1. INTRODUCTION
Biosurfactants are amphipathic molecules produced by various microorganisms including bacteria, fungi, and yeast as part of the cell membrane or extracellular [1]. Biosurfactants have hydrophilic and hydrophobic domains that locate themselves preferentially at the interface between fluids of different phase polarity. The biosurfactants are resistant to environmental factors including temperature, pH, and salinity. Furthermore, they are biodegradable and less toxic [2]. Surfactant-like properties of biosurfactants make them the candidate for detergent and emulsifying agents [3]. Lesser toxicity for humans imparts various biological activities including anti-adhesion, antiviral, anti-cancer, and antimicrobial activities [4].
Recently, an eco-friendly biological method called green chemistry or green synthesis is widely used as an alternative approach to synthesize metal-based nanoparticles [5]. Ecofriendly way of nanoparticle biosynthesis has become the alternative for chemical or physical synthesis [6]. Distinguishing the stand green synthesis of nanoparticles, properties like excellent stabilizer and modifier researchers are using biosurfactant as a capping agent [7]. Biosurfactants get adsorbed on the surface of nanoparticles of metal origin preventing aggregation, distributing evenly, and maintaining nano-sized upon longer storage [1]. Different strains of Pseudomonas sp. have produced various glycolipid type biosurfactants reporting as antimicrobial properties inhibiting the growth of bacteria, mold, and fungi of pathogen origin [8]. The research communication reported the use of biosurfactant produced by newly isolated Pseudomonas aeruginosa as a capping agent for the biosynthesis of silver nanoparticles (AgNPs). AgNPs have properties like high solubility, stability, and good electronic and optical or photoelectrochemical activities. In addition, high surface-area-to-volume ratio and their quantum confinement made AgNPs an excellent nanomaterial. AgNPs can form spherical, rod, plate, triangular, and pyramidal-shaped nanoparticles with strong antimicrobial activity against multi-drug resistant bacteria and various viruses such as human immune virus and influenza [9].
2. MATERIALS AND METHODS
2.1. Microorganism Used as a Source of Biosurfactant
The P. aeruginosa strain LTR1 (European Molecular Biology Laboratory Nucleotide Sequence Database MF319551) used in throughout the report was previously isolated from the region of oil-contaminated soil of Latur (Maharashtra, India). The bacterial inoculum prepared in nutrient broth was transferred in minimal salt medium (MSM) broth containing 2% soybean oil as a carbon source to produce biosurfactant in batch culture [10]. The flask was incubated at 37°C for 96 hours at 150 rpm minute−1. After the completion of the incubation period, the biosurfactant was extracted using the acid precipitation method. The crude fraction was applied on silica gel G-500 column and fractions were collected by using chloroform: methanol (50:5) as elution solution. Fraction showing positive drop collapse test was dried and used for AgNPs synthesis.
2.2. Synthesis of Biosurfactant Based AgNPs
In-situ in water-in-oil microemulsion method was utilized with slight modification for biosynthesis of nanoparticles in the presence of biosurfactant [11]. 1 mg/ml biosurfactant was dissolved in25 ml n-heptane and 1.0 ml of aqueous 0.05 M AgNO3 solution with vigorous stirring for 10 minutes at room temperature. Another solution system ran simultaneously replacing AgNO3, with an equal volume of 0.1 M aqueous sodium borohydride (NaBH4) solution. An equal volume of these two reverse micelles solutions was further mixed with continuous stirring for 60 minutes. The homogeneous mixture was centrifuged at 14,000 rpm for 20 minutes to precipitate AgNPs. The precipitate was dissolved in 10 ml n-butanol and kept in a refrigerator until further use.
2.3. Characterization of Silver Nanoparticle
2.3.1. UV-visible spectrophotometer analysis
The absorption spectrum for synthesized AgNPs was recorded on UV-visible spectrophotometer (Remi, Model No. 2002). The solution was filled in 1 cm optical path quartz cuvette and recorded the spectra in the wavelength range of 300 to 800 nm.
2.3.2. Determination of stability of biosurfactants based nanoparticles
For the determination of stability of synthesized AgNPs change in UV visible spectrum was monitored for 45 days. In brief, AgNPs solution of pH 7 kept at room temperature was sequestered for a spectrum in the range of 300–800 nm on UV-visible spectrophotometer (Remi, Model No. 2002) at room temperature.
2.3.3. X-ray diffraction (XRD) analysis
XRD spectrum for biosurfactant-based AgNPs was recorded on a powder X-ray diffractometer (Goniometer Ultima IV), equipped with K-beta filter operated at voltage 40 kV and a current of40 mA, Cu-K_ X-rays of wavelength (λ = 0.1541 nm) with a crystal monochromator. The data were collected in the 2θ/θ range of 10°–80°, with a step size of 0.01° and at a continuous scan rate. Crystalline nature and mean diameter of AgNPs were resolved by standard powder diffraction card of International Centre for Diffraction Data-Joint Committee on Powder Diffraction Standards (ICDD-JCPDS) Ag pattern Silver file No. 96-110-0137 and Match! 3 open-source tool for XRD analysis
2.3.4. Scanning electron microscope analysis (SEM)
The size and morphological topographies of synthesized AgNPs were further confirmed by SEM photographs. For dis-aggregation of nanoparticles, the solution was sonicated on a probe sonicator (Sonics Vibra-Cell, USA). The homogeneous solution was applied on cleaned and grease-free glass discs of 0.3-0.5 cm diameter. Discs were air-dried and utilized for field emission SEM (FEI, Nova Nano SEM 450, and FESEM software) photography. Since field emission (FE)-SEM was equipped with energy dispersive spectroscopy [energy dispersive X-ray spectroscopy (EDX) by Bruker].
2.3.5. Electro spray ionization-mass spectroscopy (ESI-MS)
The purified fraction of biosurfactant dissolved in methanol was analyzed by ultra performance liquid chromatography (Dionex Ultimate 3000 system) coupled with high-resolution mass spectrometry (HRMS) (Impact II UHR-TOF Mass Spectrometer System, Bruker) with an electrospray ionization source. The capillary voltage was operated in positive ionization mode, with fragmentor voltage set at 4,500 V, and with an endplate offset potential of −500 Volt. The dry gas parameter was set to 7.0 l/minute at 200°C with a nebulization gas pressure of 1.7 bars. Data were collected from the scanning range at 50–1,200 m/z. The mobile phase of acetonitrile: water gradient (10%–90%) with 0.01% formic acid was used in the column. Data obtained were analyzed with Bruker compass Data Analysis 4.2 Software (Bruker: X Flash 6130 and Espirit 1.9 software), elemental analysis was also carried out.
2.4. Antimicrobial Activity of Biosurfactant and Biosurfactant Based AgNPs
The antimicrobial activity of purified biosurfactant and AgNPs was assessed against various reference strains of fungal and bacterial pathogens using the disc diffusion method. The overnight grown cultures of Escherichia coli (NCIM 2832), Proteus vulgaris (NCIM 2813), Salmonella typhimurium (NCIM 2501), Bacillus subtilis (NCIM 2635), Staphylococcus aureus (NCIM 2654) and Candida albicans (NCIM 3466) were inoculated on Mueller-Hinton and yeast extract peptone dextrose (YPD) Agar plates [12].
The concentration of biosurfactant 10 μg/disc used for the preparation of discs (6 mm). The 100 μl solutions of biologically synthesized AgNPs were used for the preparation of discs and.6 mm discs were prepared from 100 μl of 10 μg/disc of 0.5 M pure silver nitrate and used as a negative control. Discs were placed at the center of Mueller-Hinton and YPD agar plates inoculated with test organisms within 15 minutes. Standard antimicrobial discs of ampicillin [Clinical Laboratory Services (CLS) A 10 μg/disc) and amphotericin B (CLS AB 10 μg/disc) were used as a positive control. After completing the diffusion of procedure plates were incubated at 37°C. Plates were further observed/measured for the zone of inhibition (ZOI) by ruler [13]. The means of three consecutive ZOI for each sample observation were determined and tabulated.
3. RESULTS AND DISCUSSION
The P. aeruginosa strain LTR1 isolated from oil-contaminated soil of Latur region utilized for the production of a biosurfactant in optimized MSM with 2% soybean oil as a sole source of carbon. The produced biosurfactant characterized by using HRMS indicated the presence of glycolipids congeners. The most abundant pair of mono-rhamnolipid and di-rhamnolipid congeners was Rha-Rha-C10-C10 and 2 (Rha-C10-C10) at 673.85 and 1,031.25 m/z, respectively (unpublished data). Several reports indicate that microbes isolated from oil-contaminated zones are prone to degrade hydrocarbons as they are naturally tuned for hydrocarbon degradation. IsolateP. aeruginosa strain SNP0614 isolated from the oil-contaminated site of dagang from china was efficient in the production of biosurfactant utilizing crude oils and has good emulsification activity [14]. In another report, P. aeruginosa strain MKVIT3 produced a mixture of two rhamnolipid congeners including di-rhamnolipid Rha-Rha-C10-C12 and mono-rhamnolipid Rha-C10-C8 [15].
3.1. Synthesis of AgNPs
In the present study reports, the biosynthesis of AgNPs utilizing modified microemulsion phase method and capping ability biosurfactants is discussed [11]. The bottom-up approach was used for nanoparticle synthesis. In this approach, the first phase of the microemulsion was prepared by mixing biosurfactant in n-heptanes along with an aqueous AgNO3 solution. The vigorous mixing was carried out to make a homogeneous mixture of reverse micelles at room temperature. The second phase was prepared replacing aqueous AgNO3 solution with aqueous NaBH4 was used instead of aqueous AgNO3. Mixing of first and second microemulsions precipitated the AgNPs. Nanoparticles synthesis monitored the changing color from colorless to yellow-brown and remained stable for at least 24 hours. Color change indicates the formation of nanoparticles due to a shift in surface plasmon vibration of AgNPs.
Xie et al. [11] reported that the rhamnolipid which is amphipathic had obscurity to form microemulsion in water and n-heptane. For the formation of the stable microemulsion, it required a co-surfactant of varying chain length of alcohol which affects phase formation. The use of a linear chain of alcohol such as n-butanol along with microemulsion showed the formation of reverse micelles which had greater stability.
The present study reports the use of a modified microemulsion technique able to form a pseudo ternary system which can form monolayer microemulsion having hydrophilic and lipophilic solubility. Thus, it indicates the rhamnolipid used in this study that served as a stabilizing agent for the synthesis of AgNPs. The use of extracellular synthesis of AgNPs is a more expedient, simple, and easy method as compared to intracellular. It minimizes the various steps involved in the purification of AgNPs such as the ultra-sonication cycle to break down the cell and centrifugation [16].
3.2. Characterization of AgNPs
3.2.1. UV-visible spectrophotometer analysis
The qualitative examination of AgNPs was checked by measuring the optical properties of synthesized AgNPs on UV-visible spectrophotometer (Remi, Model No. 2002). The nonmetallic dispersion of synthesized AgNPs was preliminary monitored by measuring surface Plasmon resonance (SPR) at the range of 300–800 nm. The formation of AgNPs showed a broad and strong peak at 420–440 nm (Fig. 1). The time-dependent qualitative analysis for SPR showed strong absorption at 430 nm after 3 hours of synthesis.
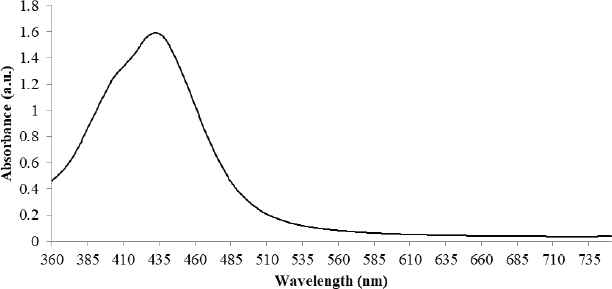 | Figure 1: UV-visible absorption spectra of AgNPs synthesized from column purified biosurfactant produced by P. aeruginosa strain LTR1. [Click here to view] |
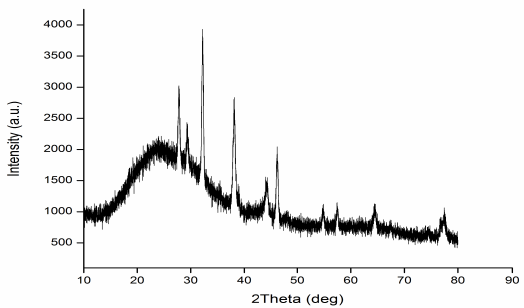 | Figure 2: XRD of AgNPs synthesized from column purified fractions of rhamnolipid produced by P. aeruginosa strain LTR1. [Click here to view] |
3.2.2. XRD analysis of biosurfactants based AgNPs
The crystal symmetry or shape of biosurfactants-based AgNPs showed a cubic reflection of silver. The 2θ values of four peaks from biosurfactants based nanoparticles were 38.15°, 44.35°, 64.52°, and 77.49° which corresponds to (111), (200), (202), and (311) planes of silver (Fig. 2).
Bragg’s law equation determined a strong peak of reflection for synthesized AgNPs matched with face-centered cubic (FCC) symmetry of pure crystal of silver metal. The miller indices (hkl) 111 showed by nanoparticle sample was an indicative high intense peak for FCC for nano-materials. Strong intensity peaks in the XRD spectra of AgNPs indicate the high crystalline nature of nanoparticles.
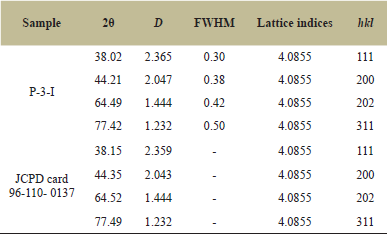 | Table 1: XRD Peak indexing of AgNPs and standard JCPD card 96-110- 0137. [Click here to view] |
The experimental diffraction angles for biosurfactant-based nanoparticles were 38.02° which showed a very negligible difference from the standard JCPDS card which has an angle of diffraction of 38.15° (Table 1). The lattice indices constant and particle density for experimental data were calculated from the most intense peak (111) of the XRD pattern which was 4.0855 Å and matching with the standard ICDD-JCPDS card. The figure of the mean (FoM) obtained XRD data of synthesized AgNPs was 0.821065 which was matching with the FoM of 0.776378 of standard (JCPDS) card. The data of XRD could validate when the value of FoM of experimental data was more than 0.60. The size of the Ag nanoparticles was determined from diffraction peaks using Debye–Scherrer’s Formula (D = 0.9λ/β Cos θ). The average particle size was 34.53 nm.
3.2.3. SEM and EDX of nanoparticles
Surface morphology, size, and elemental composition of synthesized nanoparticles were determined by FE- SEM and EDX. The size and shape of the synthesized nanoparticles depend on the structure of biosurfactants [17].
The nanoparticle shapes revealed by SEM were spherical indicative of highly crystalline and mono-dispersive (Fig. 3A). The particle size was revealed to be 36.13 nm, matching with the size calculated from XRD data. In the investigation, the morphological surface of biosurfactant-based AgNPs a small-sized spherical particle or stacking of blocks of particles with the island-sea type surface were observed.
The energy dispersive spectrometry was utilized for compositional analysis of elements in the nanoparticles. AgNPs spectral analysis showed a strong and intense peak of silver signals at 3 ke V reflected the presence of silver, carbon, and oxygen (Fig 3B and C). The quantitative data are noted in the table. An abundance of carbon and oxygen in the data revealed the existence of carbohydrate and lipid complex moiety in the nanoparticle from the biosurfactant.
 | Figure 3: A) SEM of AgNPs B) Energy dispersive X-ray analysis of AgNPs C) Quantitative distribution of elements. [Click here to view] |
Jeyaraj et al. [18] reported the synthesis of AgNPs utilizing the spent medium of P. aeruginosa culture. There was a shift in color from colorless to brown with maximum absorbance at wavelength 430–450 nm. The intense peak of SPR of AgNPs was observed at 430 nm [18]. The AgNPs synthesized from the supernatant culture of Stenotrophomonas and Pseudomonas showed maximum absorption in the range of 430–450 nm, with an intense peak at 440 nm after 72 hours. of incubation [19].
Thus, P. aeruginosa is a unique bacterial species ever reported to synthesize AgNPs in a shorter time like in 5 minutes. To understand the stability of the synthesized nanoparticles, AgNPs solution was subjected to UV-visible spectrometer to deduce the change in absorption at 430–450 for every hour. Three hours data didn’t show an increase in absorption indicating reverse micelles formed were highly stable [20].
The SEM data of AgNPs biosynthesized utilizing spent medium of Pseudomonas putida MVP2 were spherical, homo dispersive with size distribution in the range of 6 to 16 nm. The energy-dispersive XRD pattern reflects the composition containing silver, the highest peak of intensity at 3ke V [21].
3.2.4. Characterization of biosurfactants by ESI-MS
The column chromatography purified fraction of the biosurfactant was subjected to positive ion mode (M+Na+) ESI-MS to determine the accurate molecular mass and to identify the compounds. The mMass v5.5.0 software was used for the identification of glycolipid where m/z values can be compared with the library of PubChem. Up to eight homologous congeners of rhamnolipid produced by P. aeruginosa strain LTR1 carried out at 100 V by collision-induced dissociation in the scanning range of 200–1,200 m/z which explain the fatty acyl chain length, presence of rhamnose residue, units, and their structure, relative abundance as well as position in the identified rhamnolipid biosurfactants [22]. It indicates the presence of homologous congeners of rhamnolipid produced by P. aeruginosa strain LTR1 with molecular weight between 1,031.87 and 315.25. The most abundant pair of mono rhamnolipid and di-rhamnolipid congeners was Rha-Rha-C10-C10 and 2(Rha-C10-C10) at 673.85 and 1,031.25 m/z respectively (Fig. 4).
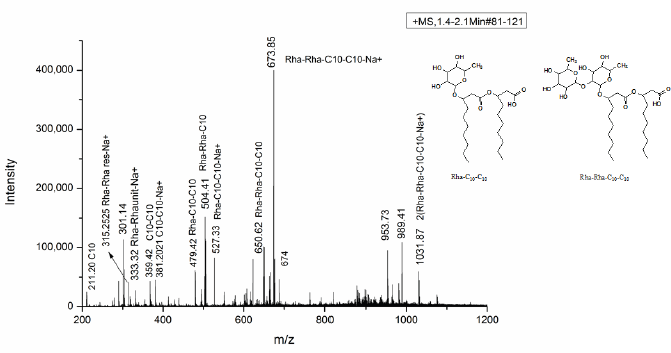 | Figure 4: ESI-MS of column purified biosurfactant produced by P. aeruginosa strain LTR1 using soybean oil as the sole carbon source. [Click here to view] |
The di-rhamnolipid at 673.85 m/z of Na+ adduct of Rha-Rha-C10-C10 showed cleavage and form two fragments with loss of terminal fatty acid chain of C10 and two rhamnose residues (292 Da) with subsequent formation of the ion at 527.33 m/z of Rha-Rha-C10 and 381.20 m/z of C10-C10, respectively. The peak of fragmentation showed product ion at peak value 315.25, confirmed linkage and presence of di-rhamnose ion of 146 Da with their Na+ adduct. The possible fragmentation also yields product ion at 650.54 m/z of Rha-Rha-C10-C10, at 504.41 m/z of Rha-Rha-C10, and 357 m/z Rha-C10 due to loss of terminal fatty acid chain of –C10H20O3 (170 Da), rhamnose ion (146 Da) along with fatty acid loss (170 Da) and loss of two rhamnose unit along with one terminal fatty acid yield fragment of C-10 at 211.20 m/z [23].
The ESI-MS spectrum of mono rhamnolipid showed two (Rha-C10-C10) at 1,030.25 m/z. The intense peak of mono rhamnolipid at 479.42 m/z indicates the presence of monorhamnolipid of Rha-C10-C10 due to the rupture of ester linkages between rhamnolipid. The ion product presence at 333.32 m/z indicated the release of one terminal fatty acid of C10 and the congeners were Rha-C10:1. Structural analysis of rhamnolipid fraction P-II produced by LTR1 showed an intense peak of the sodiated adduct of di-rhamnolipid of Rha-Rha-C10-C10 with their major relative abundance of Rha-Rha-C10-C10 (25.16%), Rha-Rha-C10 (37.97%), and mono rhamnolipid of Rha-C10-C10 (20.61%), 2 molecules of Rha-C10-C10 (14.89%) with less abundance of di- rhamnolipid unit (16.07%), as well as free fatty acid (6.28%) with the length of C10 totally, were found in the purified biosurfactant. The structure of identified rhamnolipid was drawn by using chem Draw ultra 8.0.
3.3. Antimicrobial Activity of Purified Biosurfactant and Their AgNPs
The column purified fraction showed higher antimicrobial activity against standard strains of bacterial human pathogens (Fig. 5). The maximum ZOI was recorded against S. aureus (17 mm), followed by E. coli (16 mm) and minimum ZOIs were observed for B. subtilis and P. vulgaris (11 mm). It also showed higher antifungal activity against standard strains of C. albicans. The values of the ZOI were recorded against C. albicans (19 mm). On the other hand, biosurfactant-based nanoparticles were shown to enhance antimicrobial activity compared to the only biosurfactant. AgNPs showed growth inhibition with significant ZOI (values in brackets) E. coli and S. typhi (18 mm), P. vulgaris (20 mm). S. aureus (19 mm), and B. subtilis (16 mm). Synthesized AgNPs also inhibited the growth of C. albicans, the opportunistic fungal pathogen of humans with the19 mm ZOI at a concentration of 10 μg/disc [24].
4. CONCLUSION
Pseudomonas aeruginosa was found to be an excellent biosurfactant producer in a sufficiently large amount. The omnipresent nature of degradation of various carbon skeletons ranging from hydrocarbons, xenobiotic compounds, and complex carbohydrates like cellulose can be utilized for removing waste and generating value-added products like biosurfactants. Pseudomonas aeruginosa strain LTR1 has been successfully utilized to produce biosurfactants and synthesis of biosurfactant-based AgNPs. Biosurfactant itself showed antimicrobial activity against human pathogens like E. coli, P. vulgaris, S. typhi, B. subtilis S. aureus, and C. albicans. Furthermore, antimicrobial activities were found enhanced for the biosurfactant-based AgNPs. Cytotoxic effect of biosurfactant and its nanoparticles against animal cells work is in progress.
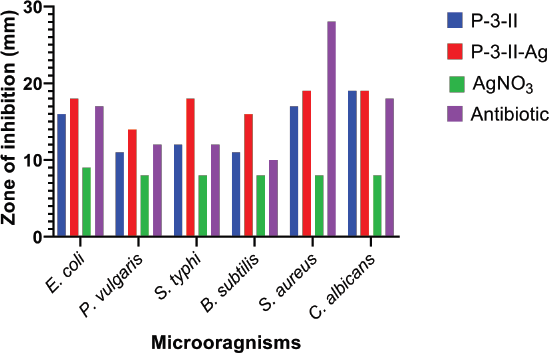 | Figure 5: ZOI of purified biosurfactant and its AgNPs against bacterial and fungal human pathogens. [Click here to view] |
5. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
6. FUNDING
There is no funding to report.
7. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
REFERENCES
1. Chen CY, Baker SC, Darton RC. The application of a high throughput analysis method for the screening of potential biosurfactants from natural sources. J Microbiol Methods 2007;70(3):503–10; http://doi.org/10.1016/j.mimet.2007.06.006
2. Eswari JS, Dhagat S, Mishra P. Biosurfactant assisted silver nanoparticle synthesis: a critical analysis of its drug design aspects. Adv Nat Sci Nanosci Nanotechnol 2018;9(4); http://doi.org/10.1088/2043-6254/aaec0e
3. Greek BF. Sales of detergents growing despite recession. Chem Eng News 1991;69(4):25–52; http://doi.org/10.1021/cen-v069n004.p025
4. Rodrigues L, Banat IM, Teixeira J, Oliveira R. Biosurfactants: potential applications in medicine. J Antimicrob Chemother 2006;57(4):609–18; http://doi.org/10.1093/jac/dkl024
5. Joanna C, Marcin L, Ewa K, Gra?yna P. A nonspecific synergistic effect of biogenic silver nanoparticles and biosurfactant towards environmental bacteria and fungi. Ecotoxicology 2018;27(3):352–9; http://doi.org/10.1007/s10646-018-1899-3
6. Thakkar KN, Mhatre SS, Parikh RY. Biological synthesis of metallic nanoparticles. Nanomedicine 2010;6(2):257–62; http://doi.org/10.1016/j.nano.2009.07.002
7. Kiran GS, Sabu A, Selvin J. Synthesis of silver nanoparticles by glycolipid biosurfactant produced from marine Brevibacterium casei MSA19. J Biotechnol 2010;148(4):221–5; http://doi.org/10.1016/j.jbiotec.2010.06.012
8. Reddy AS, Chen CY, Baker SC, Chen CC, Jean JS, Fan CW, et al. Synthesis of silver nanoparticles using surfactin: a biosurfactant as stabilizing agent. Mater Lett 2009;63(15):1227–30; http://doi.org/10.1016/j.matlet.2009.02.028
9. Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv 2009;27(1):76–83; http://doi.org/10.1016/j.biotechadv.2008.09.002
10. Deshmukh N, Kadam D. Isolation, characterization and identification of biosurfactant producing bacteria from oil contaminated soil. Int J Recent Sci Res 2019;10(11):35896–901.
11. Xie Y, Ye R, Liu H. Synthesis of silver nanoparticles in reverse micelles stabilized by natural biosurfactant. Colloids Surf A Physicochem Eng Aspects 2006;279(1–3):175–8; http://doi.org/10.1016/j.colsurfa.2005.12.056
12. Cavalieri SJ, Harbeck RJ, McCarter YS, Ortez JH, Rankin ID, Sautter RL, et al. Manual of antimicrobial susceptibility testing. American Society for Microbiology, Pan American Health Organization, Washington, DC, 2005.
13. Bauer AW, Kirby WMM, Sherris JCT, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 1966;45(4):493–6. CrossRef
14. Liu WJ, Duan XD, Wu LP, Masakorala K. Biosurfactant production by Pseudomonas aeruginosa SNP0614 and its effect on biodegradation of petroleum. Appl Biochem Microbiol 2018;54(2):155–62; http://doi.org/10.1134/S0003683818020060
15. Das M, Patowary K, Vidya R, Malipeddi H. Microemulsion synthesis of silver nanoparticles using biosurfactant extracted from Pseudomonas aeruginosa MKVIT3 strain and comparison of their antimicrobial and cytotoxic activities.IET Nanobiotechnol 2016;10(6):411–8; http://doi.org/10.1049/iet-nbt.2015.011916.
16. Singh P, Ravindran S, Suthar JK, Deshpande P, Rokhade R, Rale V. Production of biosurfactant stabilized nanoparticles. Int J Pharma Bio Sci 2017;8(2):701–7; http://doi.org/10.22376/ijpbs.2017.8.2.b701-707
17. Das R, Ali E, Abd Hamid SB. Current applications of X-ray powder diffraction-a review. Rev Adv Mater Sci 2014;38(2):95–109.
18. Jeyaraj M, Varadan S, Anthony KJP, Murugan M, Raja A, Gurunathan S. Antimicrobial and anticoagulation activity of silver nanoparticles synthesized from the culture supernatant of Pseudomonas aeruginosa. J Ind Eng Chem 2013;19(4):1299–303; http://doi.org/10.1016/j.jiec.2012.12.031
19. Malhotra A, Dolma K, Kaur N, Rathore YS, Ashish, Mayilraj S, et al. Biosynthesis of gold and silver nanoparticles using a novel marine strain of Stenotrophomonas. Bioresour Technol 2013;142:727–31; http://doi.org/10.1016/j.biortech.2013.05.109
20. Tade RS, Nangare SN, Patil PO. Agro-industrial waste-mediated green synthesis of silver nanoparticles and evaluation of its antibacterial activity. Nano Biomed Eng 2020;12(1):57–66. CrossRef
21. Gopinath V, Priyadarshini S, Loke MF, Arunkumar J, Marsili E, MubarakAli D, et al. Biogenic synthesis, characterization of antibacterial silver nanoparticles and its cell cytotoxicity. Arab J Chem 2017;10(8):1107–1; http://doi.org/10.1016/j.arabjc.2015.11.011
22. Behrens B, Engelen J, Tiso T, Blank LM, Hayen H. Characterization of rhamnolipids by liquid chromatography/mass spectrometry after solid-phase extraction. Anal Bioanal Chem 2016;408(10):2505–14. CrossRef
23. Pereira JF, Gudiña EJ, Dória ML, Domingues MR, Rodrigues LR, Teixeira JA, et al. Characterization by electrospray ionization and tandem mass spectrometry of rhamnolipids produced by two Pseudomonas aeruginosa strains isolated from Brazilian crude oil. Eur J Mass Spectrom 2012;18(4):399–406. CrossRef
24. Kulkarni P, Chakraborty R, Chakraborty S. Biosurfactant mediated synthesis of silver nanoparticles using Lactobacillus brevis (MTCC4463) and their antimicrobial studies. IJPSR 2019;25:1753–9.