1. INTRODUCTION
DNA barcoding is an indicator used to build databases of DNA sequences that can be further exercised as DNA markers intended for “authentication” of the plant, along with studying the plant characters. DNA barcoding was originally based on differences in the mitochondrial Cytochrome c oxidase (cox-I) gene sequences, which successfully enabled the recognition of various flora and fauna [1]. However, it was later realized a need to establish other genes as barcodes as the cox-I gene was found to be less effective in identifying flowering plants [2]. Plant working group (PWG) has approved matK + rbcL as primary barcodes with ITS2 as one of the additional loci for distinguishing plants at the Third International Barcoding Conference, the Consortium for the Barcode of Life [3].
The family Solanaceae consists of a wide range of flowering plants with approximately 2,500 different plant species with significant benefits for human beings in terms of cost and productivity. This family includes many commercially valuable plants, Nicotiana tobacco, Capsicum annuum, Solanum lycopersicum, and Solanum melongena which are a few of them [4]. The genus Capsicum comprises the five most domestically cultivated species out of the overall 37 known species. These includes C. annuum L., C. frutescens L., C. chinense Jacq.,C. pubescens., and C. baccatum L [5]. The identification of all these species is usually made on the basis of the differences in their morphological characteristics, such as the shape of the fruit, color of the corolla, length of pedicel, and the number of growth at each pedicel [5,6]. Unfortunately, the Capsicum species annuum, frutescens, and chinense appear similar in their physical characters, making it difficult to distinguish between them morphologically. As most of the plants, physical characteristics depend on various environmental factors. The three species are commonly known as the “annuum-chinense-frutescens” group due to their close association [1,5]. Various attempts have been made to characterize different Capsicum species utilizing their morphological traits, enzyme loci, restriction fragment length polymorphism, random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism, direct or directed amplification of minisatellite region DNA amplified using the polymerase chain reaction (DAMD-PCR), cleaved amplified polymorphic sequence, simple sequence repeat length polymorphism, and inter-simple sequence repeats to determine genetic variations between and within different Capsicum species [7]. An effort was also made to distinguish C. frutescens from C. chinense using RAPD markers, where it was suggested C. frutescens and C. chinense to be different species [8]. However, a strong base describing the molecular diversity between these three closely resembling Capsicum species employing different markers used in DNA barcoding is yet to be established. Along with this, a strong genetic base that can be used to identify C. frutescens is still missing and needs to be addressed. With this as an objective, the present research is carried out to characterize C. frutescens using five different DNA markers.
The current research aimed to amplify matK, rbcL, ITS, rpoB, and trnH- psbA loci from C. frutescens and identify the best suitable barcode for identification of C. frutescens. To construct, a phylogenetic tree of the sequences obtained and comparing with the sequences of other plants belonging to Capsicum species and analyzing the delimitation strength of these barcodes. The study will assist in authenticating C. frutescens, along with its identification. It will also give an idea about the usefulness of these five genetic loci in differentiating between the three Capsicum species: annuum, frutescens, and chinense.
2. METHODS AND MATERIALS
2.1. Sample Collection
The seeds of C. frutescens were collected from Thiruvananthapuram, Kerala (8° 31’ 26.9004’’ N 76° 56’ 11.8968” E) India. The seeds were then grown in the Institute of Science Garden Mumbai, and fresh leaves were utilized for DNA isolation.
2.2. DNA Extraction
Genomic DNA extraction was carried out from young leaves using the cetyltrimethylammonium bromide (CTAB) method with few modifications [9]. The leaves were pulverized in a mortar using liquid nitrogen, and the ground leaves were then mixed with a CTAB isolation buffer. The quality of DNA extracted was, then, checked in 0.8% agarose gel. The extracted DNA was then quantified using NanoDrop Spectrophotometer (Thermo Fischer).
2.3. PCR Amplification and Sequencing
PCR was performed to amplify five genetic markers from C. frutescens which included two intergenic spacer regions (trnH-psbA, rpoB–trnCGAR), two plastid gene regions, namely, Ribulose 1, 5 bisphosphate (RbcL), and maturase kinase (matK) and a nuclear region, namely, inter transcribed spacer (ITS). The PCR was carried out using 25 μl reaction volume with each reaction mixture containing 90–100 ng of DNA template, 2.5 U Taq. Polymerase enzyme (ThermoFischer), 2.5 mM MgCl2, 1X Taq. DNA polymerase buffer, 0.2 mM dNTPs, and 0.5 μM primers ordered from Eurofins Genomics India Pvt. Ltd. Primer sequence used for amplifying different DNA regions of the gene is mentioned in Table 1, and their annealing temperatures are given in Table 2. The amplified genes were then separated on 1.5% agarose gel stained with 0.1 μg/ml ethidium bromide using 1X TAE buffer. The separated amplicons were later purified further to remove any possible contamination using Qiagen PCR clean-up kit. The samples were, then, sent for sequencing at Eurofins Pvt. Ltd. (India).
Table 1: List of primers used for PCR amplification.
| Gene | Primer sequence | Primer reference |
|---|---|---|
| ITS | ||
| Forward primer | TCCGTAGGTGAACCTGCGG | [12] |
| Reverse primer | TCCTCCGCTTATTGATATGC | |
| matK | ||
| Forward primer | CGATCTATTCATTCAATATTTC | [13] |
| Reverse primer | TCTAGCACACGAAAGTCGAAGT | [14,15] |
| rbcL | ||
| Forward primer | ATGTCACCACAAACAGAGACTAAAGC | |
| Reverse primer | GTAAAATCAAGTCCACCRCG | |
| trnH- psbA | ||
| Forward primer | GTTATGCATGAACGTAATGCTC | [16] |
| Reverse primer | CGCGCATGGTGGATTCACAATCC | |
| rpoB | ||
| Forward primer | CKACAAAAYCCYTCRAATTG | [17] |
| Reverse primer | CACCCRGATTYGAACTGGGG |
Table 2: PCR reaction conditions with respective annealing temperatures utilized for DNA amplification.
| Locus | Length of amplicons (base pairs) | Tm (degree) | Cycle conditions |
|---|---|---|---|
| ITS | 548 | 58°C | 95°C/1 |
| matK | 862 | 54°C | 32.8 |
| rbcL | 993 | 58°C | 72°C/7 |
| trnH- psbA | 553 | 58°C | 4°C/O |
| rpoB | 183 | 58°C |
2.4. DNA Sequencing and Sequence Analysis
Post amplification, the samples were analyzed on agarose gel electrophoresis and were sequenced using the Sanger sequencing method. The nucleotide sequences obtained for C. frutescens were then compared to sequences available in NCBI using basic local alignment search tool (BLAST). The sequences obtained were later aligned using multiple sequence alignment with hierarchical clustering [10] and edited using Finch TV.
Nucleotide sequence divergences were calculated pair-wise using the Kimura-2-parameter model, and the distance matrices were generated using a maximum likelihood (ML) tree with 1000 bootstrap replications and also parameters such as conserved sites, variables sites, parsimony informative sites, and GC content of the sequences were estimated using molecular evolutionary genetic analysis (MEGA) X software [11].
3. RESULTS
3.1. PCR Result and Quality Analysis of DNA Sequences
All the five regions of C. frutescens were successfully amplified by PCR and found to be good quality. The result indicates that the primer set utilized for the study was able to amplify respective regions and gave a sharp band on Agarose gel electrophoresis, which is required for successful DNA sequencing. All the five nucleotide sequences obtained for C. frutescens were submitted to the NCBI Gene Bank database. Their accession numbers are given in Table 3.
Table 3: List of characteristics features to all five genetic markers from C.
| Barcode | Accession number | Number of conserved sites | Number of variable sites | Number of parsimony informative sites | Number of singleton sites | G+C % |
|---|---|---|---|---|---|---|
| ITS | MT643918.1 | 458 | 105 | 17 | 86 | 64 |
| matK | MN528466.1 | 845 | 16 | none | none | 32.8 |
| rbcL | OK085708 | 989 | 4 | none | none | 43.3 |
| psbA | OK663603 | 553 | none | none | none | 27.4 |
| rpoB | ON368037 | 175 | 9 | none | 9 | 33.3 |
3.2. Sequence Analysis
All the five genes nucleotide sequences (matK, rbcL, ITS, rpoB, and trnH- psbA) obtained for C. frutescens were aligned with the retrieved sequences of Capsicum annumm and Capsicum chinense (see Supplementary material) obtained from NCBI database. It was found that ITS was able to differentiate between all the three species of Capsicum most efficiently. The retrieved sequences varied in length from 548 bp for C. frutescens to 653 bp for C. chinense and 636 bp for C. annuum. After alignment, it was observed that both C. frutescens and C. annuum showed gaps from position 100–107 and from position 111–115 along with a gap was also observed at position 254 when compared to C. chinense. However, interestingly, at position 251, all the three species of Capsicum showed different nucleotides, where frutescens showed the presence of guanine, while adenine was found for chinense, whereas annuum showed presence of cytosine nucleotide. It was seen that ITS sequence of C. frutescens showed 64% of G + C content. Conserved sites for ITS were found to be 458; variables were 105, parsimony informative sites were 17, and singleton sites were 17.
Sequence alignment of matK showed certain dissimilarities in many regions, but there were two specific regions at positions 7 and 913, where C. frutescens showed different nucleotides compared to the other two species. At position 7, C. frutescens showed substitution of thymine nucleotide, while other two species showed presence of cytosine nucleotide, while at position 913 frutescens showed the presence of cytosine, while the other two species showed a gap at the same position. Alignment of the matK sequences also showed that species frutescens and chinense are closely related as they both showed gaps at similar locations, while annuum did not show any gaps at these locations. matK sequence of C. frutescens showed 845 conserved sites and 16 variable regions, while no parsimony informative sites or singleton sites were observed with 32.8% of G + C content.
The aligned sequences of rbcL showed frutescens to have a substitution at position 573 with guanine nucleotide, while the other two species showed the same nucleotide adenine at that position. While the alignment also showed three other positions, where C. chinense showed substitutions. rbcL sequence of C. frutescens showed no parsimony-informative and singleton sites, while it had 989 conserved sites and four variable sites with 43.3% G + C content.
Next barcode analyzed was trnH- psbA; the aligned trnH- psbA nucleotide sequence of all the three Capsicum species displayed a notable change at certain positions. It was observed that C. frutescens and C. chinense showed a gap at position 360 while, interestingly, the annuum did not show any gap at this position. C. frutescens trnH- psbA sequence showed 27.4 % G + C content and had no variable sites, parsimony informative sites or singleton sites with 553 conserved sites. While nucleotide alignment of rpoB showed a gap at position 5 in C. frutescens, while the other two species showed that the presence of adenine nucleotide along with this C. frutescens also showed substitution of nucleotides at four more positions, while chinense and annuum showed similar nucleotides at the same position. rpoB sequence of C. frutescens showed the presence of 175 conserved sites, nine variable sites, and singleton sites with no parsimony informative sites with 33.3% G + C content.
3.3. Phylogenetic Analysis
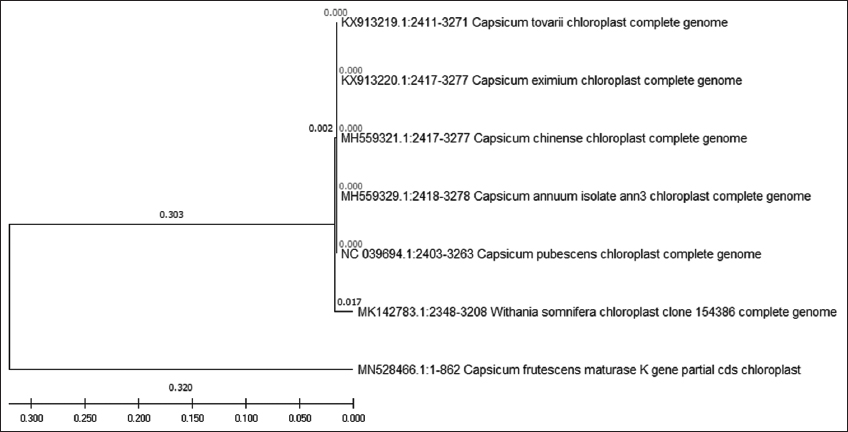
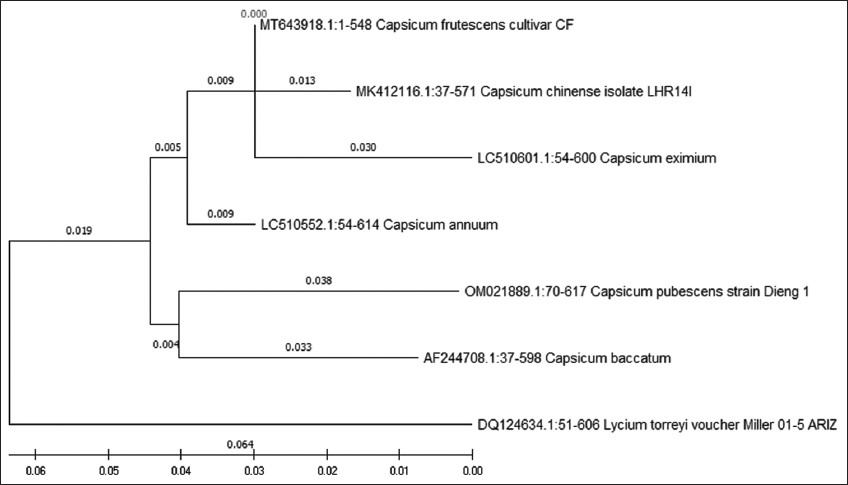
A BLAST was performed on all the five genetic marker sequences obtained for C. frutescens and were aligned using Multiple Sequence Comparison by Log-Expectation in the MEGA X. The nucleotide sequences of Capsicum genus were used to evaluate the evolutionary relationship using ML method with a 1000 replications bootstrap resampling. Outgroup belonging to the Solanaceae family were selected for all analyses. The first genetic marker studied was matK [Figure 1]. All the Capsicum species and the outgroup selected Withania somnifera were clustered together in one group, while C. frutescens was present in a separate monophyletic group. The next analysis performed was of ITS [Figure 2], which showed different clusters; the first cluster showed species frutescens, annuum, chinense, and eximium to be closely related and the other cluster had pubescens and baccatum species, while the outgroup Lycium torryei was seen to be present in a separate monophyletic clade. The result obtained was in correlation with previously observed results for species differentiation in Capsicum using ITS genetic marker, where it was noted ITS to be insufficient in differentiating among different Capsicum species, mainly between C. frutescens and C. chinense [18].
 | Figure 1: Phylogenetic relationship among the Capsicum species and outgroup taxa: The ML phylogenetic tree was reconstructed using the matK sequence data. The tree with the highest log likelihood (–2299.78) is shown. [Click here to view] |
 | Figure 2: Phylogenetic relationship among Capsicum species and outgroup taxa: The ML phylogenetic tree was reconstructed using the ITS sequence data. The tree with the highest log likelihood (–1398.41) is shown. [Click here to view] |
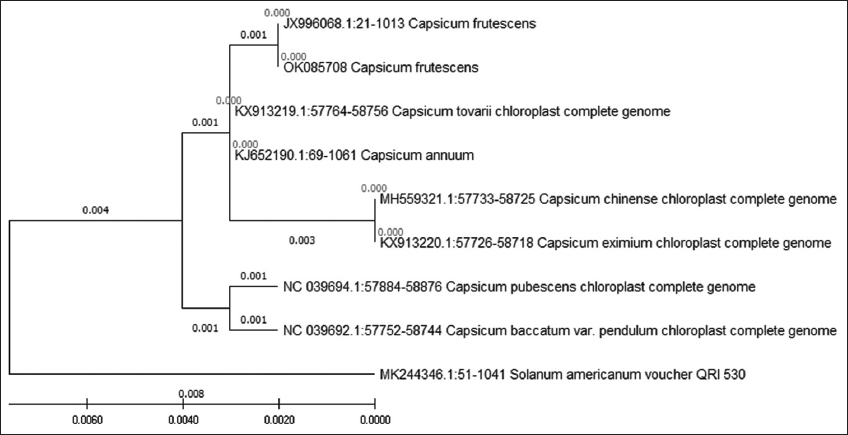
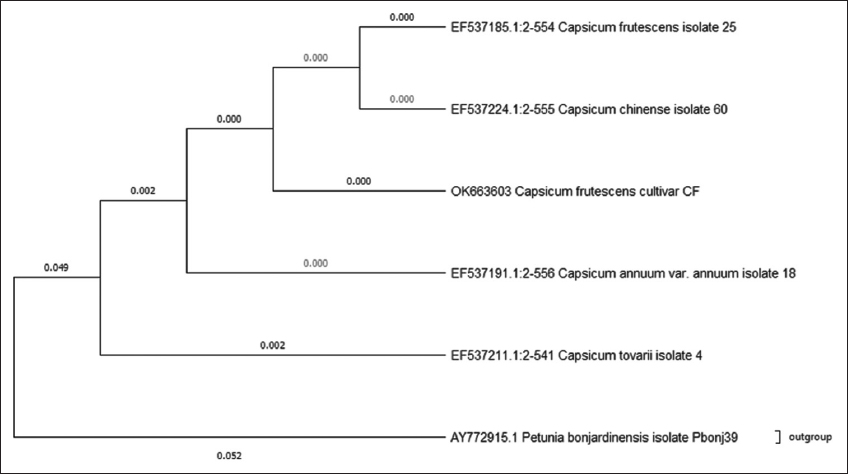
Phylogenetic analysis of rbcL [Figure 3] nuleotide sequences showed a well- resolved tree, where C. frutescens belonged to a monophyletic group, while Capsicum annumm belonged to a separate cluster, along with Capsicum toverii and the third cluster was made up of Capsicum chinense and eximium. It also showed a fourth cluster with Capsicum pubescens and baccatum, while the outgroup Solanum Americanum showed a separate cluster. Phylogeny analysis of trnH- psbA sequences [Figure 4] showed a tree with only two clusters, the first cluster contained all the Capsicum species, including annumm, chinense, and frutescens, while the outgroup formed a monophyletic cluster. Dendrogram representation of rpoB [Figure 5] showed a phylogenetic tree with three monophyletic clusters, where the first cluster had all the Capsicum species selected for analysis except C. frutescens which belonged to the second cluster, whereas the third cluster showed the outgroup that was selected.
 | Figure 3: Phylogenetic relationship among Capsicum species and one outgroup taxa: The ML phylogenetic tree was reconstructed using the rbcL sequence data. The tree with the highest log likelihood (–1502.33) is shown. [Click here to view] |
 | Figure 4: Phylogenetic relationship among Capsicum species and one outgroup taxa: The ML phylogenetic tree was reconstructed using the psbA sequence data. The tree with the highest log likelihood (–1099.29) is shown. [Click here to view] |
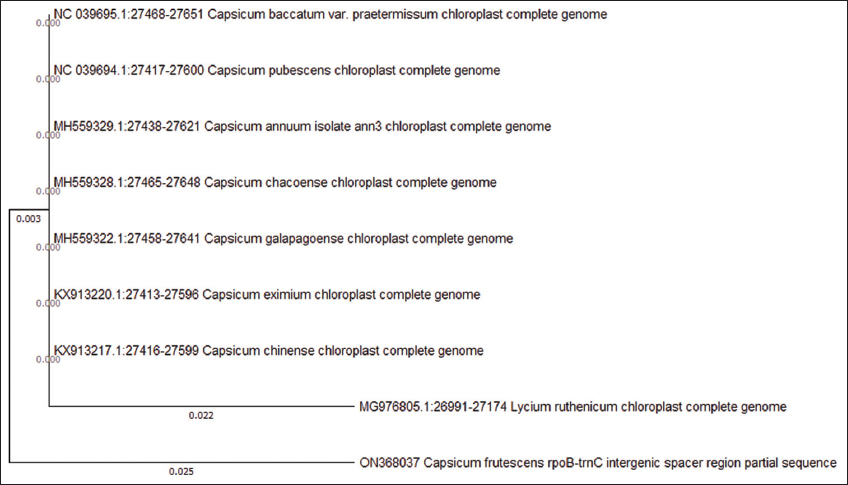 | Figure 5: Phylogenetic relationship among Capsicum species and one outgroup taxa: The ML phylogenetic tree was reconstructed using the rpoB sequence data. The tree with the highest log likelihood (–306.93) is shown. [Click here to view] |
The phylogenetic tree of matK shows it to be effective in differentiating C. frutescens from other Capsicum species as C. frutescens formed its separate branch with a length of 0.320, while all the other Capsicum species had a branch length of 0.303. It also showed 845 conserved sites and 16 variable regions with a GC content of 32.8%, while sequence analysis of ITS showed presence of highest number of variable sites with 64% GC content, while dendrogram analysis reveals that ITS was able to classify all the three Capsicum species in a separate clade and while they share the same ancestor and formed sister clades, they also showed branch length that were very close to one another suggesting them to be closely related.
The phylogenetic analysis of rbcL sequences gave very good results in terms of the conserved site with only four variable sites and 43.3% GC content. rbcL also proved to be a very good barcode in terms of its ability to differentiate between the three closely related sisters Capsicum species. The resulting phylogenetic tree placed frutescens, annuum, and chinense species all in three different clades making it effectively suitable to be used as a potential barcode. This again coincides with the results obtained in previous study, where phylogenetic analysis of rbcL marker placed C. frutescens in a monophyletic clade [19]. Whereas trnH- psbA did not resolve any Capsicum species as all the species were placed in the same clade with the same branch length, while the outgroup was placed in a different branch. While the phylogenetic tree of rpoB showed, C. frutescens has a wide difference when compared to other Capsicum species and placed frutescens under separate clade while keeping other Capsicum species and the outgroup species in the same cluster. It also showed that all the species involved in phylogenetic analysis shared a common ancestor. From the results obtained, it can be said that barcodes ITS and rbcL could be used as markers for differentiating the three Capsicum species (annuum, frutescens, and chinense), while matK and rpoB markers can be used as potential barcodes for species identification of C. frutescens.
4. DISCUSSION
Identification and discrimination of species are the first step in plant taxonomy, and DNA barcoding provides a rapid and efficient method to discriminate species [20]. In the present study, we have investigated five potential DNA barcoding loci, namely, matK, rbcL, ITS, rpoB, and trnH- psbA, to identify C. frutescens, and to examine the discriminating ability of these genetic loci in distinguishing the three morphological similar Capsicum species. In the present study, five genetic loci from C. frutescens have been sequenced and compared with the available sequences of different Capsicum species in the GeneBank database of NCBI. The potential of the loci as a barcode was determined using multiple sequence alignment and their phylogenetic analysis. Sequence analysis of different loci from C. frutescens showed psbA to possess the highest number of conserved regions with zero variable sites; this was followed by rbcL and rpoB, which showed four and nine variable sites, respectively. High conserved sites suggest a low level of evolution in these sequences; this can also be observed in the phylogenetic tree that was created, where psbA and rpoB did not separate different Capsicum species into clusters, but rpoB was able to separate C. frutescens from other Capsicum species. This result observed was contradictory to the result observed by Jarret (2008), where rpoB placed frutescens species together with all other Capsicum species in the dendrogram [1]. Whereas rbcL dendrogram showed differentiation of Capsicum species into separate clusters, the branch length was close, suggesting the species to be closely related. The Consortium for Barcode of life PWG has recommended a combination of two loci matK + rbcL, to be used as a barcode for plants as a single barcode could not be identified as a universal barcode [21]. The result obtained in the present study proved to be in alignment with this conclusion as the matK locus was also able to place C. frutescens into a separate cluster interestingly, it was not able to differentiate among the remaining Capsicum species and was all placed in the same clusters.
Sequence analysis of matK correspondingly showed 16 variable sites for C. frutescens. Multiple sequence alignment of matK and rbcL also gave substantial results, where C. frutescens showed substitutions and gaps at specific positions. The highest number of variable sites was observed for ITS which gave a well-resolved phylogenetic tree. ITS successfully placed the three morphologically similar Capsicum species into three different clusters, proving it to be an effective barcode for analysis of Capsicum species and also identification of C. frutescens along with matK + rbcL markers. After evaluating the barcodes, the combination of three barcode markers matK + rbcL + rpoB can be proposed as a suitable barcode for identifying C. frutescens. In a previous China Plant BOL Group study, it was observed that the combination of rbcL + matK + ITS barcode markers gave 77.4% species discrimination [22]. The present study combination of rbcL + ITS loci was observed to be successful in distinguishing the three closely related Capsicum species of annuum-frutescens-chinense.
5. CONCLUSION
The present research attempts to analyze five different DNA markers as a way to authenticate C. frutescens. The present study employs DNA barcoding approaches and provides significant findings for determining the phylogeny and connection among different Capsicum species. The current study attempts to show the possibility of differentiating the three Capsicum species, namely annuum, frutescens, and chinense using five different gene markers as they are morphologically very similar. The results indicate that specific differences exist between the three Capsicum species. It was observed that matK, rbcL, and rpoB were useful in identifying C. frutescens, whereas rbcL and ITS successfully differentiated the three morphologically similar Capsicum species. This knowledge will not only help taxonomists to identify C. frutescens but also will provide essential insights into the differences existing in the Capsicum genus on the molecular level.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article and the sequences are available online in the NCBI database.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Jarret RL. DNA barcoding in a crop genebank:The
2. Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haider N,
3. Pang X, Song J, Zhu Y, Xu H, Huang L, Chen S. Applying plant DNA barcodes for
4. Rosario LH, Padilla JO, Martínez DR, Grajales AM, Reyes JA, Feliu GJ,
5. Hassan NM, Yusof NA, Yahaya AF, Rozali NN, Othman R. Carotenoids of
6. Aguilar-Melénde A, Morrell PL, Roose ML, Kim SC. Genetic diversity and structure in semiwild and domesticated chiles (
7. Ince AG, Karaca M, Onus AN. Genetic relationships within and between
8. Baral JB, Bosland PW. Unraveling the species dilemma in
9. Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus 1990;12:13-5. [CrossRef]
10. Corpet F.Multiple sequence alignment with heirachial clustering. Nucl Acids Res 1988;16:10881-90. [CrossRef]
11. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9. [CrossRef]
12. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal Rna genes for phylogenetics PCR Protoc 1990;1:315-22. [CrossRef]
13. Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW. Molecular phylogenetics of
14. Levin RA, Wagner WL, Hoch PC, Nepokroeff M, Pires JC, Zimmer EA,
15. Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants:The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One 2007;2:e508. [CrossRef]
16. Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of
17. Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J,
18. Shiragaki K, Yokoi S, Tezuka T. Phylogenetic analysis and molecular diversity of capsicum based on rdna-its region. Horticulturae 2020;6:1-13. [CrossRef]
19. Walsh BM, Hoot SB. Phylogenetic relationships of
20. Duan H, Chen F, Liu W, Zhou C, Zhou Y. Research and applications of DNA barcode in identification of plant species. Res Plant Biol 2014;4:29-35.
21. Ghahramanzadeh R, Esselink G, Kodde LP, Duistermaat H, van Valkenburg LC, Marashi SH,
22. Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ,