1. INTRODUCTION
Many emerging bacterial infectious diseases (EBIDs) are a significant burden on global economies and public health [1,2]. Around thirty percent of fatalities worldwide is mainly because of bacterial infections leading to human dismalness and mortality. Understanding the global temporal and spatial patterns of EBIDs becomes a priority for the current scientific fraternity. One such organism of immediate attention for surveillance and epidemiological research is Pseudomonas aeruginosa, a gram -ve, encapsulated, facultatively aerobic rod-shaped bacterium, which is one among the six ESKAPE pathogens with a growing rate of virulence in humans and animals [3,4]. However, P. aeruginosa has evolved newer escape mechanisms to bypass chemical antibacterial agents bacteriostatic and/or bacteriocide [5,6]. This armored bacterial pathogen has developed resistance due to its switching abilities in physicochemical growth patterns of its niche between planktonic (free-floating) and biofilm (sessile) forms [7]. The biofilm development in pseudomonas involves a series of well-orchestrated steps starting from locating an adhesive surface, reversible adsorption on the substratum, production of exopolysaccharides, or exopolymeric substances (EPS), maturation, detachment, and co-infection [8-10].
During the biofilm stage an elemental phenomenon that occurs in bacteria, especially in P. aeruginosa, is quorum sensing that is cell-to-cell, density-dependent community cross-talk. It is observed that quorum sensing is responsible to facilitate thiol-disulfide redox metabolism of the bacterium [11-13]. Pseudomonas are prone to become resistant to antibiotics due to the formation of biofilms that complicates any treatment or infections. Further, 4-quinolones and N-acyl-homoserine lactones are involved as the major quorumone in signaling cascades [14,15]. Further, The main essential function of the prokaryotic cell is to transport proteins to the environment, compartments of cells, and other bacteria cells in pure culture or co-culture and mixed culture populations [16]. This mechanism is executed by an effector protein-mediated cascade known as the protein secretion system. There are different types of secretion systems, including Type 1 Secretion System, (T1SS), T2SS, T3SS, T4SS, T5SS, T6SS, and T7SS. In P. aeruginosa the T3SS, is mainly involved in virulence triggering oxidative stress (by generating reactive oxygen/nitrogen species) and could be majorly targeted to block bacterial signal trafficking. The bacterial flagellum and the injectisome are composed of conserved machinery known as the type III secretion system. This T3SS is the primary virulence determinant of P. aeruginosa that promotes tissue destruction and escapes phagocytosis by secreting numerous toxins that are translocated into host cells [17,18]. Type-3 cytotoxins play a crucial role during P. aeruginosa quorum-signaling, very importantly bi-functional cytotoxins ExoS and ExoT, along with ExoU have a cascade effect [19]. Electrostatic bonding determines the domain-substrate (ADP-ribosyltransferase) specificity in exo-cytotoxin protein modeling [20]. Henceforth, computational biology is employed to have a better insight into the complex fitting pieces of the puzzle. Tactically targeting P. aeruginosa T3SS-Exo system could be one of the possible ways to combat this pathogen.
With this background, it has been a priority to develop alternative approaches either preventive or therapeutic. Various methods are prescribed in classical, traditional plant-based Indian medicine for the development of effective drugs with lesser side effects but lack drug-target-based mechanistic design. The current study attempts to employ plant extracts from Indian herbs to effectively manipulate and percolate the thick EPS barrier ultimately targeting the bacterial strength or polysaccharide-mediated cell signaling. The details of the plants under investigation are listed in Table 1. A thorough literature meta-data analysis of the phytochemicals present in 20 selected plants revealed the presence of phyto-antibiofilm contributors such as quercetin, catechin, allicin, and eugenol which has previously been studied for antibacterial/antibiofilm activity [21-24]. However, the interaction of these phytochemicals was not evaluated against T3SS Exo-cytotoxin cascade. With this rationale, the phytomolecules were screened using In silico docking for their effective binding against T3SS targets (ExoS, ExoT, and ExoU) with corresponding binding energy i.e., Atomic Contact Energy (ACE) or Gliding Energy indicating their inhibition [25]. Polyphenolics, the class of secondary metabolites are proven to exhibit antistress and antibacterial activity against pseudomonas [5] and profiling was evaluated along with various free-radical scavenging activities. Further, an in vitro experimental microtitre plate anti-adhesive model was set up to assess the anti-biofilm activity of selected twenty plant extracts (hydroethanolic extract 70:30 v/v). In silico modeling was also performed to understand the molecular-level interactions of the potent phyto-principles against T3SS cascade. The results of the current study infer to identify herbaceuticals that can eradicate P. aeruginosa biofilms ultimately leading to better health and hygiene.
2. MATERIALS AND METHODS
The chemicals, solvents, and reagents considered for the study were procured from Himedia Pvt. Ltd, Mumbai, India and were of extra pure analytical grade.
2.1. Plant Material
The medicinal plants considered for the current study are listed in Table 1. The leaves were collected with a time duration between March and July 2018 from Mysuru, Karnataka, India. For authentication on identification, the specimens were prepared and are deposited at the herbarium, Department of Studies in Botany, University of Mysore, Karnataka, India. The leaves of Clerodendrum phlomidis (Agnimantha), Abutilon indicum (Atibala), Sida cordifolia (Bala), Coccinia grandis (Bimboshta), Oxalis corniculate (Changeri), Lantana camara (Chaturangi), Plumbago zeylanica (Chitraka), Acalypha indica (Haritamanjari), Martynia annua (Kakanasa), Solanum xanthocarpum (Kanthakari), Mimosa pudica (Lajjalu), Centella asiatica (Mandookaparni), Cyperus rotundus (Musta), Fumaria parviflora (Parpata), Cocculus hirsutus (Patalagarudi), Boerhavia diffusa (Punarnava), Osmium americanum (Tulasi), Zanthoxylum acanthopodium (Tumbura), Vetiveria zizanoides (Ushira) and Trachyspermum ammi (Yavani) were cleaned and rinsed for surface sterilization with 0.5% sodium hypochlorite solution. All samples were air-dried under gentle sunlight for 48 h and the powder was stored until further use in zip-locked polythene containers.
 | Table 1. List of plants having potent antibiofilm activity on Psuedomonas species. [Click here to view] |
2.1.1. For extraction
Briefly, around 25 g of the leaves from the selected 20 plants were individually mixed with 100 mL hydroethanolic solvent (70:30 v/v) i.e., ethanol and water, respectively. Soxhlet apparatus was used with sample processer under 10 psi pressure using a vacuum flash evaporator [26,27]. The semi-solid residual photo-cocktail was filtered using a double-layered muslin cloth and centrifuged at 8000 rpm for 10 min. The supernatant extract was separated and was stored at 8°C until further.
2.2. Plant Extracts Profiling and Antioxidant Activity
Twenty hydroethanolic leaves extracts were subjected to total polyphenolic estimation. The free radical scavenging assay was assessed using 1,1-diphenyl-2picrylhydrazyl (DPPH), metal ion chelating activity using ferrozine, and reducing power activity with ferric chloride was performed respectively.
2.2.1. Estimation of total phenolic content (TPC)
The twenty leaves extracts were subjected to polyphenolic estimation using Folin-Ciocalteau method using gallic acid as standard (1 mg/mL) equivalents with 10 μL volume [26,28]. The optical density was read at 640 nm using a spectrophotometer. The TPC concentration was considered for all further quantitative estimation and bioactivity.
2.3. Evaluation of Radical Scavenging Activities of Selected Plant Extract
2.3.1. DPPH free radical scavenging assay
Quantitatively, the plant extract was subjected at 5 μg/mL, 10 μg/mL, 15 μg/mL, 20 μg/mL, and 25 μg/mL with gallic acid as a standard reference to assess the scavenging activity against DPPH free radical [26] and the optical density was read at 517 nm using a spectrophotometer. The percentage activity was calculated using the formula;
2.3.2. Metal ion chelating assay
Gradient concentrations of plant extracts were evaluated for their metal ion chelating efficacy against ferrous iron-ferrozine complex, as previously described [26,28]. The optical density was read at 562 nm using a spectrophotometer. Ethylenediaminetetraacetic acid (EDTA) was considered as a standard reference and was calculated as per the above-mentioned equation.
2.3.3. Reducing power assay
All plant extracts were tested for their reducing potential using iron (III) as previously illustrated [26,29] and gallic acid (1 mg/mL) equivalents were used as standard. The optical density was read at 700 nm using a spectrophotometer.
2.4. Anti-biofilm activity
All twenty hydroethanolic crude extracts were subjected for assessment of the anti-biofilm activity of P. aeruginosa for their anti-adhesive, biofilm strength, and EPS inhibition using the microtitre plate method with crystal violet and ruthenium red staining as previously designed by our lab [7,8].
2.4.1. Biofilm production
The selected strain of P. aeruginosa was cultured in Luria-Bertani broth media with 0.6% yeast extract as nitrogen source and 0.5% glucose as an energy source under optimized parameters of pH and temperature as per Zameer et al. [5]. Samples were drawn at regular intervals and monitored for optical density to assess the growth curve of P. aeruginosa. Biofilm cultivation was carried out onto polystyrene 96 well microtitre plates. The plates were sterilized by the standard procedure described by Zameer et al. [7,8]. Further, 200 μL actively grown starter culture in microtitre wells were inoculated. The same was used to quantitate biofilm strength, EPS-using crystal violet, and ruthenium red staining respectively [30].
2.5. Quantification of Biofilm
2.5.1. Biofilm strength assay
The strength of biofilm was quantified as per the protocol described by Djordjevic et al. [31], with minor modification using the gradient concentrations of the plant extract inhibitors at 25 μg/mL. Briefly, the staining of adherent bacteria onto the cultivating surfaces was carried out using polystyrene 96 well microtitre plates. The surfaces were washed thrice with 250 μL of PBS (pH 7.4) during which non-adherent bacteria were washed out. The plates were dried in an inverted position at 42°C for 30 min. The growing biofilms were scraped using a sterile scalpel and were collected in sterile vials. Further, 150 μL of 1% crystal violet solution was added to each well and further incubated at 37°C for 30 min. Later, samples were carefully rinsed with water to decant the excess dye. 200 μL of 95% ethanol was used as dye solubilizer with subsequent incubation at 4°C for 45 min. Samples were drawn from individual wells and were monitored at 595 nm using a microtiter plate reader [7,8].
2.5.2. EPS inhibition assay
The production of EPS was carried out using the ruthenium red staining technique according to Borucki et al. [30], using the gradient concentrations of the plant extracts inhibitors at 25 μg/mL. Briefly, biofilms were stained for matrix EPS with the addition of 200 μL of 0.1 % ruthenium red to all the vials and wells which were incubated for 45 min at RT. Further, the media obtained from the wells was quantitated at 450 nm using a microtiter plate reader. The amount of dye reduced was compared and calculated with an average measurement of the blank [7,8] and the corresponding percentage inhibition was calculated.
2.6. Molecular Docking
Structure-activity relationship studies provide insight into the target-ligand interactions [32,33]. The proposed study considers three major effector proteins of T3SS in P. aeruginosa responsible for their role in pathogenesis and acute infections [34,35]. The crystal structure of the targets, namely, ExoS (Protein Data Bank [PDB]: 1HE9), ExoT (PDB: 6GNN), and ExoU (PDB: 4AKX) were retrieved from PDB. The ligand molecules were docked onto the active site of the receptors considering phytochemical evidence and abundance of the plant extracts from the previous studies in the selected plants. Allicin (PubChem: 65036), Catechin (PubChem: 9064), Eugenol (PubChem: 3314), and Quercitin (PubChem: 5280343) were taken into account depending on their various bioactivities. These ligands are currently been proven for their antibacterial properties. Molegro software package was used and the results were analyzed considering the ACE and gliding values [25,36]. The negative values indicate higher binding efficacy in the active site pockets between the ligand and the target indicating active inhibition [37].
2.7. Statistical Analysis
All data were processed for statistical validation using GraphPad Software, Inc., (version 6.0, California, USA). The experiments were designed in triplicates and three independent experiments were performed. The results were expressed in mean ± standard deviation (n = 3) with standard error bars indicated in the graph. One-way analysis of variance, followed by Duncan’s multiple range test was performed with P < 0.05. The percentage of inhibition for the phytoextracts was determined in terms of IC50 with regression correlation studies considering the polyphenolics.
3. RESULTS AND DISCUSSION
Microbes have co-evolved and have become an integral component of the host system and humans are no exception. This ubiquitous ecological interaction might vary in their role plays from being symbionts, mutualists, and antagonists. Microbes are fundamentally involved in the homeostasis of the metabolic, physiological, and immune systems regulations. However, these tiny-complex organisms have another detrimental phase as pathogens, and their interaction in the host-microbe association is not well understood, which could essentially affect the survival of the host leading to death. Recent advancements in drug design, newer antimicrobials, vaccine therapy, anti-viral therapy, anti-microbial peptide path-blockers, membrane disruptors, and host-defense reinforcement have made a significant impact in combating pseudomonas-related infections. Further, continuous implications of antibiotics have provoked the emergence of extensively drug-resistant, multidrug-resistant, extended-spectrum β-lactamase, and carbapenemase-producing pseudomonal strains which have imposed a serious therapeutic challenge. These events render even the most effective drugs ineffective and hence pose a threat to food, meat, and dairy industries extrapolating their infectious existence in humans causing cystic fibrosis, urinary tract, and nosocomial infections. Hence, understanding the phytomolecules in terms of their polyphenolics, antioxidant, and anti-biofilm potency becomes the prime focus of the current study.
3.1. Plant Extract Screened in the Current Study
The study aims to explore complementary and alternative medicine as a holistic approach to screen plants as anti-biofilm contributors for P. aeruginosa. Hence most of the plants considered for the study are the common weed [Table 1] found in Karnataka, India throughout the year without much seasonal variation and mentioned Indian traditional medicine for their curative function. In any disorder/infection, free radicals play a pivotal role in intensifying pathogenesis and T3SS is no exception. Henceforth, in the current study, we have attempted in exploring the photobiological for their antioxidant potency and antibiofilm efficiencies using hot hydroethanolic leaves extracts [38,39].
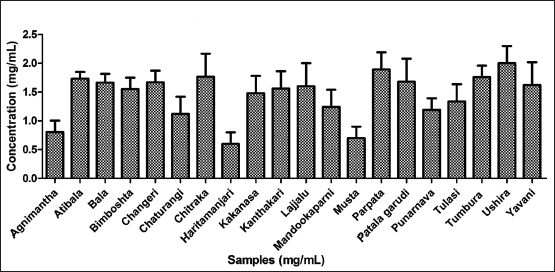 | Figure 1. Total phenolic content of twenty hydroethanolic plant extracts. [Click here to view] |
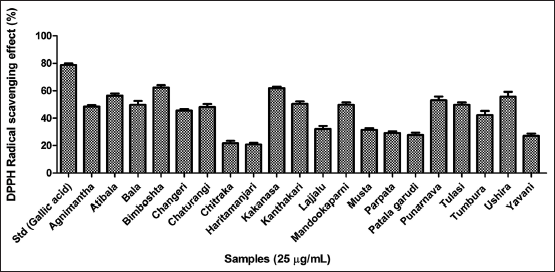 | Figure 2. Antioxidant assay of twenty hydroethanolic plant extracts using DPPH free radical scavenging assay. [Click here to view] |
3.2. Evaluation of Polyphenolics
Polyphenols are the group of secondary metabolites which are produced by the plants as defensins to avoid infections from rhizosphere intruders. These include the wide class of alkaloids, flavonoids, tannins, saponins, and terpenoids at large. In this study, the twenty plant leaves extracts were estimated for their total phenolics for understanding the abundance of the total phenolic content (TPC) present in the plant leaves, and the same is further used as a quantitative measure in all estimations and assays. Among the plants screened Ushira, Parpata, and Chitraka were the top three plants with the highest TPC as depicted in Figure 1.
3.3. Assessment of Antioxidant Efficacy
Three major antioxidant scavenging assays were performed for all the twenty leaves extracts. The results of the DPPH free radical scavenging assay inferred the highest activity by Bimboshta, Kakanasa, and Atibala respectively as reflected in Figure 2 and compared to gallic acid as standard. Further, for the metal ion chelating efficiency Atibala, Parpata, and Mandookaparni exhibited better activity compared to EDTA as standard [Figure 3]. Further, reducing power potency Atibala, Tulasi and Ushira exhibited maximum activity as mentioned in Figure 4. The results of all three antioxidant assays reflect the ability of plant polyphenols to effectively function as antioxidant contributors [40].
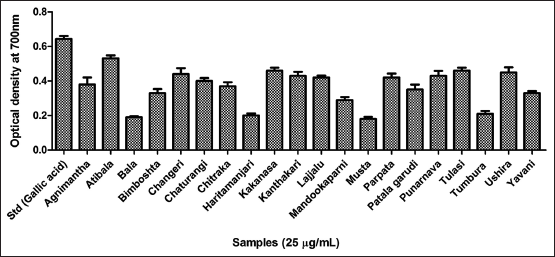 | Figure 3. Antioxidant assay of twenty hydroethanolic extracts using metal ion chelating assay. [Click here to view] |
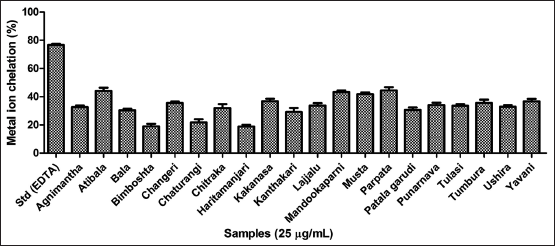 | Figure 4. Antioxidant assay of twenty hydroethanolic extracts using reducing power assay. [Click here to view] |
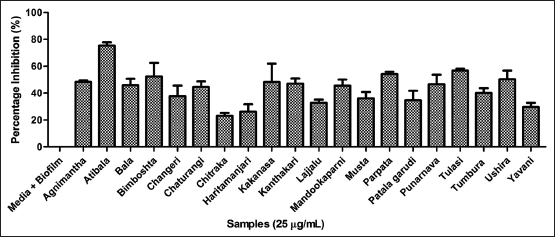 | Figure 5. Assessment of biofilm strength using crystal violet method microtitre plate method. [Click here to view] |
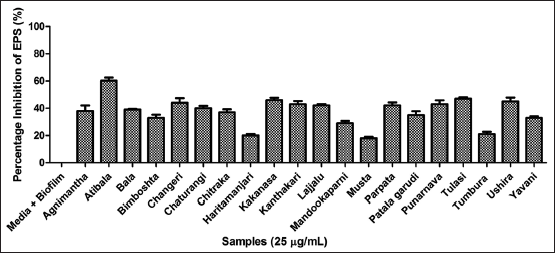 | Figure 6. Assessment of EPS using ruthenium red method microtitre plate method. EPS: Exopolymeric substances. [Click here to view] |
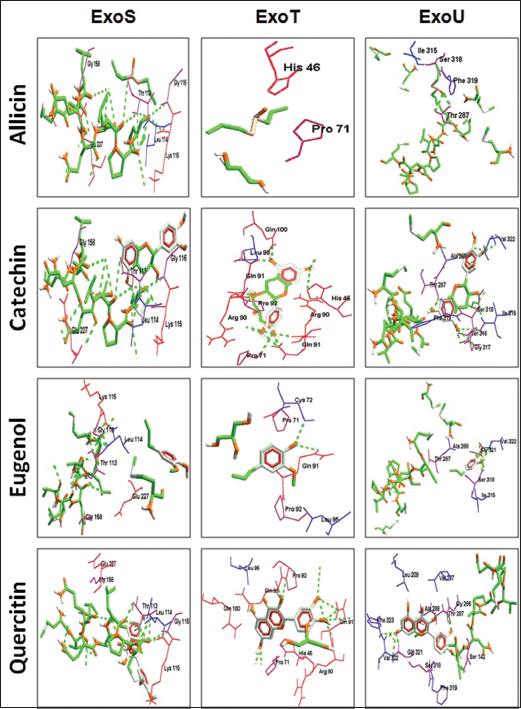 | Figure 7. Molecular docking of the potent phyto-ligands with the target for understanding structure-activity relationship studies. [Click here to view] |
3.4. Assessment of Antibiofilm Potency
The phenolics can function as anti-quorumone molecules against bacterial biofilms [41]. Henceforth, the plant extract was further analyzed for its efficacy to inhibit biofilm strength and EPS. The 96 well plate method was performed to analyze the anti-adhesive activity of P. aeruginosa biofilms. Among twenty extracts Atibala, Tulasi and Parpata exhibited maximum activity as reflected in Figure 5. The EPS was inhibited by Atibala, Tulasi, and Kakanasa extracts respectively [Figure 6].
3.5. Molecular Docking of the Phyto-attributes on the T3SS-Exo System
For better insights into the mode of action phytomolecules (ligands) under investigation namely allicin, catechin, eugenol, and quercitin were docked on the T3SS effector proteins ExoS, ExoT, and ExoUwhich were not previously studied against the selected ligands. The results are depicted in Figure 7 as docking poses and Table 2 illustrates the atomic contact energy values and gliding values [37]. The result indicated that quercetin and catechin could inhibit ExoS and the latter inhibits both ExoT and ExoU very effectively with the highest ACE values [42]. From Figure 8, it is very evident that these phytochemicals are effective against the oxidative species regulating pyocyanin, elastase, and superoxide dismutase (SOD1) as well as exotoxin A, ExoS, and ExoU, their effect is observed on pro-inflammatory cytokines (TLR4 and TLR5). These phytomolecules could be further explored for their nanoparticles to facilitate higher activity [34,43-45] and better percolation into the biofilm cluster facilitating insights into the mechanistic understanding of the host-pathogen interactions [46-49].
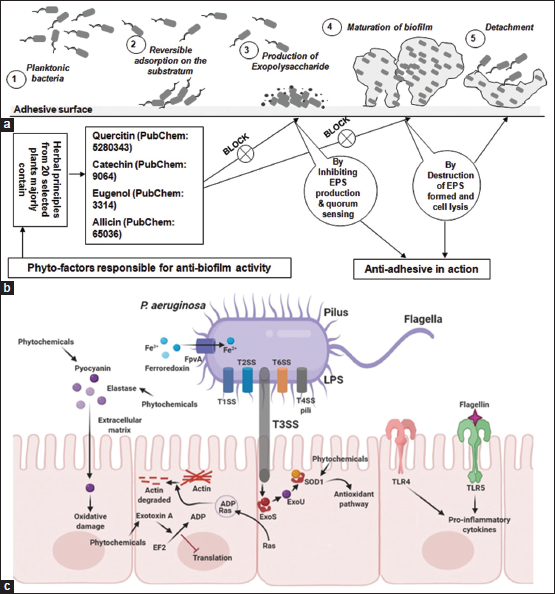 | Figure 8. Overview of the probable mode of action. Panel A: Indicates the various stages of biofilm development in Pseudomonas aeruginosa. Panel B: Depicts the mode of action of phyto-contributors in inhibiting EPS and biofilm strength and lysis. Panel C: Cellular level interaction of phytochemicals with the potential targets of T3SS-Exo cytotoxic cascade. EPS: Exopolymeric substances, T3SS: Type-3 secretion system. [Click here to view] |
 | Table 2. Molecular docking and SAR of the four potent phytomolecules namely allicin, catechin, eugenol, and quercetin were docked with the target molecule of T3SS namely ExoS, ExoT and ExoU respectively. [Click here to view] |
4. CONCLUSION
The results reflect the efficacy of the 20 selected plant extracts to exhibit potent antioxidant and antibiofilm activity against P. aeruginosa. Further studies have to be explored for having better insights into the characterization of the pure compounds and to understand the mechanistic action on the T3SS cascade at a cellular and molecular which could be efficient in developing newer drug targets against P. aeruginosa biofilms in specific and bacterial infections in general.
5. ACKNOWLEDGMENTS
Mr. Avinash MG would like to thank the Indian Council of Medical Research (ICMR) for the award of Senior Research Fellow (SRF) Award Letter Number - File no. AMR/Fellowship/17/2019 ECD-II dated 28.6.2019 and the University of Mysore for the smooth execution of the work. Further, FZ extends gratitude towards the management and office bearers of Dayananda Sagar University, Bengaluru, Karnataka, India, for their constant inspiration, motivation, and encouragement to pursue scientific research.
6. AUTHORS CONTRIBUTION
Avinash MG carried out the experiments, assimilated data, and drafted the manuscript. Farhan Zameer designed the protocol and Shubha Gopal analyzed data and edited the manuscript.
7. FUNDING
Monetary support in the form of Senior Research Fellow (SRF) was awarded to Mr. Avinash MG vide Award Letter Number - File no. AMR/Fellowship/17/2019 ECD-II dated 28.6.2019 from Indian Council of Medical Research (ICMR), New Delhi, Govt. of India.
8. CONFLICT OF INTEREST
The authors express no conflict of interest.
9. ETHICAL APPROVALS
?Not Applicable.
10. DATA AVAILABILITY
?Not Applicable, raw data files could be provided upon request to the authors.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature 2008;451:990-3. CrossRef
2. Abebe E, Gugsa G, Ahmed M. Review on major food-borne zoonotic bacterial pathogens. J Trop Med 2020;2020:4674235. CrossRef
3. Tummler B. Emerging therapies against infections with Pseudomonas aeruginosa. F1000Res 2019;8:F1000 Faculty Rev-1371. CrossRef
4. Navidinia M. The clinical importance of emerging ESKAPE pathogens in nosocomial infections. J Paramed Sci 2016;7:2008-4978.
5. Zameer F, Rugmangada MS, Chauhan JB, Khanum SA, Kumar P, Devi AT, et al. Evaluation of adhesive and anti-adhesive properties of Pseudomonas aeruginosa biofilms and their inhibition by herbal plants. Iran J Microbiol 2016;8:108-19.
6. Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front Microbiol 2019;10:539. CrossRef
7. Zameer F, Gopal S, Krohne G, Kreft J. Development of a biofilm model for Listeria monocytogenes EGD-e. World J Microbiol Biotechnol 2010;26:1143-7. CrossRef
8. Zameer F, Gopal S. Evaluation of antibiotic susceptibility in mixed-culture biofilms. Int J Biotech Biochem 2010;6:93-9.
9. Ashwini P, Sumana MN, Shilpa U, Mamatha P, Manasa P, Dhananjaya BL, et al. A review on Helicobacter pylori: Its biology, complications and management. Int J Pharm Pharm Sci 2015;7:14-20.
10. More VS, Ebinesar A, Prakruthi A, Praveen P, Fasim A, Rao A, et al. Isolation and purification of microbial exopolysaccharides and their industrial application. In: Microbial Polymers. Singapore: Springer; 2021. p. 69-86. CrossRef
11. Gopal S, Vanishree S, Farhan Z, Juergen K. Prediction of proteins putatively involved in the thiol: Disulfide redox metabolism of a bacterium (Listeria): The CXXC motif as query sequence. In Silico Biol 2009;9:407-14. CrossRef
12. Goo E, An JH, Kang Y, Hwang I. Control of bacterial metabolism by quorum sensing. Trends Microbiol 2015;23:567-76. CrossRef
13. Mielko KA, Jablonski SJ, Milczewska J, Sands D, Lukaszewicz M, Mlynarz P. Metabolomic studies of Pseudomonas aeruginosa. World J Microbiol Biotechnol 2019;35:178. CrossRef
14. Diggle SP, Cornelis P, Williams P, Cámara M. 4-quinolone signaling in Pseudomonas aeruginosa: Old molecules, new perspectives. Int J Med Microbiol 2006;296:83-91. CrossRef
15. Pradeep HP, Vijay GS, Shubha G. Revamping the role of biofilm regulating operons in device-associated staphylococci and Pseudomonas aeruginosa. Indian J Med Microbiol 2014;32:112-23. CrossRef
16. Chevalier S, Bouffartigues E, Bodilis J, Maillot O, Lesouhaitier O, Feuilloley MG, et al. Structure, function and regulation of Pseudomonas aeruginosa porins. Fed Eur Microbiol Soc Microbiol Rev 2017;41:698-722. CrossRef
17. Sato H, Frank DW. ExoU is a potent intracellular phospholipase. Mol Microbiol 2004;53:1279-90. CrossRef
18. Pena RT, Blasco L, Ambroa A, Gonzalez-Pedrajo B, Fernandez-Garcia L, Lopez M, et al. Relationship between quorum sensing and secretion systems. Front Microbiol 2019;10:1100. CrossRef
19. Hauser AR. The Type III secretion system of Pseudomonas aeruginosa: Infection by injection. Nat Rev Microbiol 2009;7:654-65. CrossRef
20. McMackin EA, Djapgne L, Corley JM, Yahr TL. Fitting Pieces into the Puzzle of Pseudomonas aeruginosa Type III secretion system gene expression. J Bacteriol 2019;201:e00209-19. CrossRef
21. Memariani H, Memariani M, Ghasemian A. An overview on anti-biofilm properties of quercetin against bacterial pathogens. World J Microbiol Biotechnol 2019;35:143. CrossRef
22. Liu L, Xiao X, Li K, Li X, Shi B, Liao X. Synthesis of catechin-rare earth complex with efficient and broad-spectrum anti-biofilm activity. Chem Biodivers 2020;17:e1900734. CrossRef
23. Deryabin D, Galadzhieva A, Kosyan D, Duskaev G. Plant-derived inhibitors of AHL-mediated quorum sensing in bacteria: Modes of action. Int J Mol Sci 2019;20:5588. CrossRef
24. Borlinghaus J, Bolger A, Schier C, Vogel A, Usadel B, Gruhlke MC, et al. Genetic and molecular characterization of multicomponent resistance of Pseudomonas against allicin. Life Sci Alliance 2020;3:e202000670. CrossRef
25. Satapathy P, Prakash JK, Gowda VC, More SS, Chandramohan V, Zameer F. Targeting Imd pathway receptor in Drosophila melanogaster and repurposing of phyto-inhibitors: Structural modulation and molecular dynamics. J Biomol Struct Dyn 2020;10:1-12.
26. Meghashri S, Kumar HV, Gopal S. Antioxidant properties of a novel flavonoid from leaves of Leucas aspera. Food Chem 2010;122:105-10. CrossRef
27. Prasad A, Devi AT, Prasad MN, Zameer F, Shruthi G, Shivamallu C. Phyto anti-biofilm elicitors as potential inhibitors of Helicobacter pylori. 3 Biotech 2019;9:53. CrossRef
28. Shameh S, Hosseini B, Alirezalu A, Maleki R. Phytochemical composition and antioxidant activity of petals of six Rosa species from Iran. J AOAC Int 2018;101:1788-93. CrossRef
29. Nedamani ER, Mahoonak AS, Ghorbani M, Mehdi K. Evaluation of antioxidant interaction in combined extracts of green tea (Camellia sinensis), rosemary (Rosmarinus officinalis), and oak fruit (Quercus branti). J Food Sci Technol 2015;52:4565-71. CrossRef
30. Borucki MK, Peppin JD, White D, Loge F, Call DR. Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol 2003;69:7336-42. CrossRef
31. Djordjevic D, Wiedmann M, McLandsborough LA. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol 2002;68:2950-8. CrossRef
32. Lakshmi VR, Begum AB, Naveen P, Zameer F, Hegdekatte R, Khanum SA. Synthesis, xanthine oxidase inhibition, and antioxidant screening of benzophenone tagged thiazolidinone analogs. Arch Pharm 2014;347:589-98. CrossRef
33. Satapathy P, Khan K, Devi AT, Patil AG, Govindaraju AM, Gopal S, et al. Synthetic gutomics: Deciphering the microbial code for futuristic diagnosis and personalized medicine. Methods Microbiol 2019;46:197-225. CrossRef
34. Rakesh SK, Jagadish S, Swaroop TR, Mohan CD, Ashwini N, Harsha KB, et al. Anti-cancer activity of 2, 4-disubstituted thiophene derivatives: Dual inhibitors of lipoxygenase and cyclooxygenase. Med Chem 2015;11:462-72. CrossRef
35. Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun 2004;72:6969-77. CrossRef
36. Rakesh SK, Jagadish S, Balaji KS, Zameer F, Swaroop TR, Mohan CD, et al. 3, 5-disubstituted isoxazole derivatives: Potential inhibitors of inflammation and cancer. Inflammation 2016;39:269-80. CrossRef
37. Praveen KK, Zameer F, Murari SK. Delineating antidiabetic proficiency of catechin from Withania somnifera and its Inhibitory action on dipeptidyl peptidase-4 (DPP-4). Biomed Res 2018;29:1-9. CrossRef
38. Ramu R, Shirahatti PS, Zameer F, Ranganatha LV, Prasad MN. Inhibitory effect of banana (Musa sp. var. Nanjangud rasa bale) flower extract and its constituents umbelliferone and lupeol on α-glucosidase, aldose reductase and glycation at multiple stages. S Afr J Bot 2014;95:54-63. CrossRef
39. Ramu R, Shirahatti PS, Zameer F, Dhananjaya BL, Prasad MN. Assessment of in vivo antidiabetic properties of umbelliferone and lupeol constituents of banana (Musa sp. var. Nanjangud Rasa Bale) flower in hyperglycaemic rodent model. PLoS One 2016;11:e0151135. CrossRef
40. Farhan Z, Gopal S. Transcriptional analysis of thiol disulfide redox metabolism (TDRM) genes in Listeria monocytogenes in biofilm and planktonic forms grown at different temperatures. Int J Pure Appl Sci 2010;4:21-7.
41. Beulah KC, Devi AT, Waseem K, Hedgekatte R, Prasad MN, Dhananjaya BL, et al. Phyto-antiquorumones: An herbal approach for blocking bacterial trafficking and pathogenesis. Int J Pharm Pharm Sci 2015;7:27-34.
42. Prasad A, Baker S, Prasad MN, Devi AT, Satish S, Zameer F, et al. Phytogenic synthesis of silver nanobactericides for anti-biofilm activity against human pathogen H. pylori. SN Appl Sci 2019;1:341. CrossRef
43. Khodayary R, Nikokar I, Mobayen MR, Afrasiabi F, Araghian A, Elmi A, et al. High incidence of Type III secretion system associated virulence factors (exoenzymes) in Pseudomonas aeruginosa isolated from Iranian burn patients. BMC Res Notes 2019;12:28. CrossRef
44. Lahiri D, Nag M, Dutta B, Mukherjee I, Ghosh S, Dey A, et al. Catechin as the most efficient bioactive compound from Azadirachta indica with antibiofilm and anti-quorum sensing activities against dental biofilm: An in vitro and in silico study. Appl Biochem Biotechnol 2021;193:1617-30. CrossRef
45. Ramu R, Shirahatti PS, Dhanabal SP, Zameer F, Dhananjaya BL, Prasad MN. Investigation of antihyperglycaemic activity of banana (Musa sp. Var. Nanjangud rasa bale) flower in normal and diabetic rats. Pharmacogn Mag 2017;13 Suppl 3:S417. CrossRef
46. Putta S, Yarla NS, Kilari EK, Surekha C, Aliev G, Divakara MB, et al. Therapeutic potentials of triterpenes in diabetes and its associated complications. Curr Top Med Chem 2016;16:2532-42. CrossRef
47. Aishwarya S, Kounaina K, Patil AG, Satapathy P, Devi AT, Avinash MG, et al. Nutraceutical attributes of Tamarindus indica L. Devils’ tree with sour date. Science of spices and culinary herbs latest laboratory, pre-clinical, and clinical studies. Bentham Sci Int 2020;33:33-65. CrossRef
48. Khan K, Aishwarya S, Satapathy P, Veena SM, Melappa G, Zameer F, et al. Exploration of dill seeds (Anethum graveolens): An ayurpharmacomic approach. In: Science Spices and Culinary Herbs-latest Laboratory, Pre-clinical, and Clinical Studies. Vol. 2. Bentham Sci Int: Singapore; 2020. p. 116-52. CrossRef
49. Satapathy P, Aishwarya S, Shetty MR, Simha NA, Dhanapal G, Shree RA, et al. Phyto-nano-antimicrobials: Synthesis, characterization, discovery, and advances. Front Antiinfect Drug Discov 2020;8:196. CrossRef