021 | 21 42 1.05 N | 73 13 43.16 E | Sandy | Light brown | 7.32 | 2.25 | 2 | 0 | 0 | 0 | 2 |
| 32 | PJ325 | 20.02.2021 | 21 42 1.80 N | 73 13 41.48 E | Sandy | Light brown | 7.50 | 0.15 | 3 | 0 | 0 | 1 | 4 |
| 33 | PJ326 | 20.02.2021 | 21 42 5.74 N | 73 13 37.77 E | Sandy | Light gray | 7.18 | 0.13 | 3 | 0 | 0 | 0 | 3 |
| 34 | PJ327 | 20.02.2021 | 21 42 3.95 N | 73 13 27.16 E | Clay | Black | 6.37 | 0.34 | 2 | 0 | 0 | 1 | 3 |
| 35 | PJ328 | 20.02.2021 | 21 42 5.92 N | 73 13 22.61 E | Sandy | Dull white | 3.10 | 0.77 | 1 | 0 | 2 | 0 | 3 |
| 36 | PJ329 | 20.02.2021 | 21 42 4.92 N | 73 13 25.61 E | Sandy | Brown | 2.96 | 1.02 | 1 | 0 | 0 | 0 | 1 |
| 37 | PJ330 | 20.02.2021 | 21 42 32.33 N | 73 13 55.77 E | Sandy | Gray | 6.90 | 0.23 | 2 | 0 | 0 | 0 | 2 |
| Total | 37 | 54 | 19 | 3 | 13 | 89 |
PSB: Phosphate-solubilizing bacteria, KSB: Potash-solubilizing bacteria, ZSB: Zinc-solubilizing bacteria.
Four representative samples were selected and analyzed for in-depth soil characterization based on the unique characteristics of soils. The presence of different heavy metals, such as nickel, zinc, copper, chromium, lead, barium, cobalt, manganese, and lead, was also detected in soil samples. The concentration of cobalt, nickel, copper, manganese, and chromium above 100, 100, 15, 55, and 10 ppm, respectively, may cause inhibitory or toxic effects on different plant growth [20,21]. The results of the present study depicted that coal mine spoils possessed such constraints that inhibited the plant growth. The organic matter was higher in PJ302 as compared to the other three samples. The cation exchange capacity was almost equal for PJ302 and PJ304. Barium content was highest in PJ302 and very least in PJ328. The cobalt content was highest in PJ301 and not detected or very least in PJ304 and PJ328, respectively. Manganese content was very less in PJ328 as compared to other three samples. The magnesium and potassium contents were very highest in PJ304 as compared to other samples. Nickel contents were not detected in PJ304 and PJ328. Zinc, copper, chromium, and lead contents were very least in sample PJ428. Apart from these findings, the physicochemical characteristics of PJ304 are distinct from the rest of the samples analyzed, confirming the heterogeneity of mined soil [Table 2]. These factors may have an even greater impact on plant growth.
Table 2: Physicochemical properties of representative soil samples collected from coal mine spoils.
| Parameters | PJ301 | PJ302 | PJ304 | PJ328 |
|---|---|---|---|---|
| Appearance | Black colored loamy soil | Brown colored soil | Red colored soil | Dull white colored soil |
| pH | 2.44 | 3.01 | 6.44 | 3.10 |
| EC (mS/cm) | 2.76 | 2.14 | 0.11 | 0.77 |
| Moisture content (% w/w) | 29.14 | 21.1 | 5.96 | 11.03 |
| Organic Matter (% w/w) | 1.23 | 3.10 | 0.86 | 0.92 |
| Organic Carbon (% w/w) | 0.71 | 1.80 | 0.50 | 0.54 |
| Total Nitrogen (% w/w) | 0.31 | 0.63 | 0.32 | 0.63 |
| Total Phosphorous as P2O5 (% w/w) | 0.83 | 0.58 | 0.74 | 0.84 |
| Total Soluble Sulfate (% w/w) | 0.60 | 0.52 | 1.52 | 1.114 |
| Cation Exchange Capacity (meq/100g) | 5.42 | 12.18 | 12.21 | 5.81 |
| Barium as Ba (mg/kg) | 40.21 | 208.28 | 88.02 | 0.06 |
| Cobalt as Co (mg/kg) | 117.30 | 3.16 | 0.05 | |
| Calcium as Ca (mg/kg) | 348.63 | 104.88 | 865.90 | 333.34 |
| Manganese as Mn (mg/kg) | 18.85 | 161.84 | 176.76 | 0.36 |
| Magnesium as Mg (mg/kg) | 204.38 | 666.5 | 4425.01 | 383.17 |
| Potassium as K (mg/kg) | 204.29 | 337.96 | 1006.63 | 166.53 |
| Sodium as Na (mg/kg) | 316.1 | 707.08 | 269.63 | 527.24 |
| Nickle as Ni (mg/kg) | 150.48 | 27.79 | ||
| Zinc as Zn (mg/kg) | 93.09 | 99.47 | 14.34 | 0.05 |
| Copper as Co (mg/kg) | 41.56 | 102.49 | 103.85 | 0.21 |
| Chromium as Cr (mg/kg) | 57.97 | 90.59 | 99.32 | 0.44 |
| Lead as Pb (mg/kg) | 7.81 | 52.39 | 11.74 | 0.07 |
3.2. Isolation and Screening of PGPR
The present research work was carried out with an emphasis on the isolation and characterization of PGPR populations from the rhizosphere of plants at coal mining sites. The isolates were first selected based on their morphological characteristics, then purified and further tested for the zone of solubilization [Figure 2]. A total of 89 isolates were selected for further detailed study. Among these, the putative nitrogen-fixing bacteria were most abundant (60.67%) followed by phosphate-solubilizing bacteria (21.35%), zinc-solubilizing bacteria (14.60%), and potash-solubilizing bacteria (3.37%).
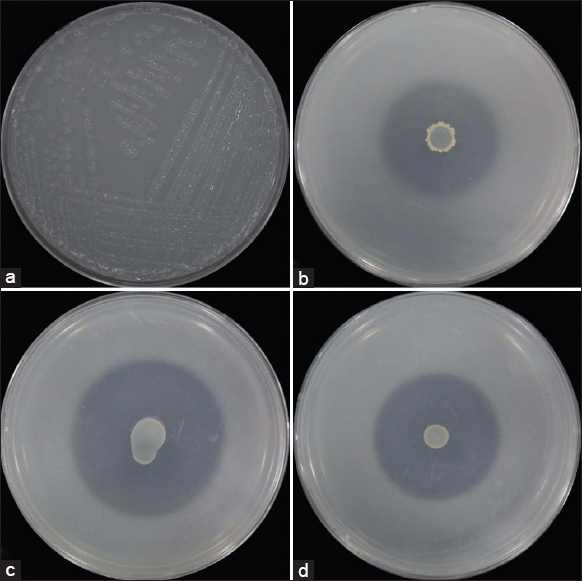 | Figure 2: Growth of (a) putative nitrogen-fixing bacteria on Jenesen agar, (b) phosphate-solubilizing bacteria on Pikovskaya’s agar, (c) potash-solubilizing bacteria on Aleksandrow agar, and (d) zinc-solubilizing bacteria on zinc solubilizing media. [Click here to view] |
Symbiotic or non-symbiotic nitrogen fixation can contribute to the crop nitrogen content in the form of ammonia [22]. Nitrogen free medium with mannitol as a carbon source supports the growth of only mannitol fermenting microbes which may or may not ferment other carbohydrates easily. Jensen’s medium contains sucrose, so all organisms which can fix nitrogen and utilize sucrose as a carbon source will grow on this medium [23]. The present study was focused on the isolation of such nitrogen-fixing bacteria, which can ferment sucrose and grow well using carbohydrates of sugar cane and other organic wastes. Therefore, screening of nitrogen-fixing bacteria was carried out using Jensen’s medium. A total of 54 isolates were able to grow in a nitrogen free medium.
Phosphate is a vital nutrient for plant growth; however, it is either lacking or poorly available to plants due to the highly complex and insoluble nature of organic and inorganic phosphates in soil. Phosphate solubilization by PGPR is mediated by the release of organic acids, anions, and protons, as well as the extracellular secretion of phosphate hydrolyzing enzymes [22]. Total 19 isolates formed a zone of solubilization on Pikovskaya’s agar. In addition to phosphorus, potassium, and zinc are also essential nutrients required for optimum growth of plants. In the present study, total 13 isolates formed zone of solubilization on medium containing ZnO and three isolates formed zone of solubilization on Aleksandrow media. The area of solubilization was measured according to equation (1). The solubilization index ranged between 2.14 to 4.67, 6.00 to 9.00, and 2.00 to 17.00 for phosphate, potash, and zinc-solubilizing bacteria, respectively [Table 3].
Table 3: Plant growth promoting traits of bacterial isolates.
| S. No. | Isolates | Solubilization efficiency (mm) | Solubilization index (SI) | Hemolytic type | |
|---|---|---|---|---|---|
| Zone diameter | Colony diameter | ||||
| Phosphate-solubilizing isolates | |||||
| 1 | PJS01 | 16.0 | 7.0 | 3.29 | γ |
| 2 | PJS02 | 11.0 | 5.0 | 3.20 | γ |
| 3 | PJS03 | 6.0 | 4.0 | 2.50 | γ |
| 4 | PJS04 | 11.0 | 6.0 | 2.83 | γ |
| 5 | PJS05 | 14.0 | 6.0 | 3.33 | γ |
| 6 | PJS06 | 14.0 | 7.0 | 3.00 | γ |
| 7 | PJS07 | 10.0 | 4.0 | 3.50 | γ |
| 8 | PJS08 | 10.0 | 5.0 | 3.00 | γ |
| 9 | PJS09 | 10.0 | 5.0 | 3.00 | γ |
| 10 | PJS10 | 7.0 | 3.0 | 3.33 | γ |
| 11 | PJS11 | 10.0 | 4.0 | 3.50 | γ |
| 12 | PJS12 | 22.0 | 12.0 | 2.83 | γ |
| 13 | PJS13 | 22.0 | 11.0 | 3.00 | γ |
| 14 | PJS14 | 7.0 | 2.0 | 4.50 | γ |
| 15 | PJS15 | 15.0 | 6.0 | 3.50 | γ |
| 16 | PJS16 | 11.0 | 6.0 | 2.83 | γ |
| 17 | PJS17 | 11.0 | 3.0 | 4.67 | γ |
| 18 | PJS18 | 8.0 | 7.0 | 2.14 | γ |
| 19 | PJS19 | 11.0 | 6.0 | 2.83 | γ |
| Potash-solubilizing isolates | |||||
| 20 | PJS20 | 20.0 | 3.0 | 7.67 | γ |
| 21 | PJS21 | 10.0 | 2.0 | 6.00 | γ |
| 22 | PJS22 | 16.0 | 2.0 | 9.00 | γ |
| Zinc-solubilizing isolates | |||||
| 23 | PJS23 | 5.0 | 3.0 | 2.67 | - |
| 24 | PJS24 | 11.0 | 2.0 | 6.50 | γ |
| 25 | PJS25 | 15.0 | 2.0 | 8.50 | γ |
| 26 | PJS26 | 21.0 | 12.0 | 2.75 | γ |
| 27 | PJS27 | 16.0 | 1.0 | 17.00 | γ |
| 28 | PJS28 | 16.0 | 10.0 | 2.60 | γ |
| 29 | PJS29 | 6.0 | 6.0 | 2.00 | - |
| 30 | PJS30 | 10.0 | 4.0 | 3.50 | γ |
| 31 | PJS31 | 11.0 | 5.0 | 3.20 | γ |
| 32 | PJS32 | 12.0 | 5.0 | 3.40 | γ |
| 33 | PJS33 | 8.0 | 4.0 | 3.00 | - |
| 34 | PJS34 | 11.0 | 6.0 | 2.83 | γ |
| 35 | PJS35 | 2.0 | 2.0 | 2.00 | - |
The efficiency of positive PGPR strains were screened based on their zone of solubilization according to the specification of biofertilizers mentioned in The Fertilizer (Control) Order 1985 [8]. The isolates showing minimum 5 mm, 10 mm, and 10 mm solubilization zones for phosphate, potash, and zinc-solubilizing bacteria in respective media having at least 3 mm thickness were further screened for pathogenicity by hemolysin test. In the present study, zinc-solubilizing isolates, namely, PJS23, PJS27, PJS33, and PJS35 showed zone of solubilization <10 mm and hence these isolates were not screened for hemolytic activity. In general, bacterial isolates having β-hemolytic activity are appraised as opportunistic human pathogens, α-hemolytic activity as partial pathogen, and γ-hemolytic activity as non-pathogens [24]. In the present study, the bacterial isolates showing γ-hemolytic activity were further identified by molecular methods. The isolates with γ-hemolytic activity lower the risk of this strain being harmful to humans.
Despite the benefits and potentials of PGPR, it is equally critical to understand the cost to benefit relationship associated with the development of any agricultural product based on the beneficial microbes. As a return on investment, farmers must have profit from innovative sustainable products to boost or sustain yields under increasing environmental stress. The zoning of industrialization around agriculture land has increased pollution levels in farmlands. Similarly, urbanization and infrastructure development, despite being important progress milestones, have their share of adverse effects. Both have led to the production of unmanageable quantities of domestic as well as industrial wastes. This has raised the pollution burden on agricultural land, thereby leading to increase stress conditions for crops. In such scenarios, the plant growth promoting traits of the rhizobacteria identified and isolated through similar studies can be beneficially employed to mitigate abiotic stress in agriculture.
3.3. Molecular Identification of PGPR
Molecular identification was carried out for morphologically different, nonpathogenic, and potent 31 isolates [Table 4]. The 16S rRNA gene sequencing approach was used to characterize potent PGPR strains at the molecular level. PGPR strains belonged to Cronobacter spp., Ralstonia spp., Serratia spp., Enterobacter spp., Staphylococcus spp., Pantoea spp., Cedecea spp., Curtobacterium spp., Burkholderia spp., Kosakonia spp., Acinetobacter spp., and Bacillus spp. The details of collections, identification, taxonomy, and the sequences of identified bacterial strains have been submitted to the public domain portal, that is, NCBI, and BOLD [Table 4]. Hence, any researcher can explore and use the information for future research. BOLD is a data retrieval tool that allows users to search information such as geography, taxonomy, molecular identification, and depository over public records.
Table 4: List of PGPR and their taxonomic affiliation based on 16S rRNA gene sequence.
| Isolates | Bold ID | Strain Code | GenBank Accession number | Molecular Identification | Class |
|---|---|---|---|---|---|
| PJS01 | ABRLM054-21 | ABRL054 | MZ496444 | Cronobacter sakazakii | γ- Proteobacteria |
| PJS02 | ABRLM055-21 | ABRL055 | OM406349 | Ralstonia pickettii | β-Proteobacteria |
| PJS04 | ABRLM056-21 | ABRL056 | OM407394 | Serratia marcescens | γ- Proteobacteria |
| PJS05 | ABRLM074-21 | ABRL074 | MZ960428 | Serratia marcescens | γ- Proteobacteria |
| PJS06 | ABRLM057-21 | ABRL057 | MZ562765 | Enterobacter cloacae | γ- Proteobacteria |
| PJS07 | ABRLM075-21 | ABRL075 | MZ960446 | Staphylococcus sciuri | Bacilli |
| PJS09 | ABRLM076-21 | ABRL076 | OM421805 | Serratia marcescens | γ- Proteobacteria |
| PJS10 | ABRLM058-21 | ABRL058 | MZ562766 | Ralstonia pickettii | β-Proteobacteria |
| PJS11 | ABRLM059-21 | ABRL059 | MZ562790 | Serratia marcescens | γ- Proteobacteria |
| PJS12 | ABRLM060-21 | ABRL060 | MZ562882 | Pantoea dispersa | γ- Proteobacteria |
| PJS13 | ABRLM061-21 | ABRL061 | MZ562892 | Cronobacter sakazakii | γ- Proteobacteria |
| PJS16 | ABRLM062-21 | ABRL062 | MZ597848 | Cedecea davisae | γ- Proteobacteria |
| PJS17 | ABRLM063-21 | ABRL063 | MZ960187 | Curtobacterium oceanosedimentum | Actinomycetia |
| PJS19 | ABRLM064-21 | ABRL064 | MZ597842 | Enterobacter cloacae | γ- Proteobacteria |
| PJS14 | ABRLM067-21 | ABRL067 | MZ597853 | Burkholderia cepacia | β-Proteobacteria |
| PJS15 | ABRLM068-21 | ABRL068 | MZ597850 | Serratia marcescens | γ- Proteobacteria |
| PJS21 | ABRLM070-21 | ABRL070 | MZ604314 | Burkholderia gladioli | β-Proteobacteria |
| PJS22 | ABRLM071-21 | ABRL071 | MZ604317 | Burkholderia tropica | β-Proteobacteria |
| PJS20 | ABRLM081-21 | ABRL081 | MZ664297 | Cronobacter malonaticus | γ- Proteobacteria |
| PJS25 | ABRLM069-21 | ABRL069 | MZ604310 | Serratia marcescens | γ- Proteobacteria |
| PJS28 | ABRLM077-21 | ABRL077 | MZ646786 | Kosakonia oryzae | γ- Proteobacteria |
| PJS27 | ABRLM078-21 | ABRL078 | MZ647475 | Acinetobacter baumannii | γ- Proteobacteria |
| PJS32 | ABRLM079-21 | ABRL079 | MZ664292 | Enterobacter asburiae | γ- Proteobacteria |
| PJS24 | ABRLM080-21 | ABRL080 | MZ664294 | Burkholderia contaminans | β-Proteobacteria |
| PJS26 | ABRLM082-21 | ABRL082 | OM421899 | Cronobacter sakazakii | γ- Proteobacteria |
| PJS36 | ABRLM083-21 | ABRL083 | MZ665050 | Enterobacter sichuanensis | γ- Proteobacteria |
| PJS37 | ABRLM084-21 | ABRL084 | MZ976768 | Pantoea dispersa | γ- Proteobacteria |
| PJS38 | ABRLM085-21 | ABRL085 | MZ976767 | Enterobacter cloacae | γ- Proteobacteria |
| PJS39 | ABRLM086-21 | ABRL086 | OM421806 | Kosakonia oryzae | γ- Proteobacteria |
| PJS40 | ABRLM088-21 | ABRL088 | OM407393 | Bacillus spp. | Bacilli |
| PJS41 | ABRLM089-21 | ABRL089 | MZ976819 | Bacillus subtilis subspp. subtilis | Bacilli |
The culturable PGPR diversity in coal mine spoils belonged to different genera representing three phyla; Proteobacteria, Firmicutes, and Actinobacteria [Figure 3a]. The taxonomic composition of the PGPR communities in coal mine spoils is dominated by members of Proteobacteria, accounting for 87.50% of the culturable PGPR diversity. At the class level, PGPR belonged to Gammaproteobacteria (68.75%) followed by Betaproteobacteria (18.75%) [Figure 3b]. Most of the PGPR were from Enterobacterales order, followed by Burkholderiales and Bacillales order [Figure 3c]. PGPR diversity was dominated by Enterobactericeae family, followed by Burkholderiaceae and Yersiniaceae family [Figure 3d].
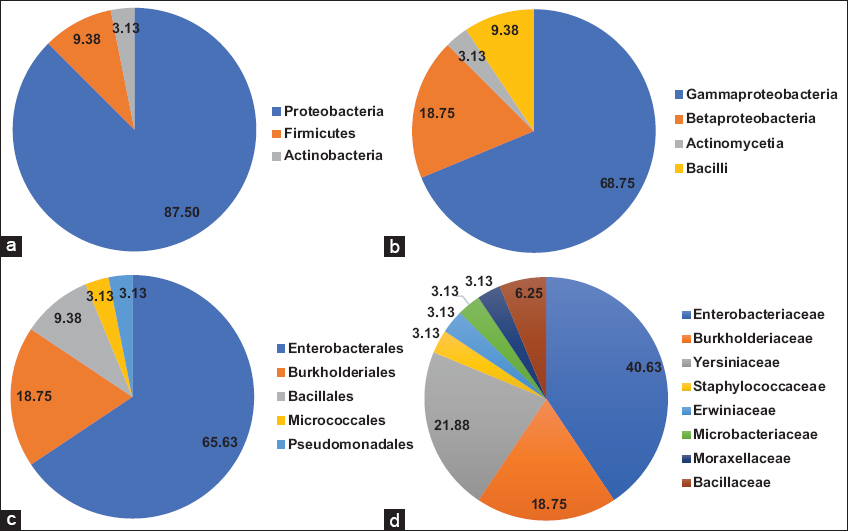 | Figure 3: PGPR diversity of coal mine. Pie charts representing the taxonomic assignment of identified PGPR: (a) phylum level; (b) class level; (c) order level; and (d) family level. [Click here to view] |
The dendrogram of phylogenetic tree depicts the evolutionary relationships between identified strains and their nearest strains [Figure 4]. According to the phylogenetic analysis, PGPR were clearly clustered into three phyla: Proteobacteria, Actinobacteria, and Firmicutes. Phylogenetic tree indicated that the genus Burkholderia and Ralstonia represented the class Betaproteobacteria. The genus Curtobacterium represented the class Actinomycetia, and the genus Bacillus and Staphylococcus represented the class Bacilli. The class Gammaproteobacteria is represented by the genus Enterobacter, Acinetobacter, Cedecea, Kosakonia, Serratia, Cronobacter, and Pantoea.
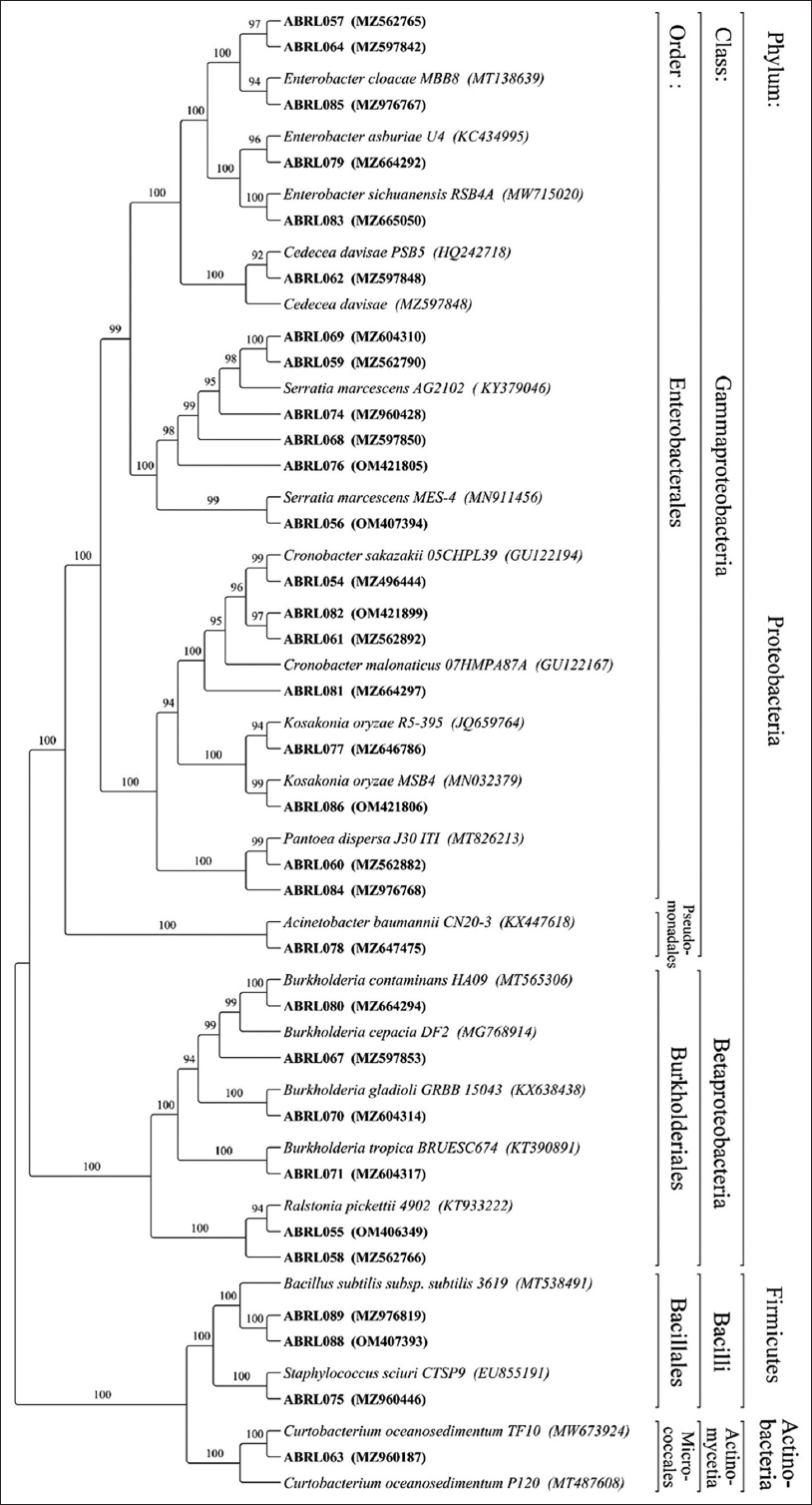 | Figure 4: Maximum likelihood phylogenetic tree based on partial 16S rRNA sequences, displaying the relationships between the identified isolates and their closest phylogenetic relatives. Bootstrap values are displayed on the tree branches. [Click here to view] |
Cronobacter sakazakii has been reported as drought stress-tolerant and salt stress-tolerant PGPR [25,26]. The zone of phosphate solubilization was reported to be 16–17 mm on Pikovskaya’s medium using Cronobacter sakazakii RF-4 by Afridi et al. [25]. In the present study, it shows zinc-solubilization in addition to phosphate-solubilization. In the presence of 2–8 percent NaCl, Cronobacter malonaticus has been found to solubilize phosphate [27]. In present study, it shows potash-solubilizing characteristics.
Ralstonia pickettii has been reported as an efficient phosphate-solubilizing rhizobacteria with a solubilization index of 3.2 [28]. The organism was reported to survive and thrive in oligotrophic (low nutrient) conditions [29]. Serratia marcescens was reported as multifarious PGPR with phosphate-solubilizing and nitrogen-fixing abilities. The bacterium has been reported to increase wheat plant growth under salt stress of up to 150–200 mM. The organism has been reported to induce systemic resistance against the Fusarium sp. by Singh and Jha [30].
Enterobacter cloacae have shown growth under osmotic stress and enabled the germination of the seed of Arachis hypogaea, Vigna mungo, and Brassica rapa [31]. The organism has been reported to be highly salt-tolerant with excellent nitrogen fixing abilities, phosphate, and potassium-solubilizing traits by Ji et al. [32]. Enterobacter asburiae has been found to be capable of phosphate-solubilization, nitrogen-fixation, and salinity stress tolerance [33,34]. The current study highlights its potential to solubilize zinc, making it a better choice for boosting plant growth. PGPR characteristic in Enterobacter sichuanensis has not been reported anywhere in the literature. Since it grew on nitrogen-free media in the present study, in-depth pot/field tests are needed to verify its efficacy in nitrogen-fixation.
Staphylococcus sciuri, a halotolerant microbe, is important for protecting tomato and rice plants from the harmful effects of salt stress [35]. It has excellent potential for phosphate-solubilization [36]. Pantoea dispersa is agriculturally important as it exhibited multiple PGPR traits under varied salt and heavy metal stress conditions [37-39]. In the present study, the solubilization index (SI) for P. dispersa is higher than reported by Singh et al. [37] (SI, 3.4). Cedecea davisae has been reported as potent phosphate and zinc-solubilizing bacteria by Mazumdar et al. [22]. Khan et al. [40] reported an increased growth of rice in high-salt environments by phytohormone producing bacterial endophyte Curtobacterium oceanosedimentum.
Burkholderia cepacia is a promising candidate to be used as a biofertilizer since it can withstand up to 5% salt stress, has antifungal activity against nine pathogenic fungi, and can solubilize phosphate. It has been demonstrated that the organism promotes growth of both healthy and infected maize plants [41]. Zhao et al. [42] reported Burkholderia gladioli with secretion of phytohromne and phoshoate-solubilizing trait. The authors also reported its antifungal potential against Sclerotium rolfsii, Alternaria solani, and Fusarium oxysporum. Moreover, it also improved the growth of jowar, maize, bajra, and wheat plants. In present study, B. gladioli showed potash-soulibilization which added one additional plant growth promoting trait to the species. Reis et al. [43] and Bolívar-Ring et al. [44] reported Burkholderia tropica as a novel nitrogen-fixing and phosphate-solubilizing bacteria. In present study this organism showed potash-solubilizing potential. A multi-trait Burkholderia contaminans was reported for enhancement of maize seedling growth and regulation of bands leaf and sheath blight. Furthermore, phosphate-solubilization, zinc-solubilization, nitrogen-fixation, and antifungal properties were discovered in B. contaminans [45].
Kosakonia oryzae was found in rice plant with nitrogen-fixing abilities [46]. In the present study, it has been found to exhibit zinc-solubilizing ability in addition to nitrogen-fixation, which further proves its efficacy to be used as PGPR. Acinetobacter baumannii has not been previously reported as PGPR. In present study, A. baumannii showed zinc-solubilizing trait. Bacillus subtilis was reported as potent nitrogen-fixing and salt tolerant bacteria by Satapute et al. [47].
When the data obtained in the present study and related research in literature are compared, it becomes evident that isolated PGPR are distinct and site-specific compared to those reported in literature. Therefore, the understanding of PGPR diversity may be specifically useful in preparing the bio-inoculants applicable for reclamation of coal mine. However, in-depth studies are required for the on-site reclamation process. The inoculum of such identified novel strains can also be suitable for plantations carried out in solid waste disposal sites because it is robust to various stress and extreme environments. The plant growth promoting traits of these rhizobacteria can be beneficially employed to mitigate abiotic stress in agriculture caused due to varied environments, such as in deserts area, salt affected land, wasteland, or even due to extreme seasonal variations (drought, floods, etc.). In addition to culturable PGPR diversity, it is important to study unculturable microbes as well as symbiotic microorganisms such as mycorrhiza as a part of biological rejuvenation activities. Therefore, it is key to explore the arbuscular mycorrhizal status of mine spoils in order to develop a better reclamation approach and apply the technology to be used at field level.
As per the biosafety data published by American Type Culture Collection ATCC, it was observed that all the isolated rhizobacteria (76.9%) except C. davisae, B. cepacian, and A. baumannii belonged to biosafety level 1 (BSL-1). Even though most of the identified bacterial strains belonged to BSL-1 category, opportunistic pathogenic reports are available in the literature for most of the identified PGPR [48-61]. According to the reports, the bacterial strains cause disease in patients who have a strong proclivity for illness, particularly those who are immunocompromised, severely debilitated, or have cystic fibrosis. These could most likely be due to coexistence or secondary infections arising from compromised health conditions of the host. Therefore, relying on such reports to assess PGPR as an opportunistic pathogen is not recommended for distinction between innocuous and hazardous bacteria.
Furthermore, the pathogenicity of organisms is also host-specific, and virulence depends on the serotype. Thus, it cannot be considered that bacterial strains isolated from the rhizosphere may be as pathogenic as the same strains isolated from clinical samples unless the serotype matches. Apart from frequently reported opportunistic pathogens, most PGPR cannot be classified as unsafe for agriculture application. To evaluate the agriculturally important microorganisms, reliable biosafety level data published by well-known government authorities namely, ATCC, German Collection of Microorganisms and Cell Cultures (DSMZ), and National Biological Resource Centre of Japan should be taken into consideration. However, it becomes necessary to carefully identify opportunistic pathogens through further research studies, if the objective is to further commercialize the approach of bioinoculants. Therefore, a risk assessment of PGPR must be affirmed by qualified and experienced institute before the commercial use of these strains as bioinoculants.
4. CONCLUSION
The present study has provided an insight into the PGPR community present at coal mine site. The isolated rhizobacterial strains were able to withstand high metal concentrations and salt stress with multiple plant growth promoting traits. The results of the present investigations and available biosafety, toxicity, and infectious reports of isolated rhizobacteria implies that they can be used as bioinoculants in microbial assisted reclamation projects in Amod coal mining area and for management of abiotic stress in agriculture and farmland as well. The PGPR diversity of Amod lignite mine was studied for the first time so that the site-specific inoculants can be utilized for effective land reclamation at the Amod lignite mine.
5. ACKNOWLEDGMENTS
The authors greatly acknowledge team of ABRL, Panoli, Ankleshwar, Gujarat for conducting research successfully. Authors also acknowledge Gujarat Mineral Development Corporation Limited, Lignite Project Rajpardi, Tal. Jagadia, Dist. Bharuch Gujarat for giving permission to collect soil samples for research.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
This work was funded by M/s. Pushpa J. Shah.
8. CONFLICTS OF INTEREST
This work was supported and funded by M/s. Pushpa J. Shah. All authors are employees of M/s. Pushpa J. Shah, with no stock options or incentives. Co-author Neil J. Shah is the Managing Partner of M/s. Pushpa J. Shah.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
The data generated and analyzed in this research article are included and the references are cited in the reference section.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Thavamani P, Samkumar RA, Satheesh V, Subashchandrabose SR, Ramadass K, Naidu R,
2. Feng Y, Wang J, Bai Z, Reading L. Effects of surface coal mining and land reclamation on soil properties:A review. Earth Sci Rev 2019;191:12-25. [CrossRef]
3. Dubey K, Singh VK, Mishra CM, Kumar A. Use of biofertilizer for reclamation of silica mining area. Makal. Disampaikan Pada Billings Reclam Symptom;2006. 4-8. [CrossRef]
4. Benidire L, Madline A, Pereira SI, Castro PM, Boularbah A. Synergistic effect of organo-mineral amendments and plant growth-promoting rhizobacteria (PGPR) on the establishment of vegetation cover and amelioration of mine tailings. Chemosphere 2021;262:127803. [CrossRef]
5. Indian Bureau of Mines. Reclamation of Land Damage by Mining. Karnataka:Indian Bureau of Mines. Available from:https://www.ibm.gov.in [Last accessed on 2021 Jun 09].
6. Chandini KR, Kumar R, Prakash O. The impact of chemical fertilizers on our environment and ecosystem. In:Research Trends in Environmental Sciences;2019. p. 69-86.
7. Shah NJ, Borada DN, Patel R, Patel A. Study on the functional diversity of plant growth-promoting rhizobacteria in Bharuch, Gujarat, India. Biosci Biotechnol Res Commun 2021;14:1338-44. [CrossRef]
8. FAI. The Fertiliser (Control) Order 1985. Shaheed Jit Singh Marg. New Delhi, India:The Fertiliser Association of India;2019.
9. Gujarat Mineral Development Corporation. Ahmedabad:Gujarat Mineral Development Corporation. Available from:https://www.gmdcltd.com [Last accessed on 2021 Jun 09].
10. Gujarat Mineral Development Corporation. Development of Agricultural Land on 10 Ha. Backfilled area for Agricultural use at GMDC Amod(G-19 Extension) Lignite mining project, Taluka:Jhagadia, District:Bharuch (Gujarat). Ahmedabad:Gujarat Mineral Development Corporation. Available from:https://www.gmdcltd.com/download/tenders [Last accessed on 2021 Jun 09].
11. Elias F, Woyessa D, Muleta D. Phosphate solubilization potential of rhizosphere fungi isolated from plants in Jimma Zone, Southwest Ethiopia. Int J Microbiol 2016;2016:5472601. [CrossRef]
12. Fan D, Smith DL. Characterization of selected plant growth-promoting rhizobacteria and their non-host growth promotion effects. Microbiol Spectr 2021;9:e0027921. [CrossRef]
13. Lane DJ. 16S/23S rRNA sequencing. Nucleic acid Techniques in Bacterial Systematics;1991. 115-75.
14. Leung MY, Blaisdell BE, Burge C, Karlin S. An efficient algorithm for identifying matches with errors in multiple long molecular sequences. J Mol Biol 1991;221:1367-78. [CrossRef]
15. Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics 2003;1:2-3. [CrossRef]
16. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9. [CrossRef]
17. Lanfear R, Calcott B, Ho SY, Guindon S. Partitionfinder:Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 2012;29:1695-701. [CrossRef]
18. Silvestro D, Michalak I. raxmlGUI:A graphical front-end for RAxML. Org Divers Evol 2012;12:335-7. [CrossRef]
19. Hammam AA, Mohamed ES. Mapping soil salinity in the East Nile Delta using several methodological approaches of salinity assessment. Egypt J Remote Sens Sp Sci 2020;23:125-31. [CrossRef]
20. Arif N, Yadav V, Singh S, Ahmad P, Mishra RK, Sharma S,
21. Wakeel A, Xu M. Chromium morpho-phytotoxicity. Plants (Basel) 2020;9:564. [CrossRef]
22. Mazumdar D, Saha SP, Ghosh S. Isolation, screening and application of a potent PGPR for enhancing growth of chickpea as affected by nitrogen level. Int J Veg Sci 2020;26:333-50. [CrossRef]
23. Jensen HL. Nitrogen fixation in leguminous plants. II. Is symbiotic nitrogen fixation influenced by Azotobacter. In Proc Linn Soc NSW 1942;57:205-12.
24. Al-Garni SM, Khan MM, Bahieldin A. Plant growth-promoting bacteria and silicon fertilizer enhance plant growth and salinity tolerance in
25. Afridi MS, Mahmood T, Salam A, Mukhtar T, Mehmood S, Ali J,
26. Zia R, Nawaz MS, Yousaf S, Amin I, Hakim S, Mirza MS,
27. Bhatt PV, Vyas BR. Production and characterization of biopolymer by plant growth promoting bacterial strain
28. Kailasan NS, Vamanrao VB. Isolation and characterization of
29. McAlister MB, Kulakov LA, O'Hanlon J, Larkin MJ, Ogden KL. Survival and nutritional requirements of three bacteria isolated from ultrapure water. J Ind Microbiol Biotechnol 2002;29:75-82. [CrossRef]
30. Singh RP, Jha PN. The multifarious PGPR
31. Panigrahi S, Mohanty S, Rath CC. Characterization of endophytic bacteria
32. Ji C, Liu Z, Hao L, Song X, Wang C, Liu Y,
33. Jetiyanon K. Multiple mechanisms of
34. Mahdi I, Fahsi N, Hafidi M, Allaoui A, Biskri L. Plant growth enhancement using rhizospheric halotolerant phosphate solubilizing bacterium
35. Taj Z, Challabathula D. Protection of photosynthesis by halotolerant
36. Akram MS, Shahid M, Tariq M, Azeem M, Javed MT, Saleem S,
37. Singh TB, Sahai V, Ali A, Prasad M, Yadav A, Goyal T,
38. Panwar M, Tewari R, Gulati A, Nayyar H. Indigenous salt-tolerant rhizobacterium
39. Ghosh A, Pramanik K, Bhattacharya S, Mondal S, Ghosh SK, Ghosh PK,
40. Khan MA, Asaf S, Khan AL, Adhikari A, Jan R, Ali S,
41. Zhao K, Penttinen P, Zhang X, Ao X, Liu M, Yu X,
42. Gunjal AB, Kapadnis BP.
43. Reis VM, Los Santos PE, Tenorio-Salgado S,Vogel J, Stoffels M, Guyon S,
44. Bolívar-Ring HJ, Contreras-Zentella ML, Teherán-Sierra LG.
45. Tagele SB, Kim SW, Lee HG, Kim HS, Lee YS. Effectiveness of multi-trait
46. Meng X, Bertani I, Abbruscato P, Piffanelli P, Licastro D, Wang C,
47. Satapute PP, Olekar HS, Shetti AA, Kulkarni AG, Hiremath GB, Patagundi BL,
48. Elkhawaga AA, Hetta HF, Osman NS, Hosni A, El-Mokhtar MA. Emergence of
49. Ryan MP, Pembroke JT, Adley CC.
50. Khanna A, Khanna M, Aggarwal A.
51. Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. Multidrug-resistant
52. Chen S, Wang Y, Chen F, Yang H, Gan M, Zheng S. A highly pathogenic strain of
53. Mehar V, Yadav D, Sanghvi J, Gupta N, Singh K.
54. Ismaael TG, Zamora EM, Khasawneh FA.
55. Ragupathi NK, Veeraraghavan B. Accurate identification and epidemiological characterization of
56. Imataki O, Kita N, Nakayama-Imaohji H, Kida JI, Kuwahara T, Uemura M. Bronchiolitis and bacteraemia caused by
57. Deris ZZ, Van Rostenberghe H, Habsah H, Noraida R, Tan GC, Chan YY,
58. Alsonosi AM, Holy O, Forsythe SJ. Characterization of the pathogenicity of clinical
59. Lee CR, Lee JH, Park M, Park KS, Kim YB, Cha CJ,
60. Zhu B, Wang S, Li O, Hussain A, Hussain A, Shen J,
61. Power RF, Linnane B, Martin R, Power N, Harnett P, Casserly B,