Pseudomonas aeruginosa has emerged as a serious nosocomial threat with a high morbidity and mortality rate, especially in immunocompromised patients. Its pathogenicity is attributed to the virulence modulating – quorum sensing system. The coexistence of Chlorella emersonii KJ725233, a novel microalga with P. aeruginosa, pointed to its subterfuge to alter the pathogenicity of its partners. The study was, thus, aimed at determining the ability of CEK to modulate the pathogenesis of the aggressor P. aeruginosa. With a MIC of 0.5 mg mL−1, the methanolic extract of C. emersonii KJ725233 was able to inhibit the synthesis of pyocyanin (62.48 ± 1.11%), protease (84.72 ± 1.11%), elastase (73.47 ± 0.11%), pyochelin (69.95 ± 7.12%), rhamnolipid (86.76 ± 0.48%), and polysaccharide (44.72 ± 1.58%) which are established virulence factors of P. aeruginosa. RT-PCR studies indicated the downregulation of its quorum sensing genes, lasI (84.63 ± 5.98%), lasR (85.56 ± 3.45%), rhlI (88.33 ± 3.56%), rhlR (88.73 ± 2.91%), and pqsA (72.61 ± 1.91%) which are known to play a crucial role in the pathogenesis of the organism. The presence of phytol in the methanolic extracts was indicated by its GC-HRMS analysis. With an ability to effectively incapacitate the virulence system of P. aeruginosa, C. emersonii KJ725233 presents itself as an efficient disarming agent with a potential use in pharmaceutical formulations.
Desai SS, Singh RD, Ghosh SB, Kelkar V. A novel strategy for disarming quorum sensing in Pseudomonas aeruginosa-Chlorella emersonii KJ725233. J App Biol Biotech. 2020;8(05):78-83. DOI: https://dx.doi.org/10.7324/JABB.2020.80510.
Pseudomonas aeruginosa, an opportunistic pathogen known to readily develop antibiotic resistance, has been widely implicated in nosocomial infections [1]. The CDC ESKAPE lists this as a priority pathogen not only due its clinical relevance in various disease and conditions such as cystic fibrosis, cancer, immunocompromised individuals, burns, and implanted medical devices [2,3] but also due to its association with high morbidity and mortality rates in such individuals [4]. P. aeruginosa is known to regulate its pathogenicity using the cell to cell communication called quorum sensing (QS) [1]. QS permits these bacteria to scrutinize their cell population through the release of signaling molecules called autoinducers [5] and establish infection.
QS systems in P. aeruginosa are systematized hierarchically with las system consisting of LasI and LasR as the synthase and regulator, respectively, at the top [6,7]. It uses N-3-(oxo-dodecanoyl)-L-homoserine lactone (OdDHL) as a signaling molecule and further regulates the rhl and the pqs systems. The rhl system consists of RhlI and RhlR as the synthase and regulator, respectively. It uses N-butanoylhomoserine (BHL) lactone as a signaling molecule, whereas the pqs system uses 2-heptyl-3-hydroxy-4-quinolone (PQS) as a signaling molecule [7]. During bacterial growth, these autoinducers (OdDHL, BHL, and PQS) accrue in the culture environment till it attains the threshold concentration that instigates the signaling necessary for the regulation of virulence genes expression [5]. QS has, thus, become a primary target in therapeutics [8].
At the source, freshwater microalga is known to exist in symbiotic relationship with the bacterial inhabitants. C. emersonii KJ725233 is one such microalga that coexisted with bacterial population in an artificial pond in the Western regions of Maharashtra. It has been isolated, identified by 18s rDNA sequencing, and has exhibited the presence of phenolics such as flavonoids [9,10]. The present study is one of the first to report the anti-quorum sensing potential of a microalga against P. aeruginosa. The methanolic extract of C. emersonii KJ725233 has displayed anti-pseudomonas activity [9] making it worthwhile to determine its anti-quorum sensing potential. The study for the 1st time reports the downregulation of quorum sensing system of the priority pathogen P. aeruginosa by any microalga so far to the best of our knowledge.
The present study was undertaken after identifying coexistence of C. emersonii KJ725233 with P. aeruginosa. This observation necessitated the investigation of the defensive stratagem of this microalga with pathogenic bacterial neighbors.
C. emersonii KJ725233 was isolated from Western regions of Maharashtra, India [10]. C. emersonii KJ725233 was mass cultured in BG-11 media with 12h:12 h light:dark conditions and with 12 h of photoperiod for a period of 30 days.
After 30 days of incubation, the biomass was harvested by centrifugation at 5000 rpm for 20 min and dried at 60°C for a period of 24 h. The extract was prepared by sonicating (LABMAN sonicator – Digital Ultrasonic Cleaner LMUC Series, India) dried biomass in methanol at a concentration of 0.1 g mL−1 for 40 min. These suspensions were centrifuged at 5000 rpm for 20 min and the supernatant was transferred to a crucible. The extraction was repeated thrice and all the supernatants were pooled together. These were dried at 28 ± 1°C and reconstituted in dimethyl sulfoxide at a concentration of 5 mg mL−1.
The antimicrobial potential of C. emersonii KJ725233 against P. aeruginosa MTCC1688 was determined by agar well diffusion method [9]. The MIC of the methanolic extract was determined by broth microdilution method [11] using an inoculum density of 1*104 cells mL−1. The methanolic extract was used at concentrations from 0.1 to 1 mg mL−1, whereas quercetin was used as a positive control at concentrations of 2–20 μg mL−1. To determine whether the MIC was bactericidal or bacteriostatic, the culture from both the MIC tube of the extract and quercetin was plated on sterile LB agar plates.
A 3 mL of 1*106 cells mL−1 were inoculated with varying concentrations, namely, 0.042, 0.083, 0.125, 0.166, and 0.333 mg mL−1 of the methanolic extract and 3.33 μg mL−1 of quercetin in glycerol supplemented nutrient broth medium [glycerol (3%), meat extract (0.1%), yeast extract (0.2%), peptone (0.5%), and sodium chloride (0.5%)] for pyocyanin inhibition assay, CYKN [casein (1%), dipotassium dihydrogen phosphate (0.2%), yeast extract (0.5%), and sodium chloride (0.5%)] for protease and elastase inhibition assay; LB broth for pyochelin inhibition and in tryptone soy broth [tryptone (1.7%), peptone (0.15%), sodium chloride (0.5%), dipotassium hydrogen phosphate (0.25%), dextrose (0.25%), and malt extract (0.15%)] for polysaccharide and biofilm inhibition. These were incubated at 37°C for 24 h. At the end of incubation, the supernatant was obtained by centrifugation at 5000 rpm for 20 min and processed as earlier described for pyocyanin [12-14], protease [14-17], elastase [18], rhamnolipid [19], pyochelin [20,21], polysaccharide [22], and biofilm formation [23,24]. Alterations in quorum sensing gene expression of P. aeruginosa KJ725233 by quantitative real-time polymerase chain reaction (qRT-PCR)
To determine the alterations in the quorum sensing genes of P. aeruginosa MTCC1688, 1*106 cell mL−1 were incubated with 1 mg mL−1 of CEME at 37°C for 24 h. The total RNA was extracted using TRI reagent (Sigma, USA) by following the manufacturer’s protocol. cDNA was synthesized from RNA using Thermo Fisher Scientific – RevertAid First Strand cDNA synthesis kit. The PCR reaction was performed on LightCycler® 480 Instrument II – Roche (Switzerland) using LightCycler® 480 SYBR Green I master mix. PCR conditions employed were –94°C for 5 min for 1 cycle, 40 cycles at 94°C for 30 s, 59°C for 30 s, and 72°C for 30 s using primers mentioned in Table 1. The primers were designed using Pseudomonas genome database (https://www.pseudomonas.com/) and Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi). RpoD was used as a housekeeping gene. Melting curves analysis was performed at the end of amplification to confirm data quality. Fold change in target gene expression was translated into critical threshold cycle (CT) calculated by delta-delta CT algorithms [25].
Table 1: Primer sequences for RT-PCR
| Primer | Sequences (5’–3’) | Amplicon size (bp) |
|---|---|---|
| RpoD forward primer | CGATCGGTGACGACGAAGAT | 176 |
| RpoD reverse primer | GTTCATGTCGATGCCGAAGC | |
| LasI forward primer | GCGCGAAGAGTTCGATAAAA | 68 |
| LasI reverse primer | GCTCCTTGAACACTTGAGCA | |
| LasR forward primer | CTGTGGATGCTCAAGGACTAC | 210 |
| LasR reverse primer | ACCGAACTTCCGCCGAAT | |
| RhlI forward primer | GCTCTCTGAATCGCTGGAAG | 153 |
| RhlI reverse primer | GTTTGCGGATGGTCGAACTG | |
| RhlR forward primer | TCGGAAATGGTGGTCTGGAG | 219 |
| RhlR reverse primer | GGTCAGCAACTCGATCATGC | |
| PqsA forward primer | CCGGACCTACATTCTCTCCC | 156 |
| PqsA reverse primer | GACTTGGGATTGATCACGGC |
GC-HRMS analysis was carried out using GC (Agilent Technologies, USA) equipped with Accutoff MS with HP-5 MS capillary column 30 m in length with an internal diameter of 0.32 mm and film thickness of 0.25 μm containing 5% phenyl polysiloxane [9].
All the experiments were performed in triplicates. Data were analyzed by one-way ANOVA and are presented as mean±SD. P < 0.05 was considered less significant.
The antibacterial activity of the methanolic extract of C. emersonii KJ725233 against P. aeruginosa MTCC1688 is reported earlier [9]. To determine the MIC of the methanolic extract, broth dilution method was used and the MIC for the methanolic extract of C. emersonii KJ725233 was found to be 0.5 mg mL-1 whereas that of quercetin was 6 μg mL−1 [Figure 1]. However, the MIC of the methanolic extract (0.5 mg mL−1) was found to be bacteriostatic whereas that of quercetin (6 μg mL−1) was found to be bactericidal in nature.
.png) | Figure 1: Determination of minimum inhibitory concentration of A. Methanolic extract of Chlorella emersonii KJ725233 (0–1 mg mL−1); B. Quercetin (0–20 μg mL−1) against Pseudomonas aeruginosa MTCC 1688 [Click here to view] |
The present study dealt with the spectrophotometric determination of inhibition of P. aeruginosa virulence factors, namely, pyocyanin, protease, elastase, pyochelin, rhamnolipid, polysaccharides, and biofilm production. CEME at 0.33 mg mL−1 exhibited 62.48 ± 1.11% pyocyanin inhibition, 84.72 ± 1.11% protease inhibition, 73.47 ± 0.11% elastase inhibition, 69.95 ± 7.12% pyochelin inhibition, 86.76 ± 0.48% rhamnolipid inhibition, 44.72 ± 1.58% polysaccharide inhibition, and 83.77 ± 6.48% biofilm inhibition. On the other hand, quercetin at 3.33 μg mL−1 exhibited 42.72 ± 2.04% pyocyanin inhibition, 59.05 ± 3.33% protease inhibition, 30.12 ± 1.81% elastase inhibition, 67.88 ± 7.83% pyochelin inhibition, 50.62 ± 2.20% rhamnolipid inhibition, 28.57 ± 2.58% polysaccharide inhibition, and 26.86 ± 0.52% biofilm inhibition [Figure 2].
 | Figure 2: Percent inhibition of the virulence factors of Pseudomonas aeruginosa MTCC1688 by varying concentrations of the 0.042–0.33 mg mL−1 methanolic extract and quercetin (3.33 μg mL−1) [Click here to view] |
The relative expression of the target genes of P. aeruginosa was determined by comparing them with the reference rpoD gene. Relative expression levels of these genes in P. aeruginosa treated with CEME were compared with that of the untreated P. aeruginosa and expressed in terms of fold change by 2-??Ct method. Percent reduction in gene expression was determined by 1-FC*100 [26] [Figure 3]. Although CEME was used at a concentration 100 folds higher than that of quercetin; quercetin was found to affect las, rhl, and pqs system 13.93–27.03 times more as compared to CEME [Figure 3].
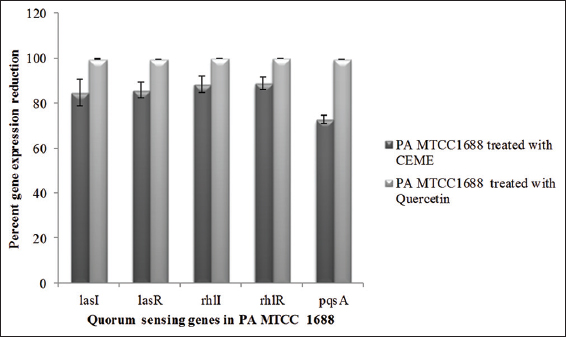 | Figure 3: Percent reduction in expression of quorum sensing genes of Pseudomonas aeruginosa MTCC1688 on treatment with methanolic extract of CEK (0.33 mg mL−1) and with quercetin (3.33 μg mL−1) [Click here to view] |
GC-HRMS analysis enabled the identification of the bioactives present in the methanolic extract of C. emersonii KJ725233. Phytol was one of the major compounds identified at 21.51 min with a peak area of 64.77%. In addition to phytol, 1-docosene, hexadecane, bis-cyclohexylidene-2-oxocyclohexyl methane, pentadecanoic acid, 14-methyl, methyl ester, and 9,12-octadecadienoic acid methyl ester were also identified in CEME [9].
A serious concern about P. aeruginosa infections is the higher mortality rate in immunocompromised patients (70–80%) compared to other bacterial infections such as Staphylococcus or other Gram-negative bacteria [27]. The ability of pathogenic bacteria such as P. aeruginosa to induce a diseased condition depends on its potential to synthesize agents such as toxins and adhesion molecules collectively termed as “virulence factors” that violently destroy the host immune system [28]. P. aeruginosa possesses a multitude of virulence factors such as pyocyanin, proteases, siderophores, rhamnolipids, polysaccharides, and biofilm [29]. Pyocyanin acts as a virulence factor primarily because of its ability to generate ROS by oxidizing glutathione with simultaneous reduction of oxygen as well as by interfering in the electron transport by accepting an electron from NAD or NADP and transferring it to oxygen [12,30,31]. It is produced by different strains of P. aeruginosa responsible for pulmonary and extrapulmonary infections as well as in environmental strains. It promotes P. aeruginosa colonization in the respiratory tract by significantly affecting tracheobronchial mucous velocity and hence can be found at concentrations as high as 100 μM in the lungs of cystic fibrosis patients [32,33]. P. aeruginosa subspecies secrete an array of proteases such as protease IV, alkaline protease, LasA protease, and LasB elastase that target the host proteins such as plasminogen, fibrinogen, α1-proteinase inhibitor, cytokine, and complement factors. Pyocyanin is known to disable protease inhibitors, thereby affecting the protease-antiprotease balance subsequently resulting in lung epithelial injury. Siderophores, further, aid in iron acquisition that ultimately helps the bacteria in colonization of the host. They are known to induce oxidative damage with a continual inflammatory response [34-36]. Rhamnolipids, on the other hand, are known to play a role in host immune cell and erythrocyte destruction while imparting twitching, swarming motility, and inducing biofilm formation [37]. They are known to inactivate tracheal cilia in the mammalian cell and are found in the sputa of the cystic fibrosis patients indicating their role in virulence [38]. Polysaccharides aid in attachment of the bacterial cells to the surface enabling it to develop thick antibiotic-resistant biofilms and provide protection against dehydration, antibiotics, and predators [39]. Biofilms known to cause chronic infections are structured consortium of bacterium embedded in matrix consisting of polysaccharide, protein, and extracellular DNA. Alginate forms a major part of the biofilms in cystic fibrosis patients. These biofilms, therefore, need to be prevented to avoid the chronic inflammation caused due to a lung tissue damage [40]. The expression of genes responsible for the synthesis of such virulence factors is controlled by the quorum sensing signal molecules regulated by the quorum sensing systems [41]. Antibacterial strategies are thus directed against these virulence factors with an aim to disarm the pathogenicity of the bacteria, thereby allowing the immune system with an improved chance of lessening the severity of infection [28].
Algae are reported to synthesize an array of compounds that are non-antibiotic quorum quenchers that interfere and disrupt the bacterial quorum sensing [31,42-44]. Phenolics such as flavonoids and tannins are widely reported for their quorum quenching potential [45]. Teplitski et al., 2004, have earlier reported the synthesis of quorum sensing mimic compounds in Chlamydomonas mutabilis, Chlorella vulgaris, and Chlorella fusca that regulate luminescence in Vibrio harveyi [44]. In addition, Natrah et al., 2011, have reported that the extracts of freshwater Chlorella saccharophila CCAP211/48 inhibit violacein production in Chromobacterium violaceum as well as interfere with the bioluminescence in Vibrio harveyi [43]. The halogenated furanones from red alga Delisea pulchra have been reported to inhibit biofilm formation by targeting the las and rhl systems [46].
The presence of phenolics and flavonoids in C. emersonii KJ725233 as described earlier [9] may have contributed to its anti-quorum sensing potential in terms of pyocyanin, elastase, protease, pyochelin, rhamnolipid, polysaccharide, and biofilm formation inhibition as seen in the present study. Phytol a quorum sensing inhibitor reported to inhibit biofilm formation, pyocyanin production, and motility of P. aeruginosa was identified in the methanolic extract of C. emersonii KJ725233 [9,47]. The quorum sensing inhibitors present in the methanolic extract of C. emersonii KJ725233 were found to downregulate all the three QS systems [Figure 3]. In addition, the oxidative stress reportedly induced by pyocyanin can also be counteracted by the strong antioxidant potential exhibited by the methanolic extract of C. emersonii KJ725233 [9]. However, though methanolic extract of C. emersonii KJ725233 was used at a concentration 100 times higher than that of quercetin; quercetin was found to affect las, rhl, and pqs system 13.93–27.03 folds more. The downregulation of the lasI and rhl genes of P. aeruginosa by natural compounds such as quercetin, halogenated furanones, baicalin, and terpinen-4-ol either alone or in combination with an antibiotic has only been reported so far [48-51].
C. emersonii KJ725233 interferes with both the AHL and the PQS quorum sensing systems in P. aeruginosa. Since these QS systems regulate virulence in P. aeruginosa, C. emersonii KJ725233 can prove to be a potential biocontrol agent that aids in disarming the pathogenicity of such a virulent microbe. It exhibits itself as a probable candidate for use as an additive in an anti-virulence drug formulation which, however, requires in vivo evaluation. Since there are more than 30,000 identified microalgal species with over 15,000 bioactives compounds, the study, thus, sets the podium to unravel another reservoir of pharmaceutically significant quorum sensing disarmer from the sea of untapped microalgal species.
Authors declared that there are no conflicts of interests.
None.
1. Maisuria VB, Santos YL, Tufenkji N, Deziel E. Cranberry-derived proanthocyanidins impair virulence and inhibit quorum sensing of Pseudomonas aeruginosa. Sci Rep 2016;6:1-12. [CrossRef]
2. Paczkowski JE, Mukherjee S, McCready AR, Cong JP, Aquino CJ, Kim H, et al. Flavonoids suppress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J Biol Chem 2017;292:4064-76. [CrossRef]
3. Aboushieb HM, Omar HM, Abozahra R, Eisheredy A, Baraka K. Correlation of quorum sensing and virulence factors in Pseudomonas aeruginosa isolates in Egypt. J Infect Dev Ctries 2015;9:1091-9. [CrossRef]
4. Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 2003;22:3803-15. [CrossRef]
5. Jaramillo-Colorado B, Olivera-Verbel J, Stashenko EE, Wagner-Dobler I, Kunze B. Anti-quorum sensing activity of essential oils from colombian plants. Nat Prod Res 2012;26:1075-86. [CrossRef]
6. Kim B, Park JS, Choi HY, Kwak JH, Kim WG. Differential effects of alkyl gallates on quorum sensing in Pseudomonas aeruginosa. Sci Rep 2019;9:7741. [CrossRef]
7. Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015;6:26-41. [CrossRef]
8. Hurley MN, Camara M, Smyth AR. Novel approaches to the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. Eur Respir J 2012;40:1014-23. [CrossRef]
9. Sawant SS, Kelkar-Mane V. Nutritional profile, antioxidant, antimicrobial potential, and bioactives profile of Chlorella emersonii KJ725233. Asian J Pharm Clin Res 2018;11:220-5. [CrossRef]
10. Sawant SS, Joshi AA, Bhagwat AM, Kelkar-Mane V. Tapping the antioxidant potential of a novel isolate-Chlorella emersonii. World J Pharm Res 2014;3:726-39.
11. Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimum inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 2008;3:163-5. [CrossRef]
12. El-Fouly MZ, Sharaf AM, Shahin AA, El-Bialy HA, Omara AM. Biosynthesis of pyocyanin pigment by Pseudomonas aeruginosa. J Radiat Res Appl Sci 2015;8:36-48. [CrossRef]
13. Tan LY, Yong WF, Chan KG. Silencing quorum sensing through extracts of Melicope lunu-ankenda. Sensors 2012;12:4339-51. [CrossRef]
14. Rahman PK, Pasirayi G, Auger V, Ali Z. Production of rhamnolipid biosurfactants by Pseudomonas aeruginosa DS10-129 in a microfluidic bioreactor. Biotechnol Appl Biochem 2010;55:45-52. [CrossRef]
15. Iiyama K, Takahashi E, Lee JM, Mon H, Morishita M, Kusakabe T, et al. Alkaline protease contributes to pyocyanin production in Pseudomonas aeruginosa. FEMS Microbiol Lett 2017;364:1-7. [CrossRef]
16. Côelho DF, Saturnino TP, Fernandes FF, Mazzola PG, Silveira E, Tambourgi EB. Azocasein substrate for determination of proteolytic activity: Reexamining a traditional method using bromelain samples. Biomed Res Int 2016;2016:1-7. [CrossRef]
17. Najafi MF, Deobagkar D, Deobagkar D. Potential application of protease isolated from Pseudomonas aeruginosa pd100. Electron J Biotechnol 2005;8:197-203. [CrossRef]
18. Galdino AC, Branquinha MH, Santos AL, Viganor L. Pseudomonas aeruginosa and its arsenal of proteases: Weapons to battle the host. In: Chakraborti S, Dhalla NS, editors. Pathophysiological Aspects of Proteases. Singapore: Springer; 2017. p. 381-97. [CrossRef]
19. Tahzibi A, Kamal F, Assadi MM. Improved production of rhamnolipids by a Pseudomonas aeruginosa mutant. Iran Biomed J 2004;8:25-31.
20. Harjai K, Gupta RK, Sehgal H. Attenuation of quorum sensing controlled virulence of Pseudomonas aeruginosa by cranberry. Indian J Med Res 2014;139:446-53.
21. Arnow LE. Calorimetric determination of the components of 3, 4-dihydroxyphenylalanine tyrosine mixture. J Biol Chem 1937;118:531-8.
22. Gupta RK, Setia S, Harjai K. Expression of quorum sensing and virulence factors are interlinked in Pseudomonas aeruginosa: An in vitro approach. Am J Biomed Sci 2011;3:116-25. [CrossRef]
23. O'Toole GA. Microtiter dish biofilm formation assay. J Vis Exp 2011;47:2437. [CrossRef]
24. Perez LR, Barth AL. Biofilm production using distinct media and antimicrobial susceptibility profile of Pseudomonas aeruginosa. Braz J Infect Dis 2011;15:301-4. https://doi.org/10.1590/S1413-86702011000400001 [CrossRef]
25. Peng LY, Yuan M, Cui ZQ, Wu ZM, Yu ZJ, Song K, et al. Rutin inhibits quorum sensing, biofilm formation and virulence genes in avian pathogenic Escherichia coli. Microb Pathog 2018;119:54-9. [CrossRef]
26. Sarabhai S, Sharma P, Capalash N. Ellagic acid derivatives from Terminalia chebula Retz. Downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS One 2013;8:e53441. [CrossRef]
27. Choi Y, Paik JH, Kim JH, Han SB, Durey A. Clinical predictors of Pseudomonas aeruginosa bacterimeia in emergency department. Emerg Med Int 2018;2018:7581036. [CrossRef]
28. Morkunas B, Galloway WR, Wright M, Ibbeson BM, Hodgkinson JT, O'Connell KM, et al. Inhibition of the production of the Pseudomonas aeruginosa virulence factor pyocyanin in wild-type cells by quorum sensing autoinducer-mimics. Org Biomol Chem 2012;10:8452-64. [CrossRef]
29. Strateva T, Mitov I. Contribution of an arsenal of virulence factors to pathogenesis of Pseudomonas aeruginosa infections. Ann Microbiol 2011;61:717-32. [CrossRef]
30. Jazayeri JA, Nguyen K, Kotsanas D, Schneiders F, Tan CH, Jazayeri M, et al. Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from cystic fibrosis patients. J Med Microbiol Diagn 2016;5:1-6. [CrossRef]
31. Ganin H, Yardeni EH, Kolodkin-Gal I. Biofilms: Maintenance, development, and disassembly of bacterial communities are determined by QS cascades. In: Kalia VC, editor. Quorum Sensing Vs Quorum Quenching: A Battle With No End in Sight. India: Springer; 2015. p. 23-37. [CrossRef]
32. Castaneda-Tamez P, Ramirez-Peris J, Perez-Velazquez J, Kuttler C, Jalalimanesh A, Saucedo-Mora MA, et al. Pyocyanin restricts social cheating in Pseudomonas aeruginosa. Front Microbiol 2018;9:1348. [CrossRef]
33. Cheluvappa R, Shimmon R, Dawson M, Hilmer SN, Couteur DG. Reactions of Pseudomonas aeruginosa pyocyanin with reduced glutathione. Acta Biochim Pol 2008;55:571-80. [CrossRef]
34. Galdino AC, Viganor L, Ziccardi M, Nunes AP, Dos Santos KR, Branquinha MH, et al. Heterogeneous production of proteases from Brazilian clinical isolates of Pseudomonas aeruginosa. Enferm Infecc Microbiol Clin 2017;35:630-7. [CrossRef]
35. Wilson BR, Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y. Siderophores in iron metabolism: From mechanism to therapy potential. Trends Mol Med 2016;22:1077-90. [CrossRef]
36. Cornelis P, Dingermans J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol 2013;3:75. [CrossRef]
37. Laabei M, Jamieson WD, Lewis SE, Diggle SP, Jenkins AT. A new assay for rhamnolipid detection-important virulence factors of Pseudomonas aeruginosa. Appl Microbiol Biotechnol 2014;98:7199-209. [CrossRef]
38. Sharmaa S, Bahjat S. Detection of rhamnolipid production in Pseudomonas aeruginosa. J Phys Conf Ser 2019;1294:1-9. [CrossRef]
39. Irie Y, Roberts AE, Kragh KN, Gordon VD, Hutchinson J, Allen RJ, et al. The Pseudomonas aeruginosa PSL polysaccharide is a social but noncheatable trait in biofilms. mBio 2017;8:1-13. [CrossRef]
40. Hoiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol 2010;5:1663-74. [CrossRef]
41. Winzer K, Williams P. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int J Med Microbiol 2001;291:131-43. [CrossRef]
42. Osorno O, Castellanos L, Ramos FA, Arevalo-Ferro C. Gas chromathography as a tool in quorum sensing studies. In: Salih B, editor. Gas Chromatography-Biochemicals, Narcotics and Essential Oils. Croatia: InTech; 2012. p. 67-96. [CrossRef]
43. Natrah FM, Kenmegne MM, Wiyoto W, Sorgeloos P, Bossier P, Defoirdt T. Effects of micro-algae commonly used in aquaculture on acyl-homoserine lactone quorum sensing. Aquac 2011;317:53-7. [CrossRef]
44. Teplitski M. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol 2004;134:137-46. [CrossRef]
45. Kiran GS, Hassan S, Sajayan A, Selvin J. Quorum quenching compounds from natural sources. In: Sugathan S, Pradeep NS, Abdulhameed S, editors. Bioresource and Bioprocess in Biotechnology. Vol. 2. Exploring Potential Biomolecules. Singapore: Springer; 2017. p. 351-64. [CrossRef]
46. Manefield M, Nys R, Naresh K, Roger R, Givskov M, Peter S, et al. Evidence that halogenated furanones from Delisea pulchra inhibit acylated homoserine lactone (AHL)-mediated gene expression by displacing the AHL signal from its receptor protein. Microbiology 1999;145:283-91. [CrossRef]
47. Pejin B, Ciric A, Glamoclija J, Nikolic M, Sokovic M. In vitro anti-quorum sensing activity of phytol. Nat Prod Res 2015;29:374-7. [CrossRef]
48. Bose SK, Chauhan M, Dhingra N, Chhibber S, Harjai K. Terpinen-4-ol attenuates quorum sensing regulated virulence factors and biofilm formation in Pseudomonas aeruginosa. Future Microbiol 2020;15:127-42. [CrossRef]
49. Ahmed AK, Rudden M, Smyth T, Dooley JS, Marchant R, Banat IM. Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl Microbiol Biotechnol 2019;103:3521-35. [CrossRef]
50. Luo J, Dong B, Wang K, Cai S, Liu T, Cheng X, et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. Plos One 2017;12:e0176883. [CrossRef]
51. Ouyang J, Sun F, Feng W, Sun Y, Qiu X, Xiong L, et al. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J Appl Microbiol 2016;120:966-74. [CrossRef]
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Year
Month