1. INTRODUCTION
Pseudomonas aeruginosa a Gram-negative rod-shaped bacterium is inclusive of the genus Pseudomonadaceae known for its highly versatile metabolic activity [1]. P. aeruginosa is an opportunistic pathogen that can be isolated from the environmental samples of water, soil, plants, and fruits. Humans can get infected with the urinary tracts, airways, blood, burns, wound injuries, and even found in outer ear infections. The characteristic feature of this organism is its ability to survive in two different models of growth systems namely planktonic (free-floating) and biofilm (sessile) forms. P. aeruginosa has a genetic makeup of almost 5021 genes, with 4000 genes common in all the strains [2]. During the biofilm stage an elemental phenomenon that occurs in bacteria, especially in P. aeruginosa, is quorum sensing that is cell-to-cell density-dependent community cross-talk [3,4]. It is observed that quorum sensing is responsible to facilitate thiol oxidative stress-related metabolism of the bacterium [5-8]. Pseudomonas bacteria are prone to become resistant to antibiotics due to the formation of biofilms that complicates any treatment or infections. The two major signal molecules that are involved in the quorum-sensing mechanism of P. aeruginosa include 4-quinolones and N-acyl-homoserine lactones [9,10]. Initially, the planktonic form of these bacteria adheres to a solid surface forming microcolonies, followed by cell migration onto the substratum forming a flat mat-like heap of microcolonies resembling a mushroom-like structure called the biofilm. There are 140 species of Pseudomonas, of which 25 are associated with human infection. Although these rarely cause diseases, they are a major threat in hospitalized patients with cancer and cystic fibrosis [11]. The cellular morphology of P. aeruginosa is made up of three layers: the outer membrane, the peptidoglycan layer, and the inner cytoplasmic membrane, respectively [Figure 1].
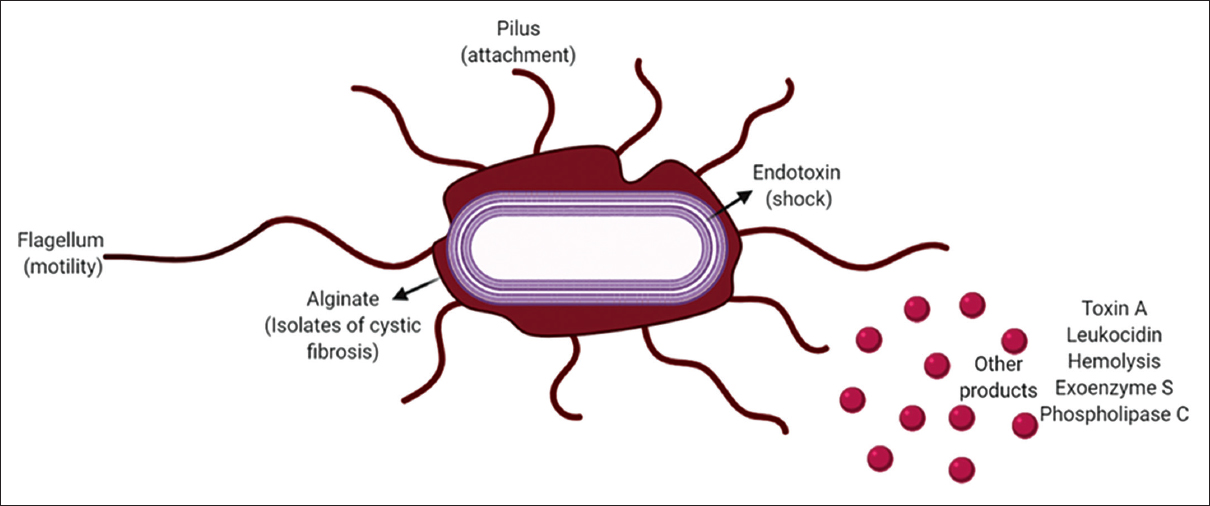 | Figure 1: Structure and pathogenic machinery of Pseudomonas aeruginosa. [Click here to view] |
The outer membrane of the bacteria is made up of protein, lipopolysaccharide, and phospholipid. The lipopolysaccharide layer is composed of 2-keto-3-deoxyoctonic acid, heptose, core polysaccharides, and hydroxyl fatty acids. Many virulence factors are produced by P. aeruginosa including endotoxin that causes terminal shocks, leukocidin that causes depression of host defenses, heat-stable hemolysis that is toxic to alveolar macrophage, toxin-A that causes lethality and inhibition of host defenses, phospholipase C that hydrolyzes lecithin, and Exoenzyme S (ExoS) that causes local and systemic toxicity [10,12,13]. The main essential function of the prokaryotic cell is to transport proteins to the environment, compartments of cells, and other bacteria cells in pure culture or co-culture and mixed culture populations [6,14]. This mechanism is executed by a protein-mediated cascade known as the protein secretion system. There are different types of secretion systems, including Type I secretion system, Type II secretion system, Type III secretion system (T3SS), Type IV secretion system, Type V secretion system, Type VI secretion system, and Type VII secretion system. In P. aeruginosa the, T3SS, plays a pivotal role in virulence and could be majorly targeted to block bacterial signal trafficking. The bacterial flagellum and the injectisome are composed of conserved machinery known as the T3SS. T3SS in the flagellum functions to transport the distal flagellar components to build the extracellular filament, whereas T3SS in the injectisome is the center of the transport machinery that helps in the formation of the extracellular needle and also to direct the transfer of substrates into the host cell from the cytosol of the bacteria. T3SS is composed of 9 highly conserved core proteins, of which 8 proteins are found in the flagellar apparatus. Substrates of T3SS are generally known as effector proteins. Pseudomonas produces effector proteins where the secretion signals are embedded. T3SS is made up of 3 major components: basal body, needle component, and the translocon [15]. The basal body is composed of cytoplasmic components that form a socket-like structure with a center rod surrounded by several rings spanning around the inner and the outer membrane. There are many systems available for epidemiological studies such as phage, pyocin, serologic, and DNA fingerprinting [13,16]. P. aeruginosa has become resistant to many antibiotics [11]; therefore, the use of phytobioactives will play a vital role in treating infections. P. aeruginosa is accepted worldwide as a public health risk because of health-care-acquired infections and its ability to acquire resistance to multiple antibiotics. This review, attempts to explore the possibilities of the application of phytobioactives to hamper the T3SS virulence mechanisms involved in P. aeruginosa at different phases of growth, survival, biofilms formation, and pathogenesis, which could be corroborative of the related genes, proteins, and/or the epigenetic machinery involved. Since herbal treatment is booming lately all the possible plants and their derivatives have been enlisted that could be used as potential blockers to inhibit this long-lasting problematic superbug. Deciphering these multi-factorial mechanisms would require a well-orchestrated approach which could lead to deeper penetration of the bioactive into the micro and macro environments primarily disrupting the signaling cascades hampering the ability of cell-to-cell crosstalk and demoralizing the bacterial community preventing pure, coculture, or mixed culture survival.
1.1. Formation of P. aeruginosa Biofilm
Acute infections, the inter-relationship between the pathogen and the host is two-way devastating since the virulent cytotoxic molecules produced by the bacteria damage the host cell response whereas the host immune system secretes reactive oxygen species (ROS), reactive nitrogen species, antimicrobial compounds, and increases phagocytic processes [17]. Motile P. aeruginosa is found to be more virulent and is easily detected by the host via the flagella or other motility apparatus. This induces inflammatory responses and triggers signaling pathways and enhances phagocytosis [10,13,18]. To survive, the bacterium opts for a sessile lifestyle with a low virulence capability so that it could avoid adverse stress conditions. They lose their motile function and adhere to surfaces to form microcolonies that are embedded in the extracellular polymeric substances to defend themselves from the fencing environment [Figure 2]. These biofilm structures are predominately mediated through complex cascading systems resembling the neural networks that confer endurance against ROS, phagocytosis, antimicrobial agents, and interspecies competitions [11,19,20]. Biofilms almost contribute to 90% of the total mass of the bacteria and these are formed on biotic and abiotic surfaces [Figure 2]. The matrix provides an intense niche to the biofilm with nutrients and energy supply for its survival during unfavorable conditions [21]. The matrix is composed of proteins, polysaccharides, lipids, and extracellular DNA as illustrated in Table 1 [22,23].
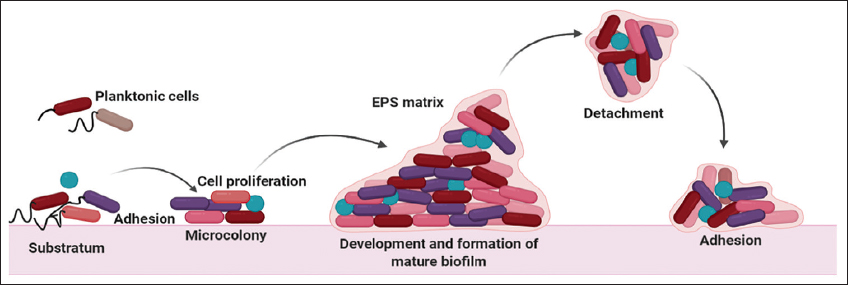 | Figure 2: Formation of Pseudomonas aeruginosa. [Click here to view] |
Table 1: Polymeric components involved in biofilm formation by P. aeruginosa.
| Name | Chemistry | Functions | Property | References |
|---|---|---|---|---|
| Psl | Exopolysaccharide | Psl explores surfaces for attachment and initiates biofilm formation by increasing cell motility, and cell-to-cell communication. They protect the cell against phagocytosis and oxidative stress during infections | Neutral charged | [18] |
| Pel | Exopolysaccharide | Pel along with Psl initiates biofilm formation and maintains cell-to-cell interaction. It is the main structural component of the biofilm stalk where it replaces Psl in its absence and cross-links with eDNA. Pel also protects the bacteria from aminoglycoside antibiotics | Positively charged | [20] |
| Alginate | Exopolysaccharide | Alginate behaves as a protective agent and provides the biofilm a slimy nature, they enhance viscoelastic character, cell density, architecture, surface adhesion, and cell aggregation | Negatively charged | [22] |
| eDNA | Nucleic acid | eDNA is a structural component of biofilms stalk that forms cations like Mg2+, Mn2+, Ca2+, and Zn2+during starvation as a nutrient source | Negatively charged | [51] |
| Type 4 pili | Multiprotein complex | Type 4 pili are important during maturation of the biofilm, they play an important role in cell adhesion in the early stage of biofilm formation | No charge | [13] |
| Flagella | Multiprotein complex | Flagella along with type 4 pili help in migration/motility necessary for biofilms formation | No charge | [23] |
1.2. Quorum Sensing in P. aeruginosa
Density-dependent, cell-to-cell communication through biochemical signals is a capability of bacteria known as quorum sensing. Several interconnected pathways are involved in regulating the social behavior of the bacteria through the quorum sensing mechanism [11,24]. Quorum sensing regulates biological processes required for bacterial adaptation and survival, expression of certain genes to a critical threshold of signaling molecules called autoinducers (AIs). Quorum sensing regulates community survival by mediating population density-dependent collective responses and increases the fitness and the chance of survival, thus enhancing their ability to escape during pathogenesis [25]. P. aeruginosa infections more than 10% of the genes are regulated by a quorum-sensing mechanism that plays a critical role in the progress of infection from acute to chronic. A cascade of genes and proteins are involved in virulence, biofilms development, regulating metabolic pathways during stress responses, and antibiotic resistance mechanisms [6,13,26]. The four major pathways that are involved in quorum-sensing dependent signaling in P. aeruginosa include Rhl, Las, IQS, and Pqs. On sensing certain AIs the first category of proteins PqsR, RhIR, and LasR are activated, these act as transcriptional activators for the second category of proteins. The second category of proteins includes the AI synthases that are PqsABCDH, Rh1I, and LasI. These proteins act as an activated circuit traveling in and out of the cell by membrane transporters, membrane vesicles, and free diffusion systems [27].
P. aeruginosa secretes specific virulence factors to evade the host immune defense system at an early stage of attachment, colonization, and acute infection. All low-molecular-weight and high-molecular-weight compounds are considered important in the progression of infection and colonization. Although these virulence factors promote bacterial development and survival they result to cause a disastrous effect on the host tissues and cause changes in the immune responses [28]. Virulence factors require community involvement and they are metabolically costly hence they are produced under the regulatory control of the quorum-sensing system [Figure 3] [29].
 | Figure 3: Regulation of virulence factors and hierarchical quorum sensing network in Pseudomonas aeruginosa. [Click here to view] |
1.3. Genome Assembly in P. aeruginosa
The genome of P. aeruginosa is 6.3 Mbp which was sequenced using whole-genome-shotgun sampling, and the genes were annotated. The open reading frames (ORFs) in the genome were found to contain 66.6% of G + C content, but in almost ten regions of 3 kilobases, there was lower G + C content. 5,570 ORFs were predicted in P. aeruginosa. The genome consists of four longest rDNA loci and one duplicated gene cluster repeat, including PA4210–PA4216 and PA1899–PA1905 which spans a few thousand base pairs. The ORFs predicted were curated individually to identify and annotate genes of P. aeruginosa with others in GenBank, to identify any similarities of these genes with other bacteria, and to identify functional motifs. For about 54.2% of the ORFs, functions were assigned. The functions assigned to the 372 ORFs of P. aeruginosa include virulence factors, lipopolysaccharide biosynthetic enzymes, exoenzymes, and proteins responsible for adhesion and motility. Since P. aeruginosa could be grown on minimal media, the genes responsible for the biosynthesis of nucleic acids, cofactors, and amino acids were identified [30]. Virulence factors such as proteases, toxins, and lipases are secreted by P. aeruginosa. Out of the four types of protein secretion pathways involved in Gram-negative bacteria, three were evident in P. aeruginosa. Type I secretion system secretes alkaline protease encoded by aprA, composed of ABC transport protein AprD, OprM-family outer membrane protein AprF, and membrane fusion protein AprE. Type II secretion systems also known as the general secretion pathway are encoded by xcp gene cluster and the unlinked pilD/xcpA gene [31]. T3SS are responsible for the contact-dependent delivery of proteins into the host cells found in plants and animal pathogens. P. aeruginosa is composed of one single T3SS PA1690-PA1725 that secretes virulence factor ExoS, Y, and T [32,33].
1.4. T3SS
T3SS is the primary virulence determinant of P. aeruginosa that promotes tissue destruction and escapes phagocytosis by secreting numerous toxins that are translocated into host cells [34]. ExsA is the primary regulator of T3SS gene expression that binds to promoters of T3SS activating transcription. AraC/XylS is a huge family of transcriptional regulators of which ExsA is a member. The structure of these proteins is composed of a conserved 100-amino-acid helix-turn-helix DNA binding domain that is positioned at the C-terminal and the N-terminal or the ligand-binding domain [35]. ExsA-dependent promoter’s transcriptional start sites are mapped with primer extension [36]. s70 dependent promoters transcriptional start sites for PexsD, Porf1, and PexoS are located downstream from near-consensus -10 (TATAAT) and -35 (TTGACA) recognition hexamers [37]. The −47 and −45 positions, ExsA consensus binding site is identified that is located in the highly conserved guanine and cytosine nucleotides [38]. Further ExsA at the -51 position consists of conserved adenine-rich areas and other conserved regions at the −35 position [39]. T3SS consists of five major families: Inv-Mxi-Spa, Ssa-Esc, and Ysc families that belong to animals, and Hrp T3SS of plant pathogens. T3SS of P. aeruginosa was first identified in 1996 whereas the structure was visualized in 2005 [Figure 4] [13,40-42].
 | Figure 4: Role of T3SS in pathogenicity. [Click here to view] |
1.5. P. aeruginosa T3SS Gene Expression
ExoS secreting strains of P. aeruginosa cause deferred apoptotic cell death whereas rapid cell lysis of host cells was caused by Exoenzyme U (ExoU). Other effector proteins include flagellar filament protein (FliC), PemA/PemB, and nuclear diphosphate kinase [Figure 4]. The T3SS expression system is controlled by various environmental factors including serum albumin/casein ratio, host cell contact, and lower concentrations of Ca2+ in the extracellular space [43]. Direct transcriptional activation by ExsA a transcription factor belonging to the family AraC controls the primary gene expression of T3SS. Many plant phenolic compounds such as salicylic acid, its analogs, and its precursors are found to activate or inhibit gene expression of T3SS [44]. T3SS is a secretion system that protects P. aeruginosa from any predators from amoebae to human neutrophils, it immediately gets expressed with host cell contact and highly applicable signals [42]. T3SS gene expression due to various environmental factors leads to an optimal bistable group of the population providing an advantage for cells that express or repress T3SS gene expression [13,45].
2. TARGETING BIOLOGICAL ACTIVITIES OF P. AERUGINOSA THROUGH PHYTO-MODULATORS
P. aeruginosa has different modes of movement including twitching, swimming, and swarming. Swarming is a motility mode for the surface movement that takes place in semi-solid media and it is one of the fastest modes of movement that involves both flagella and type IV pili [Figure 5]. Swarming bacteria are found to be more virulent and also more resistant to antibiotics [46]. Many genes and pathways are involved in the swarming motility of bacteria including quorum sensing [11,47]. Swarming cells when compared with the vegetative parts overexpress T3SS genes, and use this system to inject toxic effector proteins causing cytotoxicity in the host cell. T3SS effector proteins including ExoS, ExoT, ExoU, and ExoY are responsible for causing cytotoxicity and also induce apoptosis in P. aeruginosa [48]. Naturally obtained plant compounds like tannins, and cis-2-dodecenoic acid inhibits the swarming motility and downregulates virulent genes or effector proteins and quorum sensing genes in T3SS [49,50]. The study claims to screen plants and their derivatives to identify compounds that can inhibit the swarming motility of P. aeruginosa. Alpinia officinarum belonging to the family Zingiberaceae is widely used in India and other Asian countries as a medicinal plant due to its diverse compounds [51]. The plant is known to possess anti-bacterial [52], anticancer, antioxidant [53], and antiemetic properties [54,55]. Rhizomes of the plant were commercially obtained and P. aeruginosa MTCC 3541 was collected from a microbial-type culture collection center. Powdered rhizomes were subjected to different solvent extraction out of which methanol extract exhibited the best activity in the swarming inhibitory assay. From the results obtained it was found that A. officinarum rhizomes inhibited the swarming motility of P. aeruginosa. The phytomolecules responsible to cause the inhibition was found to be 1-(3,4-dihydroxyphenyl)-2-(methylamino)ethan-1-one. Enhanced expressions of virulent genes are connected with swarming motility therefore they analyzed the effect of the phytomolecules against the virulent genes. The virulent genes of the T3SS include exoS, exoT, gacS, vfr, fleQ, and fliC where the expression of exoT, exoS, gacS, and fleQ was downregulated significantly on exposure to the phytomolecule. The fleQ was more significantly down-regulated when compared to all other genes. fleQ is the master regulator gene of flagella synthesis that controls motility and biofilm formation. The actual mechanism of inhibition of action needs a much more comprehensive study to be determined [13,56].
 | Figure 5: Mechanistic targeting of phytochemicals on T3SS machinery and their related pathways. [Click here to view] |
Antibiotic treatment is the most usually utilized system to control pathogenic contaminations; in any case, it has added to the age of antibiotic-resistant bacteria. T3S system consists of 43 genes that include T3 effectors, regulatory functions, effector chaperones, and T3 machinery [42,57]. T3SS is transcriptionally controlled by ExsA where ExsA is regulated by three interacting proteins ExsC, ExsD, and ExsE. T3SS of P. aeruginosa is one of the significant destructiveness factors by which it secretes and moves T3 effector proteins into human host cells. Since P. aeruginosa is also known to infect plants, plant-derived defense signaling phenolic compounds were screened to identify T3 inhibitors [13,58]. Out of 72 compounds that were analyzed TS103 and TS134 were identified as the best inducers of T3 on the other hand TS027 and TS101 inhibited T3. These compounds exhibited their effect by regulating the expression levels of small RNAs RsmY and RsmZ that altered exoS transcriptional activity. These compounds were found to act on these RNAs through GacSA-RsmYZ-RsmA-ExsA regulatory pathway [42,44].
P. aeruginosa quorum-sensing system is a hierarchical network that regulates the activity of rhII/R through lasI/R system [59]. The expression of protease and elastase is regulated by the las system whereas pyocyanin and rhamnolipids are produced by rhl system. Two acyl-homoserine lactone signaling molecules: N-butanoyl-L-homoserine lactone and N-(3-oxododecanoyl)-L-homoserine lactone are produced by P. aeruginosa. These two signaling molecules are responsible for activating lasI/R and rh1I/R systems in turn activating the virulent nature [42,60]. Excessive host inflammatory responses lead to harmful effects such as generalized hyperthermia, severe tissue edema, and organ dysfunction. Anti-inflammatory drugs will interrupt chronic infection therefore a huge number of plant-derived compounds are widely used in the treatment of microbial infections and decrease inflammation due to their less toxic nature [61]. This study, Andrographis paniculata belonging to the family Acanthaceae was harnessed due to its antibacterial, antiviral, antioxidant, anti-inflammatory, and hepato-stimulant activities [62]. Methanol and chloroform extract of A. paniculata exhibited significant anti-quorum sensing activity. A. paniculata extracts at a concentration lower than minimum inhibitory in a dose-dependent manner lowered biofilm formation, virulent product production, and swarming motility of P. aeruginosa without affecting its growth. The last system was found to play a role in colonizing host tissues whereas rhl system contributed to the virulent nature of the pathogen by chelating the attached iron from transferring [63]. A. paniculata extracts were found to inhibit the free planktonic cells to attach to abiotic surfaces thus preventing the first step of biofilm formation. It was concluded that the chloroform extract of A. paniculata exhibited potent anti-inflammatory activity at a very low concentration by inactivating p38- and ERK1/2-signaling pathways [13,64]. Table 2 represents plants that could be used as anti-pseudomonal agents.
Table 2: Represents plants that could be used as anti-pseudomonal agents.
| Plant name | Organism | Dosage | Mode of action | References |
|---|---|---|---|---|
| Santalum album, Bergenia ciliata, Jasminum officinale | P. aeruginosa | 100–300 µg/disc | Inhibition of bacterial growth in disc diffusion assay | [4] |
| Stereospermum kunthianum, Bridelia, Cassia tora, Acacia acuminata, Anthocephalus cadamba, Schleichera oleosa, Eugenia jambolana, Mimusops elengi, Tectona grandis, Pterocarpus santalinus | P. aeruginosa | 80 µL/disc | Minimum inhibitory concentration and minimum bactericidal concentration were determined for all the 10 timber-yielding plants | [13] |
| Aegle marmelos | P. aeruginosa, Escherichia coli, Salmonella typhii | 2–10 µg/mL | Prevention of formation of biofilm with highest activity | [13] |
| Moringa oleifera | P. aeruginosa, Staphylococcus aureus | 0.05 mg/mL | Preventing the free planktonic cells to attach to surfaces, disrupting the pre-formed biofilm, reduction of metabolic activity | [19] |
| Dendrophthoe falcata | P. aeruginosa | 1 mg/well | Anti-biofilm activity, reduction of quorum sensing mechanism | [91] |
| Anacardium occidentale, Coffee Arabica, Lycopersicon esculentum, Theobroma cacao, Curcubita | P. aeruginosa, Chromobacterium violaceum | 50 µL | Inhibition of fucophilic lectin PA-IIL, Reduces intestinal and other human infections by blocking lectin-dependent bacterial adhesion | [87] |
| Pongamia pinnata | P. aeruginosa | 100–400 mg/disc | Supression of TNF-a and IL-6 and enhanced IL-10 increasing anti-bacterial effect | [32] |
| Cassia alata | P. aeruginosa | 0.4 mg/mL | Attenuation of virulence factors such as elastase and protease. Reduction of swarming motility of the bacteria | [91] |
| Alpinia officinarum | P. aeruginosa | 250 µM | Expression of exoT, exoS, gacS, and fleQ were down-regulated | [44] |
| Andrographis paniculata | P. aeruginosa | 5 µg/mL | Inhibition of free planktonic cells to attach to abiotic surfaces thus preventing the first step of biofilm formation | [92] |
| Crataeva nurvala | P. aeruginosa | 15 µg/mL | Suppression of quorum sensing mediated virulence factors such as hemolysin, pyocyanin, protease and biofilm formation | [4] |
| Vitex negundo | P. aeruginosa | 30–60 µg/100 µL | Inhibition of bacterial growth | [91] |
P. aeruginosa: Pseudomonas aeruginosa
Biofilms are bacterial colonies that have a high tolerance to stress, antimicrobial compounds for a better adaptive habitat [65]. P. aeruginosa is one of the bacterial families that are considered the most sinister pathogen that forms biofilms in the human host [66]. Biofilms contribute eloquent barriers to several antibiotics therefore identifying new compounds, and drugs that can inhibit the attachment process are present of great interest [67]. Juglans regia is a temperate forest tree that is known for its nutritional importance and its therapeutic effects. J. regia extract is known to possess anti-diabetic, anti-carcinogenic, anti-inflammatory, anti-helminthic, and anti-diarrheic activities [68]. In this study, J. regia leaves were collected to analyze antibacterial and anti-biofilm properties. Methanolic extract of the plant was subjected to 50 different samples of P. aeruginosa. It prevented the biofilm formation exhibited antibacterial effect against P. aeruginosa. The extract was further tested against different bacterial strains including Streptococcus mutans, Staphylococcus aureus, Streptococcus salivarius, and Streptococcus sanguis. J. regia methanolic extract exhibited an antibacterial effect against all the strains except S. mutans strain [13,69,70].
3. CURRENT THERAPIES AGAINST P. AERUGINOSA INFECTION
At present, available new approaches for the development of anti-pseudomonal therapies include antibiotics, vaccines, phage therapy, hygienic measures, and enhancement of host defense [Figure 6].
 | Figure 6: Currently available therapies against Pseudomonas aeruginosa infections. [Click here to view] |
3.1. Antibiotics
Different strains of P. aeruginosa are liable to a broad spectrum of antibiotics including carbapenems, penicillins, monobactams, cephalosporins, and fluoroquinolones. The emergence of resistant organisms has called for alternatives especially due to infections in intensive care patients. An alternative treatment for P. aeruginosa was the revival of abandoned old drugs such as polymyxin B and colistin. But due to their significant side effects, and toxicity more therapeutic strategies have been developed against the multiple drug-resistant P. aeruginosa. Compounds that can overcome β-lactamase antibiotic resistance were developed as another strategy. Ceftolozane-tazobactum and ceftazidime-avibactam are the new cephalosporin-β-lactamase inhibitors introduced [55,71]. Chromosomal β-lactamase is inactivated by avibactam in P. aeruginosa AmpC. Other β-lactamase inhibitors are under clinical trial including vaborbactam [72], zidebactam [73], nacubactam [74], and relebactum [75]. Cefiderocol is structurally similar to cephalosporins, they are found to be the most potent in vitro drug against multiple drug-resistant P. aeruginosa [76]. Two fluoroquinolones that exhibit anti-pseudomonal activity delafloxacin [77] and finafloxacin have recently become available. Murepavadin is a peptidomimetic that targets the outer membrane protein it is found to be a new weapon against P. aeruginosa. Aerosolized anti-pseudomonal drugs were developed to treat chronic respiratory infections in patients with cystic fibrosis or bronchiectasis. Liposomal amikacin is one such aerosolized drug that can penetrate P. aeruginosa biofilms and within the airway secretions, long-term inhalation led to carcinogenicity therefore this drug was barred for P. aeruginosa infections [78].
3.2. Vaccines
Researchers have made an agenda to devise an effective vaccine against P. aeruginosa infections but to date, there are no licensed vaccines available. Octavalent O-polysaccharide-toxin A conjugate P. aeruginosa vaccine was developed by the Swiss Serum and Vaccine Institute in the 1990s for the immunization of healthy patients with cystic fibrosis. The vaccine significantly lowered the infection in cystic fibrosis patients due to high-affinity antibodies among immunized patients. In 2010, attenuated live Salmonella strains were administered to human volunteers through systemic, nasal, or oral routes [79]. Nasal and oral vaccines exhibited promising effects through a significant increase in IgA and IgG antibodies. The authors concluded that by inducing a specific antibody response in the lung the nasal and the oral vaccines would serve as promising applicants for the development of anti-pseudomonal immunization against P. aeruginosa. Globally, researchers are trying to explore synthetic biology using CRISPR-Cas9 to have a viable solution [80].
3.3. Phage Therapy
Antibiotic resistance is not only a major threat to human health but also affects the production of food and sustainable development. Phage therapy has regained interest but it was abandoned in several countries. Eastern European countries still develop phage therapy with centers in Georgia, Poland, Tbilisi, and Warsaw [81]. Establish phage therapy there needs to be an in-depth understanding of how there were mutual descent and conflict between the phage and the bacterium. It was observed that P. aeruginosa can remove all the phage receptors and also cystic fibrosis lung was resistant to phages after 5 years.
3.4. Hygienic Measures
Control measures including education of personnel, environment cleaning, and hand hygiene was demonstrated by the microbiological monitoring to avoid the development of resistance to P. aeruginosa [82]. Prolonged uses of antibiotics are leading to multiple drug resistance; therefore, antibiotics should be avoided when it is not necessary. In hospitals, the sinks are majorly contaminated with P. aeruginosa, which leads to contamination of hands during washing, medical devices, detergents, soaps, and aqueous solutions [83]. Installing filters prevented bacterial contamination of tap water and hands during washing [84]. Transmission of pseudomonal infections was observed in patients suffering from cystic fibrosis; therefore, P. aeruginosa positive and negative patients were separated [85].
3.5. Enhancement of Host Defense
The intensive care units P. aeruginosa infections are high risk due to the treatment of antibiotics and nosocomial lung infections. This infection is caused due to antibiotic-associated secondary IgA deficiency; treatment of IgA through the nasal route would serve as antibacterial therapy against P. aeruginosa infected patients [86,87]. Since all these therapies have one or the other side effects and disadvantages, using naturally available herbal medicine would reduce the risk of side effects and provide us better results and health stability [88-90].
4. CONCLUSION
P. aeruginosa is an opportunistic human pathogen that has evolved with multiple mechanisms enabling them to grow, survive, and infect its host with a higher success ratio. Further, it has also developed escape strategies at a molecular level such as secretion systems, oxidative cascades, advanced metabolic networking, intelligent, and alternative regulatory master transcriptional factors which will facilitate its holistic survival within and outside the host cell. Beyond this, it has developed effective quorumone molecules for cell-density-dependent signaling and microcolony trafficking. The exopolymeric substance provides a shielding effect against antibiotics, changing microenvironments, and potential anti-psuedomonal agents. Nanotechnology, green chemistry, synthetic biology, and genome editing technologies have paved the way to explore the possibilities of phytomolecules which can lead to newer drug design and development. The available research updates suffer from a major lacuna in explaining the mechanistic strategies of P. aeruginosa pathogenicity. Hence, a complete revamping of the current methods should be re-visited to understand this micro monster and its various transitional forms (planktonic and biofilm) equipped with nanotools which would ultimately lead to early detection and diagnosis leading to better health and hygiene.
5. ACKNOWLEDGMENTS
Mr. Avinash MG would like to thank the Indian Council of Medical Research (ICMR) for the award of senior research fellow Award Letter Number - File no. AMR/Fellowship/17/2019 ECD-II dated June 28, 2019, and the University of Mysore for the smooth execution of the work. The authors are thankful to Dr. Nagendra Prasad MN, Department of Biotechnology, JSS University, SJCE Campus, Mysore, Dr. Shaukath Ara Khanum, Department of Chemistry, Yuvaraja College, University of Mysore, Mysore, Dr. Farhan Zameer would like to place on records to extend gratitude towards Prof. Sunil S. More, Dean, School of Basic and Applied Sciences for long-term collaboration in understanding the bacterial microenvironment and chemico-biology of phytobioactives. All authors thank the Principal of Alva’s Ayurveda Medical College, the Director of ATMA Research Center, and the Management of alva’s education foundation for their continuous support and encouragement.
6. CONFLICT OF INTEREST
The authors express no conflict of interest.
7. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. FUNDING
Funding was provided to Mr. Avinash M.G. from the Indian Council of Medical Research (ICMR) as Senior Research Fellow (SRF) vide Award Letter Number - File no. AMR/Fellowship/17/2019 ECD-II dated June 28, 2019-2022.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
Upon request to the Corresponding author, data could be made available for non-commercial purposes only.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Chevalier S, Bouffartigues E, Bodilis J, Maillot O, Lesouhaitier O, Feuilloley MG,
2. Parkins MD, Somayaji R, Waters VJ. Epidemiology, biology, and impact of clonal
3. Farhan Z, Shubha G. Transcriptome analysis of thiol-disulfide redox metabolism genes in
4. Ali SG, Ansari MA, Khan HM, Jalal M, Mahdi AA, Cameotra SS.
5. Shubha G, Vanishree S, Farhan Z, Juergen K. Prediction of proteins putatively involved in the thiol:Disulfide redox metabolism of a bacterium (
6. Zameer F, Kreft J, Gopal S. Interaction of the dual species biofilms of
7. Goo E, An JH, Kang Y, Hwang I. Control of bacterial metabolism by quorum sensing. Trends Microbiol 2015;23:567-76. [CrossRef]
8. Mielko KA, Jab?o?ski SJ, Milczewska J, Sands D, ?ukaszewicz M, M?ynarz P. Metabolomic studies of
9. Diggle SP, Cornelis P, Williams P, Cámara M. 4-quinolone signaling in
10. Halebeedu PP, Vijay Kumar GS, Gopal S. Revamping the role of biofilm regulating operons in device-associated
11. Zameer F, Ms R, Chauhan JB, Khanum SA, Kumar P, Devi AT. Evaluation of adhesive and anti-adhesive properties of
12. Schmaljohn AL, McClain D, Baron S. Alphaviruses (
13. Avinash MG, Zameer F, Gopal S. The propensity of selected Indian plant extracts for polyphenolics, antioxidant, and inhibition of
14. Ashwini P, Sumana MN, Shilpa U, Mamatha P, Manasa P, Dhananjaya BL,
15. Abrusci P, McDowell MA, Lea SM, Johnson S. Building a secreting nanomachine:A structural overview of the T3SS. Curr Opin Struct Biol 2014;25:111-7. [CrossRef]
16. Green ER, Mecsas J. Bacterial secretion systems:An overview. Microbiol Spectr 2016;4:213-39. [CrossRef]
17. Prasad A, Devi AT, Prasad MN, Zameer F, Shruthi G, Shivamallu C. Phyto anti-biofilm elicitors as potential inhibitors of
18. Amiel E, Lovewell RR, O'Toole GA, Hogan DA, Berwin B.
19. Onsare JG, Arora DS. Antibiofilm potential of flavonoids extracted from
20. Jennings LK, Storek KM, Ledvina HE, Coulon C, Marmont LS, Sadovskaya I,
21. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol 2010;8:623-33. [CrossRef]
22. Strempel N, Neidig A, Nusser M, Geffers R, Vieillard J, Lesouhaitier O,
23. Moradali MF, Ghods S, Rehm BH.
24. LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev 2013;77:73-111. [CrossRef]
25. Grote J, Krysciak D, Streit WR. Phenotypic heterogeneity, a phenomenon that may explain why quorum sensing does not always result in truly homogenous cell behavior. Appl Enviroin Microbiol 2015;81:5280-9. [CrossRef]
26. Barr HL, Halliday N, Cámara M, Barrett DA, Williams P, Forrester DL,
27. Alcalde-Rico M, Hernando-Amado S, Blanco P, Martínez JL. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol 2016;7:1483. [CrossRef]
28. Feng L, Xiang Q, Ai Q, Wang Z, Zhang Y, Lu Q. Effects of quorum sensing systems on regulatory T cells in catheter-related
29. García-García JD, Sánchez-Thomas R, Moreno-Sánchez R. Bio-recovery of non-essential heavy metals by intra-and extracellular mechanisms in free-living microorganisms. Biotech Adv 2016;34:859-73. [CrossRef]
30. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ,
31. Bleves S, Gérard-Vincent M, Lazdunski A, Filloux A. Structure-function analysis of XcpP, a component involved in general secretory pathway-dependent protein secretion in
32. Yahr TL, Vallis AJ, Hancock MK, Barbieri JT, Frank DW. ExoY, an adenylatecyclase secreted by the
33. Poulsen BE, Yang R, Clatworthy AE, White T, Osmulski SJ, Li L,
34. Sato H, Frank DW. ExoU is a potent intracellular phospholipase. Mol Microbiol 2004;53:1279-90. [CrossRef]
35. Egan SM. Growing repertoire of AraC/XylS activators. J Bacteriol 2002;184:5529-32. [CrossRef]
36. Yahr TL, Wolfgang MC. Transcriptional regulation of the
37. Domínguez-Cuevas P, Marqués S. Compiling sigma-70-dependent promoters. In:Virulence and Gene Regulation. Boston, MA:Springer;2004. 319-43. [CrossRef]
38. Brutinel ED, Vakulskas CA, Brady KM, Yahr TL. Characterization of ExsA and of ExsA-dependent promoters required for expression of the
39. Vakulskas CA, Brady KM, Yahr TL. Mechanism of transcriptional activation by
40. Pastor A, Chabert J, Louwagie M, Garin J, Attree I. PscF is a major component of the
41. Galle M, Carpentier I, Beyaert R. Structure and function of the Type III secretion system of
42. Alhazmi A.
43. Kim J, Ahn K, Min S, Jia J, Ha U, Wu D,
44. Yamazaki A, Li J, Zeng Q, Khokhani D, Hutchins WC, Yost AC,
45. McMackin EA, Djapgne L, Corley JM, Yahr TL. Fitting pieces into the puzzle of
46. Lai S, Tremblay J, Déziel E. Swarming motility:A multicellular behaviour conferring antimicrobial resistance. Environ Microbiol 2009;11:126-36. [CrossRef]
47. Balasubramanian D, Schneper L, Kumari H, Mathee K. A dynamic and intricate regulatory network determines
48. Jia J, Wang Y, Zhou L, Jin S. Expression of
49. O'May C, Tufenkji N. The swarming motility of
50. Deng Y, Boon C, Chen S, Lim A, Zhang LH. Cis-2-dodecenoic acid signal modulates virulence of
51. Ma XN, Xie CL, Miao Z, Yang Q, Yang XW. An overview of chemical constituents from
52. Honmore VS, Rojatkar SR, Nawale LU, Arkile MA, Khedkar VM, Natu AD,
53. An N, Zou ZM, Tian Z, Luo XZ, Yang SL, Xu LZ. Diarylheptanoids from the rhizomes of
54. Shin D, Kinoshita K, Koyama K, Takahashi K. Antiemetic principles of
55. Pankaj S, Aishwarya S, Rashmi MS, Akshaya SN, Dhanapal G, Aishwarya SR,
56. Lakshmanan D, Harikrishnan A, Jyoti K, Ali MI, Jeevaratnam K. A compound isolated from
57. Frank DW. The exoenzyme S regulon of
58. Devi AT, Yashaswini N, Zameer F, Prasad MN. Experimental Urolithiasis Model to Assess Phyto-fractions as Anti-lithiatic Contributors:A Herbaceutical Approach. bioRxiv;2021. [CrossRef]
59. Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by
60. Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. The multiple signaling systems regulating virulence in
61. Sadlon AE, Lamson DW. Immune-modifying and antimicrobial effects of Eucalyptus oil and simple inhalation devices. Alter Med Rev 2010;15:33-43.
62. Gabrielian ES, Shukarian AK, Goukasova GI, Chandanian GL, Panossian AG, Wikman G,
63. Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in
64. Banerjee M, Moulick S, Bhattacharya KK, Parai D, Chattopadhyay S, Mukherjee SK. Attenuation of
65. Mashhady MA, Abkhoo J, Jahani S, Abyar S, Khosravani F. Inhibitory effects of plant extracts on
66. Das MC, Sandhu P, Gupta P, Rudrapaul P, De UC, Tribedi P,
67. Quave CL, Plano LR, Pantuso T, Bennett BC. Effects of extracts from Italian medicinal plants on planktonic growth, biofilm formation and adherence of methicillin-resistant
68. Javidanpour S, Tabtabaei SR, Siahpoosh A, Morovati H, Shahriari A. Comparison of the effects of fresh leaf and peel extracts of walnut (
69. Beulah KC, Aishwarya TD, Kamran W, Meghashri S, Raghavendra H, Nagendra PM,
70. Dolatabadi S, Moghadam HN, Mahdavi-Ourtakand M. Evaluating the anti-biofilm and antibacterial effects of
71. Van Duin D, Bonomo RA. Ceftazidime/avibactam and ceftolozane/tazobactam:Second-generation b-lactam/b-lactamase inhibitor combinations. Clin Inf Dis 2016;63:234-41. [CrossRef]
72. Zhanel GG, Lawrence CK, Adam H, Schweizer F, Zelenitsky S, Zhanel M,
73. Thomson KS, Ghani SA, Snyder JW, Thomson GK. Activity of cefepime-zidebactam against multidrug-resistant (MDR) Gram-negative pathogens. Antibiotics 2019;8:32. [CrossRef]
74. Okujava R, Garcia-Alcalde F, Haldimann A, Zampaloni C, Morrissey I, Magnet S,
75. Horner C, Mushtaq S, Livermore DM. BSAC Resistance Surveillance Standing Committee. Potentiation of imipenem by relebactam for
76. Hsueh SC, Lee YJ, Huang YT, Liao CH, Tsuji M, Hsueh PR.
77. Tulkens PM, van Bambeke F, Zinner SH. Profile of a novel anionic fluoroquinolone-delafloxacin. Clin Infect Dis 2019;68(Suppl 3):S213-22. [CrossRef]
78. Chotirmall SH, Chalmers JD. RESPIRE:Breathing new life into bronchiectasis. Eur Respir J 2018;51:1702444. [CrossRef]
79. Bumann D, Behre C, Behre K, Herz S, Gewecke B, Gessner JE,
80. Satapathy P, Khan K, Devi AT, Patil AG, Govindaraju AM, Gopal S,
81. Kutateladze M, Adamia R. Phage therapy experience at the Eliava Institute. Méd Mal Infect 2008;38:426-30. [CrossRef]
82. Liu L, Liu B, Li Y, Zhang W. Successful control of resistance in
83. Döring G. Prevention of
84. Wiehlmann L, Cramer N, Ulrich J, Hedtfeld S, Weißbrodt H, Tümmler B. Effective prevention of
85. Saiman L, Siegel JD, LiPuma JJ, Brown RF, Bryson EA, Chambers MJ,
86. Robak OH, Heimesaat MM, Kruglov AA, Prepens S, Ninnemann J, Gutbier B,
87. Tümmler B. Emerging therapies against infections with
88. Satapathy P, Prakash JK, More SS, Chandramohan V, Zameer F. Structural modulation of dual oxidase (Duox) in
89. Satapathy P, Prakash JK, Gowda VC, More SS, Muthuchelian K, Chandramohan V,
90. Patil AG, Prakash JK, More SS, Chandramohan V, Zameer F. Exploring banana phytosterols (Beta-sitosterol) on tight junction protein (claudin) as anti-urolithiasis contributor in
91. Khan K, Sree A, Satapathy P, Patil AG, More SS, Zameer F. Exploration of dill seeds (
92. Aishwarya S, Kounaina K, Patil AG, Satapathy P, Devi AT, Avinash MG,