1. INTRODUCTION
Antibiotics have been used continually by humans since their discovery and have become the most important part of the prescription. In the current situation, living without antibiotics cannot be imagined. Antibiotics restrict growth or completely kill the microorganisms; therefore, microorganisms have evolved various mechanisms to counter their lethal effects. The natural mechanism of acquiring resistance against antibiotics is known as antimicrobial resistance (AMR). AMR in humans depends on antibiotic prescription patterns, market dynamics, and patient illness conditions [1]. AMR has already become a severe public health problem, causing roughly 700,000 deaths globally each year; it is estimated that by the end of the year 2050, this figure might rise to 10 million [2]. Various classes of antibiotics, such as tetracyclines, aminoglycosides, fluoroquinolone, β-lactams, and carbapenems, are commonly prescribed to treat infections. Among these antibiotics, the β-lactam class of antibiotics is mainly used globally to treat infections [3]. The bacteria acquire AMR by producing enzyme beta-lactamases that dissolve the antibiotic’s beta-lactam ring, making them ineffective [4]. Overusing the drugs leads to the diversification of beta-lactamases, which hydrolyze antibiotics and render them ineffective. Multidrug-resistant (MDR) Gram-negative bacilli can be effectively treated using carbapenems, a class of antibiotics such as imipenem (IPM) and meropenem [5]. However, Pseudomonas aeruginosa and other bacteria have become resistant to them. These bacteria possess powerful beta-lactamases called metallo beta-lactamases (MBLs), which cause resistance. The MBLs can hydrolyze all beta-lactamases except monobactams [6]. More than twenty bacterial species, including Enterobacteriaceae, P. aeruginosa, Escherichia coli, Acinetobacter baumannii, and Klebsiella pneumoniae, are known to express MBLs [7]. Five major classes of genes-”TEM, SHV, CTX-M, OXA, and Extended-spectrum beta-lactamase enzymes (ESBL)” are synthesized beta-lactamase enzymes. Out of these, the OXA type has been mainly found in P. aeruginosa. The ESBLs are typically secreted by gram-negative bacilli, especially by the family Enterobacteriaceae. The leading causative agent in hospital and community-acquired infections is ESBL-producing Enterobacteriaceae; they cause infections such as bloodstream infections, burn infections, respiratory tract infections, urinary tract infections, wound infections, and many more [8]. Resistance to β-lactam antibiotics such as cephamycins, extended-spectrum cephalosporins, carbapenems, and monobactams is mainly caused by ampC-β-lactamases. AmpC-β-lactamases are distinguished from other β-lactamases by two characteristics: resistance to ESBL inhibitors such as clavulanate and the capacity to hydrolyze cephamycins such as cefoxitin and cefotetan [9,10].
Beta-lactamase enzymes such as MBL pose a major hazard to human health. However, trials are in progress, but no MBL inhibitors have been clinically approved [11]. MBL-encoding genes can be discovered on large transferable plasmids or linked to transposons, which allow them to proliferate quickly by allowing horizontal gene transfer between various genera and species. Based on the variations in the molecular structures of their proteins, the primary five kinds of MBL genes have been discovered thus far. These genes can be broadly categorized as IMP, SIM, VIM, GIM, and SPM [12]. SPM, GIM, and SIM have only been documented in some geographical regions, but IMP and VIM variations have been reported globally [13]. Although PCR-based methods are the industry standard for detecting MBL, they can only be used in research. Culture testing and phenotypic detection are still simple, affordable, and practical approaches for the routine detection of MBLs [14]. The above beta-lactamase enzymes play an important role in acquiring resistance to the drugs; these enzymes have become a serious threat to the whole human race. For successful therapy, it is essential to identify beta-lactamase enzyme-producing organisms early to properly prescribe antibiotics. The present study aimed to detect the prevalence of ESBL, MBL, and AmpC β-lactamase-producing P. aeruginosa in patients of a tertiary care hospital in Haryana, India.
2. MATERIALS AND METHODS
A prescriptive study was carried out in the Department of Microbiology, Postgraduate Institute of Medical Sciences Rohtak (Haryana) after obtaining approval from the Institutional Human Ethics Committee, vide letter no. BREC/21/67 dated July 20, 2021, and HEC/2021/298 dated September 09, 2021. Clinical samples like blood, urine, pus/wound swabs, pleural fluids (PF), bronchoalveolar lavage (BAL), high vaginal swabs (HVS), ascetic fluid, endotracheal aspirate (ETA), sputum, and drain samples were collected and examined as per clinical and laboratory standard institute (CLSI) guidelines. Blood samples along with brain heart infusion broth were collected and incubated for 24 h at 37°C before inoculation on Blood Agar (BA) and MacConkeyAgar (MA), while other samples were inoculated directly on MA and BA culture plates. These inoculated plates were incubated at 37°C for 24 h.
2.1. Morphological and Biochemical Examinations
After incubation, culture identification was done by their morphological characteristics. Gram staining and other biochemical tests such as motility, oxidase, catalase, hemolysin production, triple sugar iron agar, and indole tests were performed. BA was used to record hemolysis. MacConkey agar for lactose non-fermenters and nutrient agar were used to observe pyocyanin pigment production. P. aeruginosa was differentiated from other species using cetrimide agar as a selective media and growth at 42°C.
2.2. Matrix-assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) Analysis
After morphological and biochemical characterizations, all samples were re-characterized by MALDI-TOF MS. It is an advanced technique that identifies the bacterium based on their structural proteins with a confidence level of 99.9% (VITEK MS BIOMERIEUX, France). For the MALDI-TOF analysis, a portion of the isolated colony was picked using a 1 μL loop and smeared on a sample spot on a clean disposable VITEK MS-DS Target slide. Then, 1 μL of VITEK® MS-CHCA matrix was added to the center of the spot. The smear was allowed to dry completely. The same procedure was also followed for the control organism (E. coli ATCC® 8739™). A fully dried sample matrix was tested within 72 h of its preparation.
2.3. Phenotypic Detection of ESBL-Producing P. aeruginosa
ESBL enzyme-producing P. aeruginosa was detected using the double disc synergy test (DDST), as it is easy, economical, and requires no a specialized bacteriological laboratory [15]. CLSI also recommends this technique. The test was performed in two stages: the first stage was a screening test, and the second stage was a confirmatory test. Screening was done as described earlier. All P. aeruginosa isolates that were resistant to 3rd generation cephalosporins, i.e., ceftazidime (CAZ), were selected and processed to confirm ESBL production. After selecting CAZ-resistant isolates, confirmatory tests were performed by DDST following CLSI recommendations. In this method, a 0.5McFarland suspension was prepared, and streaking was done using a sterile cotton swab on MHA plates. After 15 min of streaking, pairs of antibiotics containing CAZ (30 μg) and CAZ/clavulanic acid (30/10 μg) or cefixime (30 μg) and cefixime/clavulanic acid (5/10 μg) were placed on MHA plates at a distance of 20 mm apart from each other. The clavulanic acid was used as an ESBL inhibitor. These plates were incubated at 37°C for 24 h. The criteria for ESBL producer inclusion was whether the zone of inhibition was equal to or more than 5mm in the disc containing clavulanic acid.
2.4. Phenotypic Detection of MBL-Producing P. aeruginosa
Phenotypic detection of MBL-producing P. aeruginosa was performed in two stages: the first stage was the screening of antibiotic susceptibility, and the second was a confirmatory test for MBL producers. Screening for antibiotic susceptibility was done as described in the previous section. Those cultures that showed IPM resistance were selected and processed further to confirm MBL production.
A confirmatory test was done using a combined disk synergy test following the method described by Fazeli et al. [16]. The selected IPM-resistant isolate was evenly spread on the MHA plate, and two discs of IPM (10 mcg) were placed 4–5 cm apart. One disc of IPM was added with 10 μL of 0.5M EDTA solution. Then, the plate was incubated at 37°C for 18 h, and the zone of inhibition was observed. Inclusion criteria for the MBL producer were if the IPM-EDTA disc’s zone of inhibition diameter was equal to or more than 7 mm.
2.5. Phenotypic Detection of AmpC β-Lactamase Production
The AmpC β-lactamase-producing P. aeruginosa was assessed as per the method described by Vanwynsberghe et al. [17] and Fazeli et al. [16]. This test involves two steps, i.e., a screening test and a confirmatory test. Screening for antibiotic susceptibility was done as described earlier, and cefoxitin-resistant isolates were selected and proceeded for further testing for AmpC β-lactamase production. Each disc of (CAZ; 30 μg) and cefotaxime (CTX; 30 μg) was placed 20mm apart from each other on an MHA plate evenly spread with P. aeruginosa, and then the plate was inoculated at 37°C [16,17].
2.6. Identification of Resistant Genes by Biofire FilmArray using Pneumonia Panel Plus-IVD
BioFire® FilmArray® pneumonia plus panels were used to identify infecting organism(s) present in the sputum and BAL samples following the manufacturer’s instructions. Briefly, 200 μL samples from positive blood cultures were collected and then lysed with 1000 μL sample buffer in a sample vial. The lysed sample vial was then injected into a pre-rehydrated (with Hydration Solution) pneumonia plus panel cartridge. The Biofire FilmArray system scanned the loaded cartridge to identify the culture.
3. RESULTS
A total of 163 P. aeruginosa species were isolated from the patient; 108 were male and 55 were female. Antibiotic susceptibility tests revealed that 78 isolates were resistant to CAZ, 65 isolates were resistant to IMP antibiotics, and almost all were resistant to cefoxitin. The IMP, CAZ, and cefoxitin-resistant isolates were selected and further evaluated for ESBL, MBL, and Amp Cenzyme production.
3.1. Phenotypic Detection of ESBL-producing P. aeruginosa
DDST of CAZ-resistant P. aeruginosa isolates revealed that out of 78 isolates, only 61 were ESBL producers [Figure 1a]. The ESBL activity of the isolates was evaluated, and their zone of inhibition was recorded [Figure 2]. Furthermore, evaluation of ESBL producers based on sample sources revealed that the maximum, i.e., 30 isolates, were from the pus sample, followed by ETA, urine, blood, PF, BAL, sputum, drain, and throat sample, which were 8, 6, 4, 4, 4, 2, 2, and 1, respectively, and no ESBL producer was found in the HVS sample [Table 1].
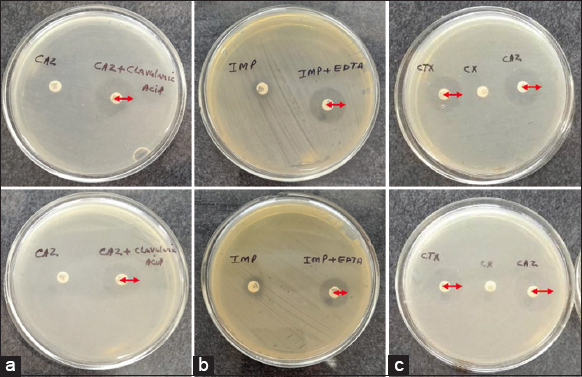 | Figure 1: Zone of inhibition by isolated cultures, Ceftazidime resistant Pseudomonas aeruginosa for Extended-spectrum beta-lactamases enzymes production (a) Imipenem resistant P. aeruginosa for MBL production (b) and AmpC beta-lactamase producing strains (c). [Click here to view] |
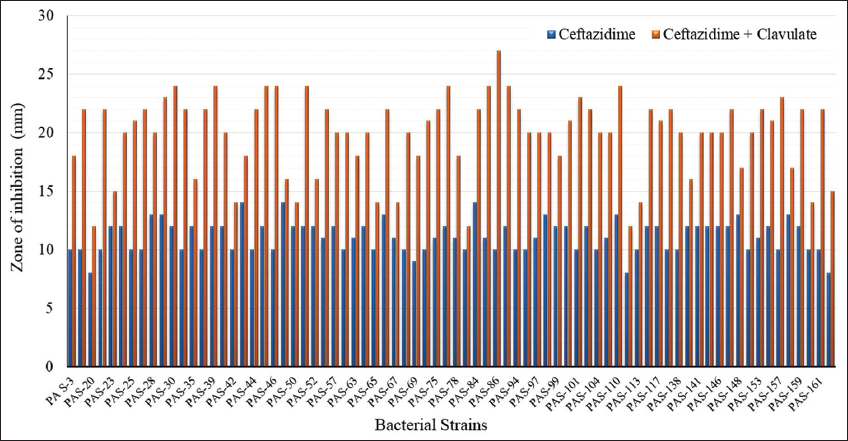 | Figure 2: The extended-spectrum beta-lactamases enzymes activity of Pseudomonas aeruginosa analyzed using the double disc synergy test method. [Click here to view] |
Table 1: CAZ resistant and ESBL producer Pseudomonas aeruginosa.
| Sample types | OPD | IPD | Total |
|---|---|---|---|
| Urine | 01 | 05 | 06 |
| Blood | 00 | 04 | 04 |
| Pus | 06 | 24 | 30 |
| Sputum | 00 | 02 | 02 |
| HVS | 00 | 00 | 00 |
| ETA | 00 | 08 | 08 |
| PF | 00 | 04 | 04 |
| BAL | 00 | 04 | 04 |
| Throat samples | 01 | 00 | 01 |
| Drain | 01 | 01 | 02 |
| Total | 09 | 52 | 61 |
3.2. Prevalence of ESBL in Outpatient Department (OPD) and Inpatient Department (IPD) Patients
Out of a total of 163 strains isolated, 48 were isolated from OPD patients, 72 were isolated from patients admitted to different hospital wards, and 43 were isolated from intensive care units (ICUs). The ESBL-producing strain isolated from wards by the DDST method was 18.75%, and 45.22% from wards and ICUs. The high prevalence of ESBL-producing strains found in IPDs is suggestive of the infection gained by the patient during his stay in the hospital. It can be minimized by adopting strict infection control measures in hospitals, especially wards and ICUs.
3.3. Phenotypic Detection of MBL-Producing P. aeruginosa
Screening of the antibiotic susceptibility of 163 P. aeruginosa isolates from different patients demonstrated that only 62 isolates were IPM-resistant [Figure 3]. A combine disk synergy test for the MBL producer was conducted on 62 isolates of P. aeruginosa, out of which only 46 were found to be MBL producers [Figure 1b]. Further analysis of sample type revealed that the maximum sample was isolated from pus, followed by ETA, BAL, urine, sputum, PF, blood, and drain [Table 2]. An analysis of the prevalence of MBL producers isolated from OPD and IPD patients was investigated. Out of 46 isolates, 36 were isolated from IPD patients, and only 16 were isolated from OPD patients.
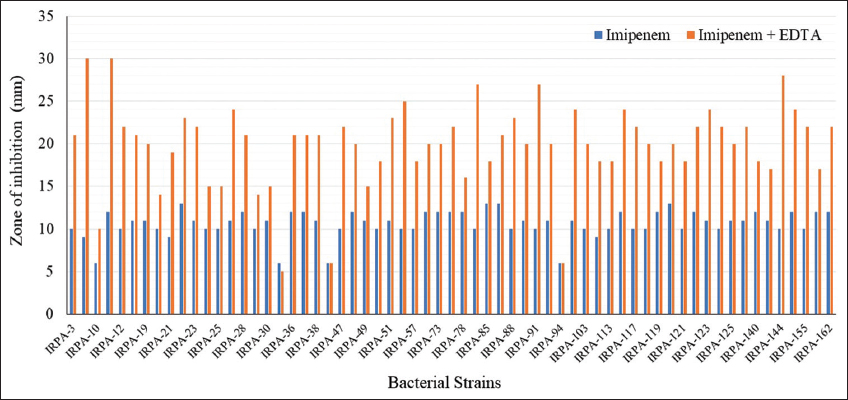 | Figure 3: Detection of imipenem resistant Pseudomonas aeruginosa using double disc synergy test method. [Click here to view] |
Table 2: Detection of MBL producer P. aeruginosa in different types of samples.
| Sample types | IMP resistant P. aeruginosa | MBL producers |
|---|---|---|
| Pus | 28 | 22 |
| ETA | 10 | 07 |
| BAL | 07 | 05 |
| Urine | 07 | 04 |
| Sputum | 05 | 04 |
| PF | 03 | 02 |
| Blood | 01 | 01 |
| Drain | 01 | 01 |
| Throat samples | 00 | 00 |
| HVS | 00 | 00 |
| Total | 62 | 46 |
P. aeruginosa: Pseudomonas aeruginosa.
3.4. Phenotypic Detection of AmpC β-lactamase Production
P. aeruginosa is ill-famed for being naturally resistant to various antimicrobial agents. It can restrict the entry of antimicrobial agents as it has a less permeable outer membrane, continuously expressing various efflux pumps and the naturally occurring chromosomal AmpC β lactamase [18].
After the screening of antibiotic susceptibility of a total of 163 P. aeruginosa isolates, it was found that all isolates were resistant to antibiotic cefoxitin, and further screening for AmpC β lactamase producers revealed that 85 isolates were positive for AmpC β lactamase [Figure 1c].
3.5. Identification of the Organism by MALDI-TOF MS Analysis
The isolated cultures were identified with MALDI-TOF MS. After analysis, the machine displayed the spectrum and results with sample position, date, and time, and identified the organism’s name with a confidence value [Figure 4].
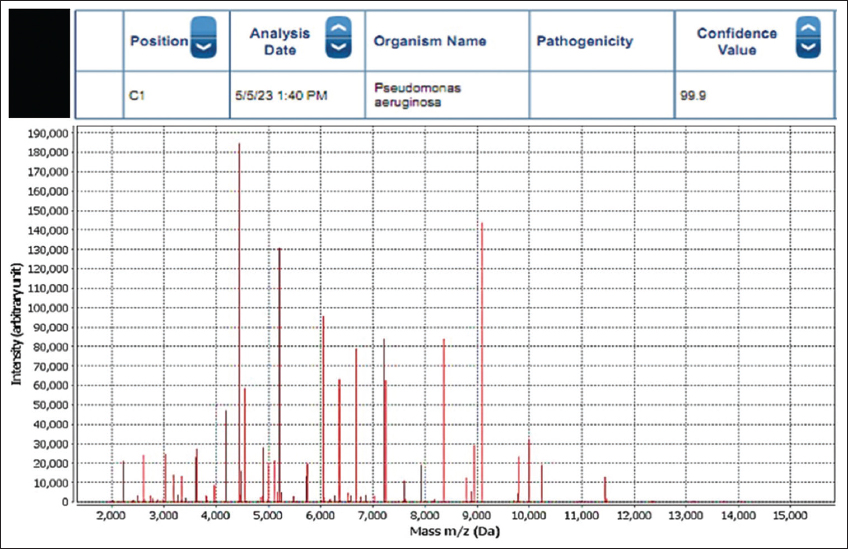 | Figure 4: Spectrum generated after colony analysis with MALDI-TOF. [Click here to view] |
3.6. Identification of Resistant Genes in the Sample Isolates by Biofire FilmArray
Biofire FilmaArray pneumonia plus-IVD Panel detected 59 specimen bacteria from 25 samples of sputum and BAL. Out of 25 samples, eight samples detected only one pathogen, i.e., P. aeruginosa, five samples detected two pathogens (P. aeruginosa and E. coli), and seven samples detected three pathogens along with P. aeruginosa (P. aeruginosa, Acinetobacter calcoaeticus-baumannii complex, K. pneumonia group). Three samples detected four pathogens along with P. aeruginosa (P. aeruginosa, Acinetobacter calcoaceticus-baumannii complex, Serratia marcescens, K. pneumoniae group), and two samples detected five pathogens along with P. aeruginosa (P. aeruginosa, Acinetobacter calcoaceticus-baumannii complex, Enterobacter cloacae complex, E. coli, Staphylococcus aureus) [Table 3].
Table 3: Resistant genes detected by biofire multiplex PCR using pneumonia plus panels.
| Sample type | Organism detected | Resistant Gene (s) detected |
|---|---|---|
| Sputum | P. aeruginosa | CTX-M |
| BAL | P. aeruginosa | CTX-M |
| Sputum | P. aeruginosa | CTX-M NDM |
| BAL | P. aeruginosa | NDM VIM |
| BAL | P. aeruginosa | None |
| BAL | P. aeruginosa | None |
| BAL | P. aeruginosa | None |
| BAL | P. aeruginosa | NDM VIM |
| BAL | P. aeruginosa ACBC | NDM VIM |
| Sputum | P. aeruginosa E. coli | CTX-M NDM OXA-48-like |
| Sputum | P. aeruginosa E. coli | CTX-M NDM |
| BAL | P. aeruginosa ACBC | NDM |
| BAL | P. aeruginosa ACBC Klebsiella pneumonia group Haemophilus influenza E. coli | CTX-M NDM |
| BAL | P. aeruginosa ACBC E. coli Haemphilus influenza Klebsiella pneumonia group | CTX-M NDM |
| Sputum | P. aeruginosa ACBC Klebsiella pneumoniae group | CTX-M NDM OXA-48-like |
| Sputum | P. aeruginosa ACBC Serratiam arcescens Klebsiella pneumoniae group | CTX-M NDM OXA-48-like |
| BAL | P. aeruginosa ACBC E. coli Haemophilus influenza | CTX-M NDM VIM |
| BAL | P. aeruginosa ACBC Klebsiella pneumonia E. coli | CTX-M IMP NDM OXA-48-like |
| BAL | P. aeruginosa ACBC Klebsiella pneumonia Serratiam arcescens | CTX-M NDM OXA-48-like |
| BAL | P. aeruginosa ACBC Klebsiella pneumonia E. coli | CTX-M IMP NDM OXA-48-like |
| BAL | P. aeruginosa ACBC Klebsiella pneumoniae group | CTX-M NDM OXA-48-like |
| Sputum | P. aeruginosa ACBC E. coli | CTX-M NDM OXA-48-LIKE |
| BAL | P. aeruginosa Moraxella catarrhalis Proteus spp. | NDM VIM |
| Sputum | P. aeruginosa ACBC Klebsiella pneumoniae group | CTX-M NDM OXA-48-like |
| BAL | P. aeruginosa ACBC Staphylococcus aureus | CTX-M NDM |
P. aeruginosa: Pseudomonas aeruginosa, E. coli: Escherichia coli.
Resistant genes for ESBL and/or carbapenemase detected were 53, including CTX-M, NDM, OXA-48-like, VIM, and IMP. The most frequently detected resistant gene was found to be NDM, followed by CTX-M and OXA-48-like, while the gene IMP was the least detected [Figure 5].
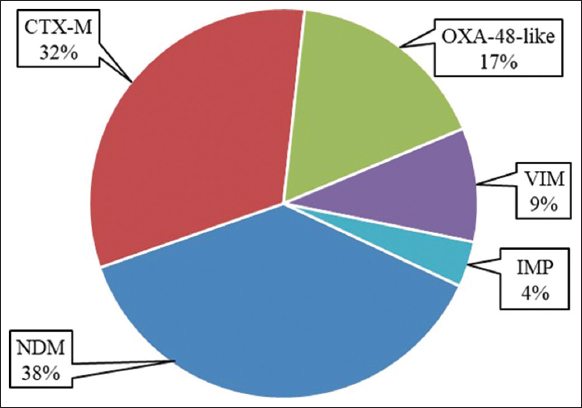 | Figure 5: Frequency of resistant genes detected from Pseudomonas aeruginosa by biofire filmarray using pneumonia plus panels. [Click here to view] |
4. DISCUSSION
More than 200 species of Pseudomonas are reported, including both pathogenic and non-pathogenic species. Pathogenic Pseudomonas species that cause human diseases are Pseudomonas maltophilia, Pseudomonas mallei, Pseudomonas pseudomallei, Pseudomonas cepacia, Pseudomonas stutzeri, Pseudomonas flourescens, Pseudomonas multiorans, Pseudomonas Putida, and P. aeruginosa. However, P. aeruginosa is the most prevalent species in hospital environments, causes severe infections in humans, and contributes to more than 50% of infections [19,20].
The prevalence of P. aeruginosa varies depending on the clinical sample source, such as pus, ETA, BAL, urine, sputum, PF, blood, drain, throat samples, and HVS. In the present study, isolates were derived from 7.4% from urine, 8.6% from blood, 42.9% from pus/wound swabs, 12.9% from sputum, 0.6% from HVS, 10.4% from ETA, 5.5% from PF, 9.8% from BAL, 0.6% from throat samples, and 1.2% from the drain. The gender-based prevalence of P. aeruginosa infection in the present study was 66.3% male patients, and 33.7% were female. Wang and Wang [21] reported that of P. aeruginosa-positive patients in China, 55.7% were male and 44.3% were female. Further, they reported sample sources: 21.6% from wounds, 28% from sputum, 13.7% from blood, 33.1% from pus, 19.1% from BAL, and 7.6% from other sources. In another study in the Iranian population, P. aeruginosa isolated from different sample sources was 41.4% from urine, 20.7% from blood, 22.4% from wounds, and 15.5% from the sample [22].
P. aeruginosa is known for its ability to resist antibiotics, and this resistance can be due to intrinsic and acquired resistance mechanisms, as well as adaptive antibiotic resistance. It possesses a high degree of innate and acquired resistance mechanisms that make it resistant to the majority of antibiotics. Low outer membrane permeability, efflux, lipopolysaccharide modification, and the bacterial enzyme AmpC are significant parts of the resistant mechanism. P. aeruginosa can acquire antibiotic resistance by acquiring resistance genes on mobile genetic elements such as plasmids. It can adapt to antibiotics by changing its gene expression, leading to resistance development [23].
Beta-lactam antibiotics, including oxymino-beta-lactams and monobactams, are inactivated by extended-spectrum beta-lactamase enzymes. ESBLs are plasmid-encoded proteins that can be easily transmitted between species. Because of its intrinsic beta-lactamase genes, P. aeruginosa is inherently resistant to beta-lactam drugs. It can contain a variety of beta-lactamases, including class A (GES), class C (AmpC), and class D (PoxB) beta-lactamases. The prevalence of ESBLs in P. aeruginosa is a major public health concern. It is critical to monitor the incidence of ESBL formation in P. aeruginosa isolates and develop antibiotic resistance strategies for this bacterial species [24].
In the present study, around 78% of CAZ-resistant isolates were ESBL producers. However, different studies have reported that ESBL production ranges from 20.4% to 88%. In a study in the Ashanti Region of Ghana, Odoi et al. (2021) reported that % of the total clinical samples collected 34% were MDR P. aeruginosa, and among them, 88% were ESBL producers [25]. A study conducted by Farhan et al. found that 54% of isolates were ESBL producers [26]. The prevalence of MDR P. aeruginosa in a multicenter study in the USA was 73%, and out of these MDR isolates, 20.4% were ESBL producers [27]. Discordance in the results indicated that this may be due to differences in environmental conditions and/or may be due to sample size.
In urine samples, only 12 isolates were P. aeruginosa, and only 9 were CAZ resistant. The prevalence of ESBL-producing isolates in urine was 50% (6/12). The result disagrees with Shaikh et al. [28], who reported 20.43% of ESBL-producing P. aeruginosa, which is quite lower than our study. Only fourteen P. aeruginosa were isolated from blood, and only four of them were ESBL producers. So, their prevalence was only 28.6% (4/14), which was double the prevalence [28].
From the pus sample, only seventy isolates were P. aeruginosa; among them, only thirty isolates were ESBL producers. The prevalence of P. aeruginosa isolates from blood in the present study was 42.85%. Saikh et al., [28] reported their prevalence at only 28.36%, which was quite lower than our results of 42.85%. From the sputum samples examined, 21 isolates were P. aeruginosa. Out of 21 strains, only two were ESBL producers, which was 2/22 = 9.52%; around 41.67% of strains were ESBL producers, which is approximately 4 times higher than our study. P. aeruginosa isolated from HVS, ETA, PF, BAL, throat samples, and drain were 01,17,9,16, 1, and 2, respectively. The prevalence of ESBL-producing strains was ETA (47%), PF (44.44%), BAL (25%), and throat samples (100%), and in drain samples, it was found to be 100%. As the number of samples examined in them was much smaller, their prevalence cannot be taken as significant.
IPM-resistant P. aeruginosa is a type of bacteria that can resist the antibiotic IPM. MBL production has been confirmed in a significant percentage of IPM-resistant P. aeruginosa isolates. In one study, 75% of IPM-resistant P. aeruginosa isolates were found to produce MBLs [29]. The genetic material for MBLs and other carbapenemases is encoded on mobile genetic elements, which can be transferred between the strains and across the bacterial species, making them more harmful to human health. A study conducted in Nepal found that 33.3% of P. aeruginosa isolates were MBL producers [30]. In the present study, a total of 163 P. aeruginosa were isolated from the different patients and samples. Among these isolates, 38% (n = 62) were found to be carbapenem (IPM) resistant, and only 74% (n = 46) isolates from them were MBL producers. Saha et al. [31] reported the prevalence of carbapenem-resistant P. aeruginosa was 39% (n = 32), and among them, 56% (18/32) were MBL producers. The prevalence of carbapenem resistance in the present study was (62/163 = 38%), similar to the findings of Saha et al. [31]. In another study, Goudarzi et al. [32] reported that carbapenem-resistant P. aeruginosa was 20% (n = 20), and among them, only 25% (n = 4) were MBL producers. Therefore, IPM-resistant P. aeruginosa can produce MBLs, making them resistant to multiple antibiotics and posing a significant public health threat.
The occurrence and spread of carbapenemases due to acquired resistance to β-lactam among MDR P. aeruginosa is a major epidemiological concern today. The MDR P. aeruginosa prevalently produces MBLs, such as VIM and IMPs. However, class A serine β-lactamases (KPC), class D serine β-lactamases (OXAtype), and other MBLs like NDM, GIM, and SPM are rarely detected in P. aeruginosa [33,34]. BAL and sputum samples were also identified using Biofire FilmArray, which revealed that the cultures were positive for the resistant genes, including the genes encoding NDM (n = 20), CTX-M (n = 17), OXA-48-like (n = 9), VIM (n = 5), and IMP (n = 2). Reports said that the prevalence of NDM and OXA-type MBLs is rare in P. aeruginosa, but in the present study, its prevalence was higher. Weber et al. [34] did not detect a gene encoding NDM in P. aeruginosa, while a gene encoding VIM was found in 81 out of 223 isolates, whereas in the present study, NDM was found in 20 out of 25 cultures. Also, the emergence of OXA is rare in P. aeruginosa, but we identified it in 9 isolates out of 25 isolates, OXA-48, which was specifically reported in India [33-35].
5. CONCLUSION
The findings of this study indicated a significant rise in ESBL-producing P. aeruginosa among patients worldwide. The ESBL prevalence was much higher in indoor patients than in outdoor patients. Antibiotics like cephalosporins, cefotaxime, and CAZ have little effect, but carbapenems and aminoglycosides are the best options for treating ESBL-producing P. aeruginosa. Drugs that are not recommended for the treatment of this pathogen are penicillin and sulfonamides like co-trimoxazoles. Strict infection control measures, careful monitoring of antibiotic administration, and routine screening for ESBL-producing strains are advised before treating the patients. Phenotypic and genotypic methods for early detection of β-lactam-resistant bacteria are important to prevent further pathogen dissemination.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be authors as per the International Committee of Medical Journal Editors (ICMJE) requirements and guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
The ethical approval from the Biomedical Research Ethics Committee, PGIMS, Rohtak, Haryana vide letter no. BREC/21/67 dated July 20, 2021, and Institutional Human Ethics Committee, Maharshi Dayanand University, Rohtak, Haryana vide letter no. HEC/2021/298 dated September 09, 2021 has been taken for the present research.
10. DATA AVAILABILITY
All the data is available with the authors and will be provided upon request.
11. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Singh KS, Anand S, Dholpuria S, Sharma JK, Blankenfeldt W, Shouche Y. Antimicrobial resistance dynamics and the one-health strategy:A review. Environ Chem Lett 2021;19:2995-3007. [CrossRef]
2. Dadgostar P. Antimicrobial resistance:Implications and costs. Infect Drug Resist 2019;12:3903-10. [CrossRef]
3. Yadav B, Tyagi RD. Development of molecular methods to detect and control emerging drug-resistance pathogens. In:Current Developments in Biotechnology and Bioengineering. Netherlands:Elsevier;2020. 377-419. [CrossRef]
4. Srivastava AK. Pharmacology:A Book of Achieving Knowledge for Drugs. Delhi:Educreation Publishing;2017.
5. Rajivgandhi GN, Alharbi NS, Kadaikunnan S, Khaled JM, Kanisha CC, Ramachandran G, et al. Identification of carbapenems resistant genes on biofilm forming K. pneumoniae from urinary tract infection. Saudi J Biol Sci 2021;28:1750-6. [CrossRef]
6. Babic M, Hujer AM, Bonomo RA. What's new in antibiotic resistance?Focus on beta-lactamases. Drug Resist Updat 2006;9:142-56. [CrossRef]
7. Curcio D. Multidrug-resistant Gram-negative bacterial infections:Are you ready for the challenge?Curr Clin Pharmacol 2014;9:27-38. [CrossRef]
8. Bassetti M, Vena A, Croxatto A, Righi E, Guery B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018;7:212527. [CrossRef]
9. Sundin DR. Hidden beta-lactamases in the Enterobacteriaceae - dropping the extra disks for detection, part II. Clin Microbiol Newsl 2009;31:47-52. [CrossRef]
10. Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa:Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009;22:582-610. [CrossRef]
11. Rotter MJ, Zentgraf S, Weizel L, Frank D, Burgers LD, Brunst S, et al. Integrating siderophore substructures in thiol-based metallo-b-lactamase inhibitors. Molecules 2023;28:1984. [CrossRef]
12. Khosravi Y, Loke MF, Chua EG, Tay ST, Vadivelu J. Phenotypic detection of metallo- b -lactamase in imipenem-resistant Pseudomonas aeruginosa. ScientificWorldJournal 2012;2012:654939. [CrossRef]
13. Zafer MM, Al-Agamy MH, El-Mahallawy HA, Amin MA, Ashour MS. Antimicrobial resistance pattern and their beta-lactamase encoding genes among Pseudomonas aeruginosa strains isolated from cancer patients. Biomed Res Int 2014;2014:101635. [CrossRef]
14. Bialvaei AZ, Kafil HS, Asgharzadeh M, Yousef Memar M, Yousefi M. Current methods for the identification of carbapenemases. J Chemother 2016;28:1-19. [CrossRef]
15. Olfa S, Mariem N, Mahjoub A, Maha M. Detection of metallo-beta-lactamase producing Pseudomonas aeruginosa using a modified IMP-lysate assay. Afr J Biotechnol 2016;15:278-83. [CrossRef]
16. Fazeli H, Sadighian H, Nasr-Esfahani B, Pourmand MR. Identification of class-1 integron and various b-lactamase classes among clinical isolates of Pseudomonas aeruginosa at children's medical center hospital. J Med Bacteriol 2012;1:25-36.
17. Vanwynsberghe T, Verhamme K, Raymaekers M, Cartuyvels R, Vaerenbergh KV, Boel A, et al.Alarge hospital outbreak of Klebsiella pneumoniae (DHA-1 and SHV-11 positive):Importance of detection and treatment of ampC b-lactamases. Open Infect Dis J 2009;3:55-60. [CrossRef]
18. Chika E, Charles E, Ifeanyichukwu I, Chigozie U, Chika E, Carissa D, et al. Phenotypic detection of AmpC beta-lactamase among anal Pseudomonas aeruginosa isolates in a Nigerian abattoir. Arch Clin Microbiol 2016;7:2.
19. Sarabhai S, Sharma P, Capalash N. Ellagic acid derivatives from Terminalia chebula Retz. Downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS One 2013;8:e53441. [CrossRef]
20. Tuon FF, Dantas LR, Suss PH, Tasca Ribeiro VS. Pathogenesis of the Pseudomonas aeruginosa biofilm:A review. Pathogens 2022;11:300. [CrossRef]
21. Wang W, Wang X. Prevalence of metallo-b-lactamase genes among Pseudomonas aeruginosa isolated from various clinical samples in China. J Lab Med 2020;44:197-203. [CrossRef]
22. Karami P, Khaledi A, Mashoof RY, Yaghoobi MH, Karami M, Dastan D, et al. The correlation between biofilm formation capability and antibiotic resistance pattern in Pseudomonas aeruginosa. Gene Rep 2020;18:100561. [CrossRef]
23. Langendonk RF, Neill DR, Fothergill JL. The building blocks of antimicrobial resistance in Pseudomonas aeruginosa:Implications for current resistance-breaking therapies. Front Cell Infect Microbiol 2021;11:665759. [CrossRef]
24. Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases:Types, epidemiology and treatment. Saudi J Biol Sci 2015;22:90-101. [CrossRef]
25. Odoi H, Boamah VE, Boakye YD, Agyare C. Prevalence and phenotypic and genotypic resistance mechanisms of multidrug-resistant Pseudomonas aeruginosa strains isolated from clinical, environmental, and poultry litter samples from the Ashanti region of Ghana. J Environ Public Health 2021;2021:9976064. [CrossRef]
26. Farhan SM, Ibrahim RA, Mahran KM, Hetta HF, Abd El-Baky RM. Antimicrobial resistance pattern and molecular genetic distribution of metallo-b-lactamases producing Pseudomonas aeruginosa isolated from hospitals in Minia, Egypt. Infect Drug Resist 2019;12:2125-33. [CrossRef]
27. Hirsch EB, Brigman HV, Zucchi PC, Chen A, Anderson JC, Eliopoulos GM, et al. Ceftolozane-tazobactam and ceftazidime-avibactam activity against b-lactam-resistant Pseudomonas aeruginosa and extended-spectrum b-lactamase-producing Enterobacterales clinical isolates from U.S. medical centres. J Glob Antimicrob Resist 2020;22:689-94. [CrossRef]
28. Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Prevalence of multidrug resistant and extended spectrum beta-lactamase producing Pseudomonas aeruginosa in a tertiary care hospital. Saudi J Biol Sci 2015;22:62-4. [CrossRef]
29. Radhika A, Lakshmi JT, Ariyanachi K, Sakthivadivel V. Detection of metallo beta-lactamase (MBL) producing Pseudomonas aeruginosa in a tertiary care hospital, Ghanpur, Medchal, India. Maedica (Bucur) 2022;17:134-42.
30. Sharma S, Devkota MD, Pokhrel BM, Banjara MR. Detection of bla(NDM-1,)mcr-1 and MexB in multidrug resistant Pseudomonas aeruginosa isolated from clinical specimens in a tertiary care hospital of Nepal. BMC Microbiol 2023;23:153. [CrossRef]
31. Saha K, Kabir ND, Islam MR, Amin MB, Hoque KI, Halder K, et al. Isolation and characterisation of carbapenem-resistant Pseudomonas aeruginosa from hospital environments in tertiary care hospitals in Dhaka, Bangladesh. J Glob Antimicrob Resist 2022;30:31-7. [CrossRef]
32. Goudarzi H, Bostanghadiri N, Riahi Rad Z, Riahi Rad Z, Yasbolaghi Sharahi J. Evaluation of b-lactamases and molecular typing of Pseudomonas aeruginosa clinical strains isolated from hospitalized children in Tehran. Arch Clin Infect Dis 2023;18:e134837. [CrossRef]
33. Yoon EJ, Jeong SH. Mobile carbapenemase genes in Pseudomonas aeruginosa. Front Microbiol 2021;12:614058. [CrossRef]
34. Weber C, Schultze T, Göttig S, Kessel J, Schröder A, Tietgen M, et al. Antimicrobial activity of ceftolozane-tazobactam, ceftazidime-avibactam, and cefiderocol against multidrug-resistant Pseudomonas aeruginosa recovered at a German University hospital. Microbiol Spectr 2022;10:e0169722. [CrossRef]
35. Borah VV, Saikia KK, Hazarika NK. First report on the detection of OXA-48 b-lactamase gene in Escherichia coli and Pseudomonas aeruginosa co-infection isolated from a patient in a tertiary care hospital in Assam. Indian J Med Microbiol 2016;34:252-3. [CrossRef]