1. INTRODUCTION
Proteases, particularly those derived from microbial sources, have gained significant popularity and dominance in the enzyme market, accounting for 45–60% of total sales worldwide [1]. Microbial proteases are preferred over animal and plant proteases due to their versatility and wide range of applications. These enzymes can be produced through various methods, with solid-state fermentation (SSF) being one of the most effective approaches [2]. SSF is advantageous as microbial proteases are typically extracellular, meaning they are secreted outside the microbial cells. In addition, this fermentation method is simple and utilizes readily available agricultural and industrial residues [3].
Proteases function by hydrolyzing the peptide bonds in proteins, breaking them down into peptides and amino acids. They are categorized as endopeptidases, such as aspartic, serine, cysteine, and threonine proteases, based on the amino acids present in their active sites or their mineral requirements [4,5]. Proteases can also be classified as endoproteases or exoproteases based on their specific actions. In general, proteases can be acidic, basic, or neutral, with acid proteases being particularly important in food processing. Fungi, including Aspergillus, Penicillium, and Rhizopus, are known for producing acid proteases, with Aspergillus producing aspergillopepsin (EC 3.4.23.18) and Rhizopus producing rhizopuspepsin (EC 3.4.23.21) [6-8].
Another type of acid protease is a renin-like acid protease, produced by molds such as Mucor, Rhizomucor, and Rhizopus. These molds produce mucorpepsin (EC 3.4.23.23) and endothiapepsin (EC 3.4.23.22) [9-11]. Acid proteases are widely used in the food industry, including applications such as cheese making, meat tenderization, protein analysis, and juice clarification. They exhibit maximum effectiveness and stability at pH 2-5 but can also function at pH 6 [12-15].
Microbial protease enzymes are favored as alternatives to animal and plant enzymes due to their ease of cultivation, ability to control production conditions, short production time, and the utilization of local resources at lower costs, such as agricultural and industrial residues like bran, husks, and waste products. The broad applications and significance of these enzymes in the food industry have led to an increased focus on their production. Therefore, this study aimed to optimize the conditions for producing aspartic proteases from locally isolated Aspergillus terreus molds, with the goal of utilizing these enzymes in various applications, including cheese manufacturing, protein hydrolyzates, and meat tenderization.
2. MATERIALS AND METHODS
2.1. Isolation, Screening, and Diagnosis of A. terreus
In a previous investigation, a collection of molds was isolated from various soil samples in the Basrah city region of Iraq using the potato dextrose agar (PDA) medium (HiMedia, India). Following this, a primary screening was conducted on the local isolates using the Skim Milk Agar medium, and the plates were inoculated with 5 mm of mycelia from the isolates. After incubation at 30°C for 2–5 days, the plates were flooded with a solution of 10% trichloroacetic acid (TCA). The clear zone around the fungal colony indicated the production of protease [2]. The local isolate exhibited the highest enzyme productivity during the secondary screening. The secondary screening followed the method described by Mamo et al. [3]. 10 g of wheat bran and 2 g of casein were moistened with 10 mL of a saline solution (at pH 5) in a conical flask. The saline solution was prepared by dissolving 2 g of potassium nitrate (KNO3), 0.5 g of magnesium sulfate heptahydrate (MgSO4.7H2O), 1 g of dipotassium hydrogen phosphate (K2HPO4), 0.439 g of zinc sulfate heptahydrate (ZnSO4.7H2O), 1.116 g of iron sulfate heptahydrate (FeSO4.7H2O), and 0.203 g of manganese sulfate heptahydrate (MnSO4.7H2O) in one litter of distilled water. The flasks were then autoclaved for 15 min at 121°C. After sterilization, the flasks were cooled and inoculated with 1 mL of suspensions containing 1 × 106 spores/mL. It was incubated at 30°C for 7 days. The most productive isolate was identified using genetic diagnosis, which includes a brief, generally universal primer (standard) used for internal transcribed spacer (ITS1 5.8S ITS2) sequence; genomic DNA was extracted from conidia, and was detected by electrophoresis using a 1.5% agarose gel. A polymerase chain reaction (PCR) technique was used to amplify the ITS gene using the primers shown in Table 1 and supplied by the integrated DNA Technologies Company in Canada. The PCR products were subjected to electrophoresis in 1% agarose gel and purified using the solution for the gel extraction kit, then sequenced by the Genetic Analyzer System (ABI-310, Applied Biosystems). The National Centre for Biotechnology Information (NCBI) database was used to obtain the closest culture sequence based on the similarity between sequences.
Table 1: The primers used in the interaction of gene ITS
| Primer | Sequence | Tm (°C) | GC (%) | Product size |
|---|---|---|---|---|
| Forward | 5′-TCCGTAGGTGAACCTGCGG-3′ | 60.3 | 50 | 500–650 base pair |
| Reverse | 5′-TCCTCCGCTTATTGATATGC-3′ | 57.8 | 41 |
ITS: Internal transcribed spacer
2.2. Preparation of Inoculum Size
The inoculum size was prepared by flooding the activated fungal isolates present on the PDA slant medium with 10 mL of sterile distilled water. The mixture was then stirred using a sterile loop to obtain a spore suspension, which was subsequently transferred to a sterile tube. The spore concentration was adjusted to 1 × 106 spores/mL using the hemocytometer [3,4].
2.3. Enzyme Extraction
After 7 days of fermentation, 100 mL of distilled water and 1 mL of Tween 80 were added to each flask of the fermented product and stirred at 30°C for 1 h at 150 rpm. The extracellular aspartic solutions were obtained by filtration using a cotton cloth, then centrifuged at 5000 rpm at 4°C for 10 min. After that, the supernatant was collected and used as a crud enzyme [16].
2.4. Enzyme Activity Assay
The acid protease was assessed following the procedure described by Hsiao et al. [17]. First, 50 μL of casein solution was mixed with 190 μL of glycine HCl buffer solution (0.05 M) and pH 3.4. Subsequently, 10 μL of the enzyme was added to the mixture, which was then incubated in a water bath at 35°C for 30 min. The reaction was stopped by adding 250 μL of TCA 5% (w/v). For the control sample, all the aforementioned components were mixed except for the addition of TCA at the beginning. The enzyme was then added, followed by incubation at 35°C for 30 min. The resulting mixture underwent centrifugation at 12000 rpm for 10 min using a Damon/IEC division centrifuge (USA). To estimate the absorption of the filtrate, a UV-UV-2900 spectrophotometer (Biotech Engineering, UK) was used at a wavelength of 280 nm. The hydrolytic activity of the enzyme was determined by measuring the increase in absorbance of 0.001 at a wavelength of 280 nm under the given reaction conditions.
2.5. Determination of Protein Concentration
The protein concentration in all experiments was determined using the method developed by Lowry et al. [18]. A standard curve was created using bovine serum albumin solution to accurately measure the protein concentration in the enzymatic solution conditions for the production of aspartic protease.
The optimal conditions for enzyme production were investigated based on the study by Othman and Sebo, [19] with some modifications.
Firstly, the best medium for enzyme production was determined by examining the effects of different media types, including wheat bran, barley bran, rice hulls, corn groats, and soybean meal. Each medium (10 g) was placed in a 250-mL conical flask, supplemented with casein, moistened with a salt solution, and sterilized at 121°C for 15 min. After cooling, inoculum was added, and the mixture was incubated at 30°C for 7 days. The crud enzyme was extracted as previously described, and the activity of protease was measured. The highest enzyme-produced substrate was chosen and tested for further optimization.
Second, the optimal percentage of the moistening solution used for enzyme production was determined. Different ratios of the moistening solution to the production medium (1:1, 2:1, 3:1, 4:1, and 5:1) were examined.
Thirdly, the best pH for enzyme production was investigated by preparing the moistening solution at different pH levels (3, 3.5, 4, 4.5, 5, 5.5, 6, 6.5, and 7). The pH of the solution was adjusted with 0.1 M HCl or 0.1 M NaOH before incorporating it into the production medium. Furthermore, different solutions were utilized for enzyme extraction, including citrate buffer (0.2 M, pH 3 and pH 4), acetate buffer (0.2 M, pH 5), sodium phosphate buffer (0.2 M, pH 6 and pH 7), and distilled water. The aim was to determine the most effective solution for enzyme extraction.
In addition, the optimal inoculum size for enzyme production was determined using various sizes (1 mL, 2 mL, 3 mL, 4 mL, 5 mL, and 6 mL) to inoculate the fermentation medium.
The study also focused on identifying the best nitrogen source for enzyme production. Different nitrogen sources, such as casein, peptone, ammonium nitrate, ammonium sulfate, and yeast extract, were added to the fermentation medium. Furthermore, the best incubation period for enzyme production was determined by incubating the inoculated medium for varying periods ranging from 2 to 7 days. Finally, the optimal temperature for enzyme production was investigated by incubating the inoculated medium at different temperatures, with a 5°C difference between each temperature, ranging from 20 to 45°C.
2.6. Statistical Analysis
Data analysis was performed using Genstat software, specifically the 2012 Edition. In the analysis, the minimum significant difference (L.S.D.) method was utilized at a probability level of 0.05.
3. RESULTS AND DISCUSSION
3.1. Isolation, Screening, and Diagnosis
The selection and isolation of microorganisms play a crucial role in studies of this nature, as they determine various aspects of the research [4]. In this study, 40 pure isolates were obtained and purified from mold using PDA medium. Various sources were utilized, including different types of rice available in local markets and soil samples collected from different areas of Basrah Governorate. The isolates were initially screened based on the formation of halos around colonies on Skim Milk Agar medium and incubated at 30°C for 48 h [2].
Figure 1 shows the primary screening process, which showed a clear halo, which indicates the ability of the isolate to produce the protease enzyme. After that, the assay of the enzyme produced from the SSF process was measured, which showed a high ability of the local isolate to produce the enzyme (258.6 units/mL and 272.2 units/mg). The isolate was further characterized visually, microscopically, and genetically. DNA was extracted from the fungal isolate, and the purity of the extract was confirmed by electrophoresis results using agarose gel at a concentration of 1.5% [Figure 2] and after amplifying the ITS gene in a PCR device, the products were separated by electrophoresis using agarose at a concentration of 2%. The results showed the appearance of clear bands with a size of 600 base pairs [Figure 3]. The results of the PCR technology were sent to the Korean company (Macrogen) for the purpose of determining the genetic sequence (the sequence of nitrogenous bases), and then these sequences were adopted in the homology search using the basic local alignment search tool program to identify the type of isolate and match it with the strains available in NCBI. Genetic analysis confirmed a 100% match with A. terreus, which has been deposited in the gene bank with the name A. terreus SFH5 (available at https://blast.ncbi.nlm.nih.gov/Blast.cgi).
 | Figure 1: Primary screening based on halo formation around isolated colonies. [Click here to view] |
 | Figure 2: Genomic DNA bands extraction from local isolate on 1% agarose gel. [Click here to view] |
 | Figure 3: Agarose gel (1.5%) electrophoresis of the products amplified with polymerase chain reaction (PCR) at 5 volt/cm2. 1× TBE buffer for 1:30 h. N: left side is DNA ladder (1500 bp), PCR product the band size 600 bp. [Click here to view] |
This study aligns with previous research that emphasizes the use of soil as a primary source for isolating different species of Aspergillus. For instance, Chimbekujwo et al. [16] isolated Aspergillus brasiliensis BCW2 from soil, and Maitig et al. [20] also employed soil as a source for isolating Aspergillus spp.
3.2. Determining the Optimal Parameters for Aspartic Protease Production
3.2.1. Optimal medium for enzyme production
To assess the efficiency of A. terreus SFH5 isolate in producing aspartic protease using SSF, various culture media, including wheat bran, barley bran, rice hulls, corn groats, and soybean meal, were employed to determine the most suitable medium for enzyme production. Statistical analysis revealed significant differences between the different media at P ≤ 0.05. Among the tested media, wheat bran exhibited the highest enzymatic activity, measuring 258.6 units/mL with a specific activity of 272.2 units/mg. On the other hand, corn groats displayed the lowest enzyme activity among the agricultural media, with an enzymatic activity of 100 units/mL and a specific activity of 104.1 units/mg, as illustrated in [Figure 4].
 | Figure 4: Effect of different agro-industrial for medium on protease enzyme production from Aspergillus terreus SFH5. [Click here to view] |
The disparity in enzyme production across different culture media can be attributed to the varying organic and mineral composition of the nutrient medium, which can impact enzyme production, activity, and stability. Corn groats’ reduced ability to absorb water and separate the moisturizing solution from the particles may hinder mold growth and enzyme production. Conversely, wheat bran’s water retention capacity and suitable composition allow for sufficient air entry, making it a commonly used medium for enzyme production.
Several studies align with the findings of this research, highlighting the use of wheat bran as an optimal medium for protease enzyme production by various Aspergillus [16] employed wheat bran for protease enzyme production from A. brasiliensis BCW2, while [21] identified wheat bran as the superior solid fermentation medium for protease enzyme production from Aspergillus oryzae MTCC 5341. Similarly, Novelli et al. [1] and da Silva et al. [22] reported wheat bran as the best culture medium for protease enzyme production from Aspergillus. Overall, these findings underscore the significance of selecting the appropriate culture medium, with wheat bran emerging as the optimal choice for protease enzyme production in SSFs.
3.2.2. Determining the optimal moistening solution percentage for enzyme production
Significant variations were observed among different ratios of the moisturizing solution at P ≤ 0.05. The optimal ratio was determined to be (3:1), which corresponds to the usage of 30 g of saline solution with 10 g of wheat bran as the production medium. This ratio resulted in an enzymatic activity of 273.3 units/mL and a specific activity of 303.6 units/mg. Conversely, the ratio of the moisturizing solution (5:1) exhibited lower activity, with an enzymatic activity of 40 units/mL and a specific activity of 42.1 units/mg. [Figure 5] illustrates the gradual increase in enzymatic activity until reaching its peak at the (3:1) ratio of the moisturizing solution, followed by a gradual decline. This effect in SSFs can be attributed to the growth and metabolism of microorganisms. An increase in the moisture level to a certain level may inhibit enzyme production. This decrease in enzyme production at higher humidity levels may be due to poor oxygen supply with increasing humidity levels [23].
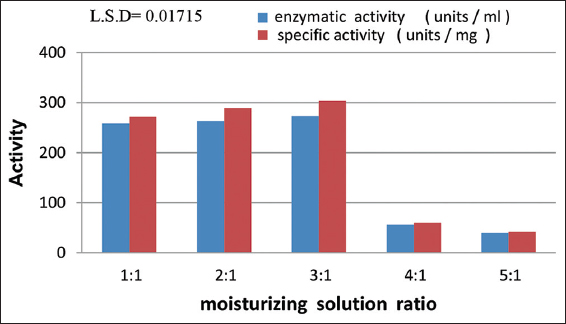 | Figure 5: Impact of moisturizing solution ratio on protease enzyme production from Aspergillus terreus SFH5. [Click here to view] |
The selection of mineral salts can significantly impact the microorganism’s performance [9]. Differences in protease activity are influenced by variations in the chemical composition and particle size, which affect porosity [2]. When the ratio of moisturizing decreases, the surface tension increases, resulting in reduced nutrient dissolution and an unfavorable environment for mold growth. Conversely, a high moisturizing ratio leads to anaerobic conditions, decreased oxygen diffusion, increased particle adhesion, and the agglomeration of wheat bran, ultimately leading to reduced enzyme productivity [23,24].
3.2.3. Optimization of pH for maximizing enzyme production
[Figure 6] illustrates the impact of pH on the production of the aspartic protease enzyme from the isolated A. terreus SFH5. Statistical analysis revealed significant differences between various pH values at P ≤ 0.05. The optimal pH for enzyme production was pH 5, resulting in an enzymatic activity of 273.3 units/mL and a specific activity of 303.6 units/mg. It is notable that enzyme productivity gradually increased within the acidic pH range of 2 to 5, but declined thereafter. The lowest enzyme activity and specific activity were observed at pH 7, measuring 78.6 units/mL and 82.7 units/mg, respectively. The influence of acidic pH on enzyme production may be attributed to its effects on the medium’s properties, such as component solubility, transferability, and ionization of vital compounds [4]. Changes in the ionic state of the substrate can lead to the formation of charged particles that may not align with the enzyme’s active ionic sites, hindering the mold’s access to the substrate [19]. These findings align with the work of Mamo et al. [3], who produced acid protease enzyme from A. oryzae DRDFS13 isolate at pH 5, as well as the findings of Radha et al. [25], who produced protease enzyme from Aspergillus ssp. isolate.
 | Figure 6: Effect of different initial pH on protease enzyme production from local isolate Aspergillus terreus SFH5. [Click here to view] |
3.2.4. Optimization of enzyme extraction methods for maximum yield
Investigating the optimal extraction solution for protease enzyme from A. terreus SFH5 Isolate. Through rigorous statistical analysis, notable differences were observed among various enzyme extraction solutions at a significance level of P ≤ 0.05. It was determined that distilled water emerged as the most effective solution for enzyme extraction, exhibiting an enzymatic activity of 280.6 units/mL and a specific activity of 311.7 units/mg. Conversely, the citrate buffer at pH 3 displayed the lowest activity, with an enzymatic activity of 126.6 units/mL and a specific activity of 137.6 units/mg, as depicted in [Figure 7]. The selection of an appropriate extraction solution plays a crucial role in maintaining enzyme activity and preventing interference in its estimation by ensuring optimal pH values. The superiority of distilled water as an extraction solution stems from its ability to dissolve substances without hindering enzyme function or binding to minerals essential for enzyme effectiveness. Conversely, the varying efficacy of buffer solutions in enzyme extraction can be attributed to differences in ionic strength and enzyme properties such as electrical neutralization point and pH stability [23]. These findings align with the results of previous studies, including Chimbekujwo et al. [16] and Mamo et al., [3] who utilized a distilled water solution in a ratio of 10:1 for protease enzyme extraction from A. oryzae DRDFS13 and A. brasiliensis BCW2 isolates, respectively.
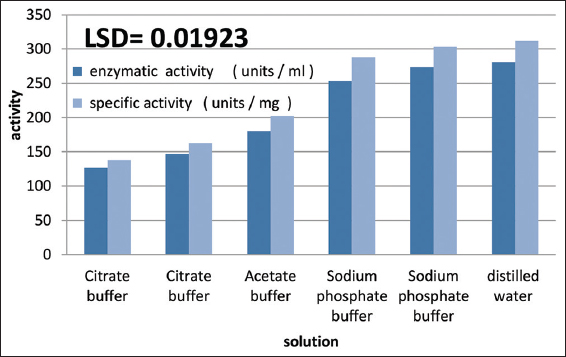 | Figure 7: Effect of buffer solutions on protease enzyme production from local isolate Aspergillus terreus SFH5. [Click here to view] |
3.2.5. Optimizing inoculum size for enhanced enzyme production
Significant variations were observed among different inoculum sizes during the statistical analysis (P ≤ 0.05 [Figure 8]. illustrates the results obtained from wheat bran medium inoculated with varying levels of inoculum sizes. The maximum production of protease enzyme from A. terreus SFH5 was achieved when using an inoculum volume of 2 mL (1 × 106 spores/10 g solid). This resulted in an enzymatic activity of 300 units/mL and a specific activity of 333.3 units/mg. Conversely, enzyme productivity decreased to its lowest levels with an inoculum size of 7 mL, where enzymatic activity dropped to 80 units/mL and specific activity reached 86.9 units/mg.
 | Figure 8: Optimizing inoculum sizes for protease enzyme production from local isolate Aspergillus terreus SFH5. [Click here to view] |
The decrease in enzyme productivity with a lower inoculum volume can be attributed to suboptimal utilization of medium components, adversely affecting the activity of metabolic materials, including enzymes. Conversely, increasing the inoculum size beyond the optimal level leads to reduced dissolved oxygen, increased competition for nutrients, cell agglomeration, and subsequently, a decline in enzyme production [26]. These findings align with previous studies conducted by [26], who recommended an inoculum volume of 1 mL for protease production from microbial isolates. Similar results were also reported by [16], where an inoculum volume of 1 mL was utilized for protease production from A. brasiliensis.
3.2.6. Optimization of nitrogen sources for enzyme production
Investigation of the impact of organic and inorganic nitrogen sources on aspartic protease enzyme production from locally isolated A. terreus SFH5. In this study, the researchers examined the influence of different organic and inorganic nitrogenous sources on the production of the aspartic protease enzyme from the isolated A. terreus SFH5. The findings presented in [Figure 9] and supported by statistical analysis, revealed significant variations among the nitrogen sources at a significance level of P ≤ 0.05. Notably, the organic nitrogen source, casein, exhibited the highest enzymatic activity, reaching 300 units/mL, with a specific activity of 333.3 units/mg. Conversely, the utilization of other nitrogen sources such as peptone, ammonium nitrate, ammonium sulfate, and yeast extract led to a gradual decline in enzyme production, with enzymatic activities recorded as (253.3, 173.3, 56.6, 42) units/mL and specific activities measured as (287.8, 192.5, 69.8, 46.6) units/mg, respectively. Nitrogen plays a fundamental role in all fermentation processes, serving as a crucial component for the genetic structure of microorganisms. Its availability is, therefore, a vital factor influencing the growth of living organisms and the synthesis of enzymes [19]. In addition, the superior production of the protease enzyme observed when employing the organic nitrogenous source (casein) can be attributed to the nature of amino acids and peptides derived from the process of protein degradation [27]. These results align with the findings of Asha and Palaniswamy [28], who identified casein as the optimal nitrogen source for protease enzyme production from the bacterial strain Bacillus cereus FT, surpassing both organic and inorganic alternatives with an enzymatic activity of up to 151 units/mL. Similarly, Othman and Sebo [19] concluded that casein serves as the most effective nitrogenous source for protease enzyme production. Overall, this study highlights the significance of selecting the appropriate nitrogen source for enhancing aspartic protease enzyme production, with casein demonstrating exceptional performance in this context.
 | Figure 9: Variation in protease production from local isolation of Aspergillus terreus SFH5 using different nitrogen sources. [Click here to view] |
3.2.7. Enhancing enzyme production by identifying the optimal incubation period
[Figure 10] demonstrates that the optimal incubation period for producing aspartic protease enzyme from the local isolation of A. terreus SFH5 is 6 days. During this period, the enzymatic activity reached 306.6 units/mL, with a specific activity of 348.4 units/mg. The enzyme production showed an increasing trend starting from the 2nd day, gradually reaching enzymatic activities of 51.3, 40, 95.3, and 130.6 units/mL, and specific activities of 16.6, 45.4, 116.26, and 167.4 units/mg, respectively. However, on the 7th day of incubation, a decrease in enzyme production was observed. This decline may be attributed to increased metabolites and nutrient depletion in the medium, resulting in reduced fungal growth and subsequent enzyme secretion outside the cell [29]. Statistical analysis confirmed significant differences among the various incubation periods at a significance level of P ≤ 0.05. These findings align with the research of Usman et al. [2], who also determined that the optimal incubation period for acid protease production from filamentous fungi was 120 h. Similarly, [30] reported that the best incubation duration for enzyme activity was 120 h, with an enzymatic activity of 181 units/mL.
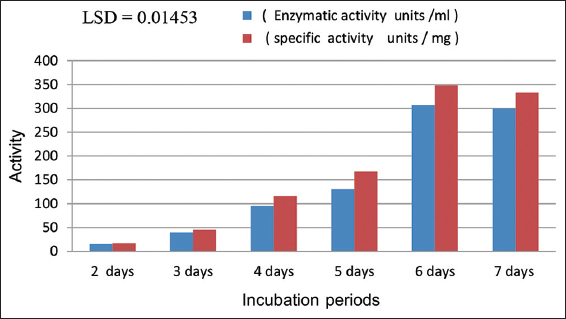 | Figure 10: Effect of the different incubation periods on protease production from local isolate of Aspergillus terreus SFH5. [Click here to view] |
3.2.8. Optimization of temperature
[Figure 11] illustrates the impact of different temperatures on aspartic protease production from the A. terreus SFH5 isolate. Statistical analysis revealed significant temperature-dependent differences at a significance level of P ≤ 0.05. The optimal temperature for enzyme production was determined to be 30°C, with enzymatic activity recorded at 306.6 units/mL and specific activity at 348.4 units/mg. At other temperatures (20°C, 25°C, 35°C, 40°C, and 45°C), the enzymatic activity and specific activity were observed to be 213.3, 253.3, 240, 174.6, 146.6 units/mL, and 250.9, 284.6, 266.6, 223.8, and 183.25 units/mg, respectively. However, exceeding the optimal temperature resulted in decreased enzyme productivity due to the elevated temperature and dryness of the fermentation medium, leading to reduced water activity (aw), limited ventilation, nutrient depletion, and impaired microbial growth. In addition, high temperatures could cause protein denaturation and enzyme inhibition. These findings are consistent with the observations of Usman et al. [2] on the optimum temperature for acid protease enzyme production and are also in line with the optimum temperature reported by Aljammas et al. [9] for protease enzyme production from Aspergillus niger FFB1 isolate. Muazu [29] similarly identified 30°C as the optimum temperature for acid protease enzyme production from Aspergillus spp. Mold.
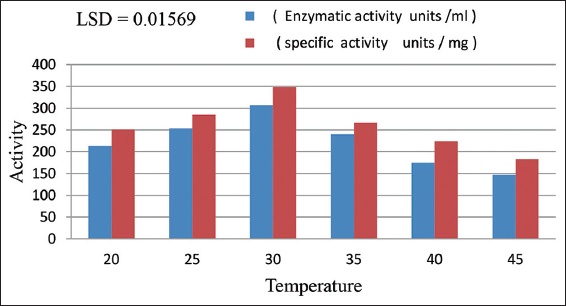 | Figure 11: Producing protease enzyme from locally isolated Aspergillus terreus SFH5 at varied temperatures. [Click here to view] |
4. CONCLUSION
The A. terreus SFH5 mold, isolated from various soil sample locations within Basrah Governorate in southern Iraq, demonstrated the capacity to produce the aspartic protease enzyme through SSF. By optimizing the isolation and growth processes, the study successfully increased the production of the enzyme. Notably, local agricultural residues were utilized as a growth medium, which significantly contributed to the enhancement of enzyme production. The results obtained from this research indicate the potential utility of local fungal isolations for the production of the aspartic protease enzyme. This enzyme can be beneficially employed in diverse food applications, such as meat tenderization, protein analysis, and natural juice clarification, among others, in the future.
5. ACKNOWLEDGMENTS
I would like to thank the staff of the Biotechnology Lab, Department of Food Science, College of Agriculture, University of Basrah, for their financial support of this study.
6. AUTHOR CONTRIBUTIONS
S.F.H., A.J.A.A., and Z.K.A.: Conceptualization; S.F.H. and A.J.A.A.: Methodology; S.F.H, and Z.K.A.: Data curation; S.F.H., A.J.A.A., and Z.K.A.: Writing—original draft preparation; A.J.A.A.: Writing—review and editing; S.F.H., and A.J.A.A.: Supervision. All authors have read and agreed to the published version of the manuscript.
7. FUNDING
The author(s) received no financial support for the research, authorship, and/or publication of this article.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All datasets were generated and analyzed in the present study.
11. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Novelli PK, Barros MM, Fleuri LF. Novel inexpensive fungi proteases:Production by solid state fermentation and characterization. Food Chem 2016;198:119-24. [CrossRef]
2. Usman A, Mohammed S, Mamo J. Production, optimization, and characterization of an acid protease from a filamentous fungus by solid-state fermentation. Int J Microbiol 2021;2021:6685963. [CrossRef]
3. Mamo J, Orellana JF, Yeleman V, Kangwa M, Fernandez-Lahore MH, Assefa F. Purification and characterization of aspartic protease produced from Aspergillus oryzae DRDFS13 MN726447 under solid-state fermentation. Catal Lett 2022;152:2033-46. [CrossRef]
4. Al-Manhel AJ. Production of exopolysaccharide from local fungal isolate.Curr Res Nutr Food Sci2017;5:338-46. [CrossRef]
5. Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS:The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 2014;42:D503-9. [CrossRef]
6. Yin LJ, Hsu TH, Jiang ST. Characterization of acidic protease from Aspergillus niger BCRC 32720. J Agric Food Chem 2013;61:662-6.? [CrossRef]
7. Patil PM, Kulkarni AA, Kininge PT. Production of milk clotting enzyme from Aspergillus oryzae under solid-state fermentation using mixture of wheat bran and rice bran. Int J Sci Res Pub 2012;2:1-12.?
8. Souza PM, Werneck G, Aliakbarian B, Siqueira F, Ferreira Filho EX, Perego P, et al. Production, purification and characterization of an aspartic protease from Aspergillus foetidus. Food Chem Toxicol 2017;109:1103-10. [CrossRef]
9. Aljammas HA, Al Fathi H, Alkhalaf W. Study the influence of culture conditions on rennin production by Rhizomucor miehei using solid-state fermentations. J Genet Eng Biotechnol 2018;16:213-6. [CrossRef]
10. Bensmail S, Mechakra A, Fazouane-Naimi F. Optimization of milk-clotting protease production by a local isolate of Aspergillus niger ffb1 in solid-state fermentation. J Microbiol Biotechnol Food Sci 2015;4:467-72. [CrossRef]
11. Daudi S, Mukhtar H, Rehman AU, Haq IU. Production of rennin-like acid protease by Mucor pusillus through submerged fermentation. Pak J Bot 2015;47:1121-7.
12. Mamo J, Assefa F. The role of microbial aspartic protease enzyme in food and beverage industries. J Food Qual 2018;3:1-15. [CrossRef]
13. Sun Q, Chen F, Geng F, Luo Y, Gong S, Jiang Z. A novel aspartic protease from Rhizomucor miehei expressed in Pichia pastoris and its application on meat tenderization and preparation of turtle peptides. Food Chem 2018;245:570-7. [CrossRef]
14. Purushothaman K, Bhat SK, Singh SA, Marathe GK, Appu Rao AR. Aspartic protease from Aspergillus niger:Molecular characterization and interaction with pepstatin A. Int J Biol Macromol 2019;139:199-212. [CrossRef]
15. Guo Y, Tu T, Yuan P, Wang Y, Ren Y, Yao B, et al. High-level expression and characterization of a novel aspartic protease from Talaromyces leycettanus JCM12802 and its potential application in juice clarification. Food Chem 2019;281:197-203. [CrossRef]
16. Chimbekujwo KI, Ja'afaru MI, Adeyemo OM. Purification, characterization and optimization conditions of protease produced by Aspergillus brasiliensis strain BCW2. Sci Afr 2020;8:e00398. [CrossRef]
17. Hsiao NW, Chen Y, Kuan YC, Lee YC, Lee SK, Chan HH, et al. Purification and characterization of an aspartic protease from the Rhizopus oryzae protease extract, peptidase R. Electron J Biotechnol 2014;17:89-94. [CrossRef]
18. Lowry OH, Resebrangh NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem 1951;193:265-75. [CrossRef]
19. Othman B, Sebo NH. Utilization of some agro wests for the production of acid protease by Aspergillus niger. J Surv Fish Sci 2023;10:4319-31.
20. Maitig AM, Alhoot MA, Tiwari K. Isolation and screening of extracellular protease enzyme from fungal isolates of soil. J Pure Appl Microbiol 2018;12:2059-67. [CrossRef]
21. Vishwanatha KS, Appu Rao AG, Singh SA. Production and characterization of a milk-clotting enzyme from Aspergillus oryzae MTCC 5341. Appl Microbiol Biotechnol 2010;85:1849-59. [CrossRef]
22. da Silva OS, de Oliveira RL, Souza-Motta CM, Porto AL, Porto TS. Novel protease from Aspergillus tamarii URM4634:Production and characterization using inexpensive agroindustrial substrates by solid-state fermentation. Adv Enzyme Res 2016;4:125-43. [CrossRef]
23. Wang F, Ni H, Cai HN, Xiao AF. Tea stalks-a novel agro-residue for the production of tannase under solid state fermentation by Aspergillus niger JMU-TS528. Ann Microbiol 2013;63:897-904. [CrossRef]
24. Luti KJ, Younis RW. An induction of undecylprodigiosin production from Streptomyces coelicolor by elicitation with microbial cells using solid state fermentation. Iraqi J Sci 2014;55:1553-62.
25. Radha S, Nithya VJ, Himakiran Babu R, Sridevi A, Prasad NB, Narasimha G. Production and optimization of acid protease by Aspergillus spp. Under submerged fermentation. Arch Appl Sci Res 2011;3:155-63.
26. Palsaniya P, Mishra R, Beejawat N, Sethi S, Gupta BL. Optimization of alkaline protease production from bacteria isolated from soil. J Microbiol Biotechnol 2012;2:858-65.
27. Medina J, Monreal CM, Orellana L, Calabi-Floody M, González ME, Meier S, et al. Influence of saprophytic fungi and inorganic additives on enzyme activities and chemical properties of the biodegradation process of wheat straw for the production of organo-mineral amendments. J Environ Manage 2020;255:109922. [CrossRef]
28. Asha B, Palaniswamy M. Optimization of alkaline protease production by Bacillus cereus FT1 isolated from soil. J Appl Pharm Sci 2018;8:119-27.
29. Muazu A. Production and characterization of extracellular protease enzyme from Aspergillus niger using different agro-industrial residues. Int J Sci Technol Res 2015;4:535-41.
30. Shivakumar S. Production and characterization of an acid protease from a local Aspergillus sp. by solid substrate fermentation. Arch Appl Sci Res 2010;4:188-99.