1. INTRODUCTION
Acute kidney injury (AKI) is a disease that concerns nations on a global scenario [1]. It is a multiphasic disease that elevates the risk of acquiring chronic kidney disease (CKD) or end-stage renal disease (ESRD) [2]. CKD is often characterized by a renal function abnormality continuously for at least a period of 3 months with an expression of abnormal renal bio-markers together with a decline in glomerular filtration rate (GFR) (<60 ml/minute per 1.73 m2) [3]. Obstructive nephropathy is one of the frequent clinical causes that is attributed to this damage. Irrespective of the underlying root cause, the pathological outcome of CKD is generally observed as the renal fibrosis that accentuates towards ESRD, including obstructive nephropathy [4].
Hemodialysis and kidney transplantation are considered as the most effective treatment strategies for CKD patients [5]. Being an overpriced course of action, the highest rates of patients are being kept away from availing the benefit of the current treatment. To confront such obstacles, highly promising therapeutic strategies have been applied to treat CKD due to the advancement of technology in the medical field. New interventional drugs like angiotensin enzyme inhibitors, sodium glucose transport inhibitors, can somehow delay the progression of the disease but are associated with troublesome side effects and the prevalence of CKD continues to escalate [6]. Therefore, with a view to serve CKD, it is necessary to penetrate into its molecular mechanism. These molecular insights are pivotal in contributing efficacious therapeutic targets for CKD patients.
Through intensive research, it has now been untangled that besides many other altered processes, CKD is often marred by inflammatory responses leading to the commencement of cytokine and chemokines secretion [4]. Apart from inflammation, apoptosis is equally responsible for damaging the kidney and intensifying CKD development [7]. Moreover, the initial phases of fibrosis are a consequence of a complicated inflammatory process and is considered as the main spark for pathogenic fibrosis.
Unfortunately, the exact mechanism by which inflammation affects CKD requires further elucidation for in depth understanding of the whole process together with finding potential solutions to disease prognosis and diagnosis [8]. Several reports have suggested that NF-kB is shown to be a key transcription molecules in humans as well as animal models of CKD [9]. Consistent with the previous reports, our work now proposes that NF-kB has a much more complicated role, being a major regulator of inflammation and fibrosis in the pathology of CKD.
In the present work, we have therefore adopted a bioinformatics approach to decode the biomolecules that can be utilized for the prognosis of the disease progression. Additionally, to corroborate, bioinformatics findings were therefore validated through an in vivo approach employing a rat model of chronic kidney injury, i.e., unilateral ureter obstruction (UUO).
2. METHODS
The present work was executed by following the methodology (Fig. 1).
2.1. In Silico Studies
2.1.1. Retrieval of rat CKD genes from national centre for biotechnology information (NCBI)
Now if we investigate the pathophysiology of CKD, Inflammation, apoptosis, and fibrosis are the major consequences of CKD regardless of the disease aetiology. Consequently, finding genes and their links allying chronic kidney injury and its main processes, viz. inflammation, apoptosis, and fibrosis becomes foremost to curb CKD, which has hardly been elucidated. Therefore, the NCBI which is an international reserve for the scientific examination of biological data was employed.
NCBI was used to retrieve the list of all the genes related to CKD for rat kidneys using keywords “CKD genes”, “Inflammatory genes in CKD”, “Apoptotic genes in CKD”, “Genes for Fibrosis in CKD”, “EMT genes in CKD”, and secured in Microsoft Excel format.
2.1.2. Search tool for the retrieval of interacting genes (STRING) analysis
To establish the Protein Interaction Network (PIN) between the proteins/genes retrieved from NCBI, the STRING database (version 11.0) was employed. The reliability score of protein interaction was maintained at 0.400 (medium confidence). Interaction sources incorporated were text mining, experiments, databases, co-expression, neighborhood, gene fusion, and co-recurrence.
2.1.3. Retrieval of hub genes through cytoscape analysis
The network retrieved from STRING was then imported to Cytoscape (version 3.8.2). Using the molecular complex detection (MCODE), cluster analysis of the PIN was performed. The parameters for the same included: MCODE score ≥4, degree cut-off = 2, node score cut-off = 0.2, max depth = 100, and k-score =2. The top-scoring module was considered as the most significant one, which was further analyzed for topological characteristics like degree (k), betweenness centrality (BC), and maximum neighborhood component (MNC) by Cytoscape plugin cytohubba.
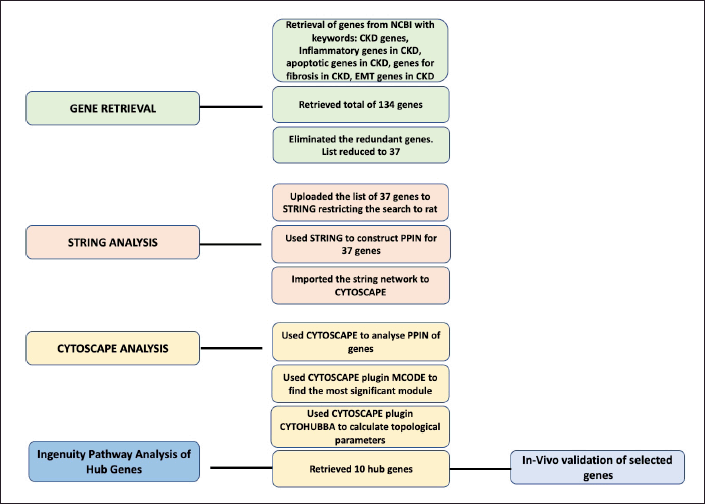 | Figure 1. Flowchart of the methodology employed to trace the significant genes involved in CKD. [Click here to view] |
Cytoscape plugin cytohubba scores each gene of a network by its topological measures based on the shortest path. In the present study, three major topological measures having both local as well as global influence, have been employed for scrutinizing hub genes viz. - Degree Centrality, BC and MNC. Genes having top 10 scores are recovered, each from above mentioned three topological measures, having a pattern of color signifying highly essential genes with red color. Among these genes, the top-ranked three genes were obtained by combining the results of the above three topological parameters viz. STAT-3, transforming growth factor alpha (TNF-α), and transforming growth factor beta (TGF-β) were then validated in vivo.
2.1.4. Qiagen ingenuity pathway analysis (IPA)
For further evaluation of the network, IPA (84978992) was used for observing any cross talk among the retrieved genes. The highest-ranked first 10 genes scrutinized from cytohubba were uploaded on Qiagen IPA software.
Core analysis of the hub genes was conducted within IPA. The IPA input was then further examined to create pathway networks aimed at identifying the primary upstream regulator among the targeted genes. Filters were applied in the upstream regulator results, specifically focussing on molecule types such as cytokines, enzymes, growth factors, nuclear receptors, transcription regulators, and transmembrane receptors. Following this filtering, the upstream regulator which targeted the remaining genes was shortlisted. Specific disease filters such as inflammation, apoptosis, and fibrosis were applied. Other parameters like species type, relation type, and model type were kept at their default settings for broader information search.
2.2. In Vivo Studies
2.2.1. Animal experimental design
The experimental work was accomplished on Sprague-Dawley rats weighing 200–250 g body weight. These were, procured from the Central Animal House of the Institute. The research proposal pursued was certified by the Institutional Animal Ethics Committee and executed following the regulations of Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA).
After 1 week’s adaptation, UUO surgery was performed under anesthesia using ketamine:xylazine (100 mg/kg:10 mg/kg) intraperitoneal injection. During the procedure, the fur from the surgical site of the animals was shaved off with a trimmer, disinfected and then they were positioned on the right lateral position on a warm pad during the course of surgery to preserve their body warmth. Through a left flank abdominal incision, the left ureter was exposed and completely ligated with a 5–0 polypropylene suture at two points and cut between the ligatures. The abdominal muscles were then sutured in layers using absorbable polyglactin suture. The skin was sutured using non-absorbable polypropylene suture. The surgical incision was cleaned with povidone-iodine solution and an aseptic dressing of the wound was done. The ureters of the Sham-operated rats were isolated but not ligated. The right kidney was left untouched.
The animals were sacrificed on postoperative days 10, 14, 16, 18, and 21 for standardization. The significant pathological changes, in the form of a dilated renal calyceal system, were found 14th day onwards. The renal fibrosis, however, was observed after the 18th postoperative day. Hence, the 14th and 18th postoperative days were selected for final analysis in the current study.
The animals were categorized into three groups established on the day of their sacrifice (n = 6 in each group): 1) Sham Group, 2) UUO 14 Group, and 3) UUO 18 Group. UUO 14 and UUO 18 group were further divided into right non-ligated contralateral kidney (RK) and left ligated kidney (LK) and were sacrificed on day 14 and day 18, respectively.
2.2.2. Evaluation of kidney injury
Evaluation of the kidney injury was performed by estimating the serum creatinine level which was carried out by employing a commercially available kit and the results were analysed using a spectrophotometer.
2.2.3. Histopathological examination
A histological examination of the kidney was carried out by fixing the kidney in 10% buffered formalin, for subsequent processing. Dehydration of the formalin-fixed tissues was then carried out in ascending grades of alcohol dilutions (30%, 50%, 70%, 90%, and 100%). The sections were cleaned in benzene and then submerged in paraffin. Microtome was used for cutting the paraffin-embedded tissues into fine sections. Following this, the hematoxylin and eosin (H and E) method was employed to stain the 5 µm thick sections of paraffin-embedded tissues. The images were then examined using a light microscope.
2.2.4. Total histological score (THS)
THS was estimated on account of degeneration, necrosis, and inflammation of the tubules. The formula used for the calculation of the histological score was—THS = TD/2+TN+TIN/2 where TD, tubular dilation; TN, tubular necrosis; TIN, tubulointerstitial inflammation. The score was marked regular between the range of 0–2, mild between 2 and 6, and above that range was considered as severe THS.
2.2.5. Quantitative real-time polymerase chain reaction (PCR)
Expression analysis of both the kidneys, right contralateral (RK) as well as left ligated (LK) was carried out by real time PCR utilizing SYBR Green master mix containing sequence-specific primers (Table 1). Data were analyzed by calculating the fold change through relative quantitation Comparative cycle threshold (CT) Method (??Ct) with GAPDH chosen as the internal control.
The reaction mixture was put together in thin-walled PCR tubes comprising of SYBER Green master mixture with forward and reverse primers, cDNA template, and nuclease free water. All the constituents were properly mixed before transferring to the PCR tray. The thermal cycles were programmed for cDNA synthesis followed by PCR amplification. The delta-delta Ct method (2-??Ct Method) also known relative Quantitation Comparative CT Method was applied in real—time PCR to calculate the relative fold change in gene expression, normalized to Ct value of the housekeeping gene.
The following formula was used to calculate the relative fold change: 2(-??Ct)
2.2.6. Immunohistochemistry
Paraffin-embedded renal sections were deparaffinized, rehydrated with descending grade of alcohol dilutions (100%, 90%, 70%, 50%, and 30%). After washing, blocking was achieved in 3% bovine serum albumin (BSA). Following blocking, incubation of the renal section was executed with the primary antibodies diluted in BSA using anti-rabbit serum against TNF-α with 1:500 dilutions and rabbit antibody to TGF-β with 1:500 dilution at 4?C overnight. The next day, goat anti-rabbit IgG secondary antibody conjugated with fluorescein isothiocyanate (FITC), diluted in BSA was used for incubation for 2 hours at room temperature. Following counterstaining the sections with 6-diamidino-2-phenylindole (DAPI), the slides were examined under a fluorescence microscope (Nikon Eclipse 80i).
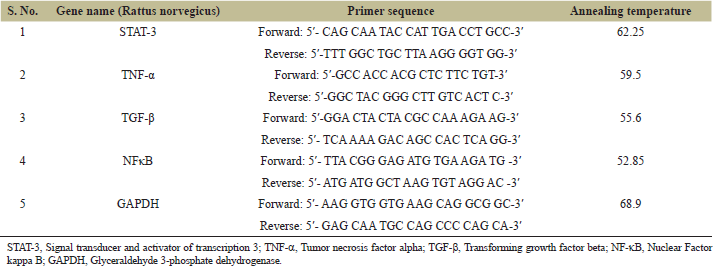 | Table 1. Primer sequences employed for the study with GAPDH as internal control. [Click here to view] |
Formalin-fixed, paraffin-embedded sections were initially heated and immersed in xylene for deparaffinization. The sections were rehydrated with descending grade of alcohol dilutions followed by immersing in distilled water. To suppress the endogenous peroxidase activity, the slides were incubated in peroxidase-blocking reagent (H2O2 in methanol). Slides were subjected to antigen retrieval in citrate buffer inside a microwave oven. Blocking of the sections was achieved in blocking media of 3% BSA in PBS (with Triton X- 100) at room temperature. After blocking, incubation of the renal sections was executed with the primary antibodies diluted in BSA using anti-rabbit serum against TNF-α with 1:500 dilution and rabbit antibody to TGF-β with 1:500 dilution at 40?C overnight in a moisture chamber. Washing of the slides with (PBST) Phosphate Buffered Saline with Tween 20 was done to get rid of unbound primary antibodies the following day. Incubation was performed with goat anti-rabbit IgG FITC conjugated secondary antibody, diluted in BSA (1:10,000 dilution) for the duration of 2 hours at room temperature in a dark and humid chamber. The tissue sections were again washed with PBST. After completion of all the requisite washing steps, the sections were counterstained with DAPI for 2–5 minutes, followed by washing with PBS to pool out excess DAPI. Finally, the sections were cover-slipped in mounting media and were examined using Nikon Eclipse 80i fluorescence microscope equipped with Northern Eclipse Imaging Elements-D (NIS-D) software.
2.3. Statistical Analysis
Statistical analysis was performed using Microsoft Excel, Office 365 (v16.0) (Microsoft, Redmond, WA). Categorical data were reported as percentages and frequency and data for continuous variables were summarized using the mean and standard deviation of three independent experiments, performed in triplicates. The variance between samples was compared using F-test Two-sample variance tool in Microsoft Excel. If the p-value in the F-test was less than 0.05, then the samples were considered not to have equal variances. Student’s t-test was used for the continuous variables with equal variance otherwise Welch’s t-test was used. The p-value of ≤0.05 was considered significant with all the statistical tests being two-tailed. To calculate the confidence interval, 95% of confidence level (α = 0.05) was taken.
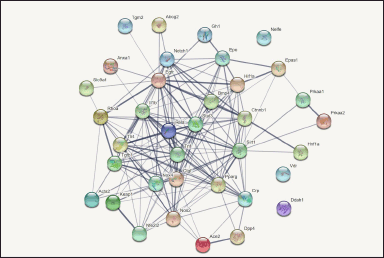 | Figure 2. The image shows protein- protein interaction network (PPIN) generated through String software tool in UUO induced chronic kidney injury. Total of 37 genes were uploaded for String analysis to generate a network having interaction score fixed to medium confidence (0.400). Lines connecting the genes indicate the interactions amongst them, with thicker lines suggesting stronger interactions between the genes. [Click here to view] |
3. RESULTS
3.1. NCBI Retrieval of Gene
Irrespective of the primary reason behind the causation of CKD, inflammation, apoptosis, and fibrosis are the main processes accountable for CKD. Therefore, in the present frame of work, these specific keywords were used to fetch genes from NCBI database. The result showed 37 candidate genes related to “CKD genes” (Supplementary File, Sheet 1), 27 “inflammatory genes in CKD” (Supplementary File, Sheet 2), 28 “apoptotic genes in CKD” (Supplementary File, Sheet 3), 27 “genes for fibrosis in CKD” (Supplementary File, Sheet 4) and 15 “epithelial mesenchymal transition genes in CKD” (Supplementary File, Sheet 5) retrieved from NCBI. The search remained restricted “rat kidney”. Thus, a total of 134 genes were screened. After eliminating the redundant genes from the list of 134 genes, the final list turned out to be 37 in number. This gene list was further analyzed for the construction of PIN (Fig. 2).
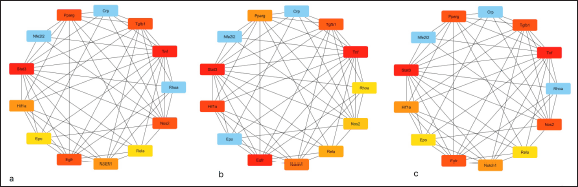 | Figure 3. The figure shows top 10 scrutinised hub genes employing Cytoscape plugin cytohubba. For scrutinization, cytohubba employed three topological measures viz. Degree Centrality, BC and MNC. Genes having top scores were recovered, each from above mentioned three topological measures, having a pattern of color signifying highly upregulated genes in CKD with red color. The shade of the colour from strong red to blue specifying the order of the genes from top to bottom. [Click here to view] |
3.2. Module Analysis and Calculation of Topological Measures to Find Hub Henes by Cytoscape Analysis
The network was then exported to cytoscape plugin MCODE, the results of which showed that the network included 2 modules. According to MCODE scores, 13 nodes, and 53 edges were present in module 1 with an 8.833 score, and 6 nodes, and 8 edges were present in module 2 with a 3.200 score. Module 1 with the maximum score of 8.833 was selected as the most influential one containing 13 genes. Now these 13 genes of modules 1 were further visualized for topological characteristics including degree (k), BC, and MNC by Cytoscape plugin Cytohubba (Fig. 3). Based on the centrality methods ten overlapping genes viz.- STAT3, TNFα, TGFβ, PPARγ, NOS2, EGFR, NOTCH1, HIF1A, EPO, and Rela were identified (Table 2). Among these 10 genes, the top-ranked three genes were obtained by combining the results of the above three topological parameters viz. STAT-3, TNF-α, and TGF-β were validated in vivo by generating a rat model of UUO model of chronic kidney injury.
3.3. Ingenuity Pathway Analysis
The master regulator of all the 10 screened hub genes was observed by implementing the core analysis approach of IPA. It has been verified in our lab that nuclear factor kappa-light-chain-enhancer of activated B cells (NF-?B) plays a major role in AKI [10,11]. In the present study also, NF-?B was identified as the master regulator of CKD genes through IPA platform. The results suggested that NF-κB complex was regulating the above-identified 10 hub genes. Upregulation of NF-κB molecule causes activation of the genes including TNF, TGFB1, NOTCH1, RELA, NOS2, HIF1A, EGFR, and EPO. The gene PPARG is getting inhibited by the regulation of NF-?B (Fig. 4). Therefore, in addition to the top three ranked genes viz. STAT-3, TNF-α, and TGF-β, the in vivo validation of NF-?B becomes highly pertinent to depict its role in CKD.
3.4. In Vivo Analysis of UUO Model in Rats
3.4.1. Renal function analysis
The biochemical analysis of the results unveiled no remarkable change in the level of serum creatinine among the Sham group and UUO 14 group (p > 0.05; CI 0.025–0.107). However, on day 18, a slight surge in creatinine level was depicted w.r.t. the Sham group (p ≤ 0.001; CI 0.188–0.340) (Fig. 5).
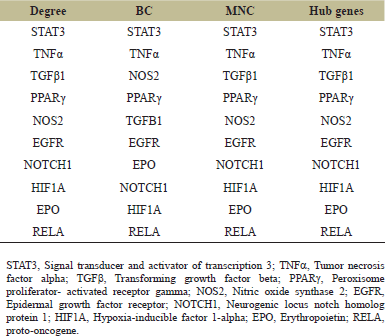 | Table 2. List of 10 genes with high degree, BC and MNC. The table shows top 10 scrutinised hub genes employing Cytoscape plugin cytohubba. For scrutinization, cytohubba employed three topological measures viz. Degree Centrality, BC and MNC. Genes having top scores were recovered, each from above mentioned three topological measures. 10 hub genes were obtained by combining the results of three topological parameters. [Click here to view] |
3.4.2. Kidney histopathology on day 14 and day 18 after ureter ligation
In the Sham control group, kidney architecture was found to be well-marked with properly organized cells, intact nuclei, normal tubules, and glomeruli. Whereas UUO 14 group was characterized by dilatation of the tubules, interstitial nephritis, and ruptured glomeruli showing widening of Bowmans’ space. These changes were exaggerated on day 18 with additional visible foci of fibrosis (Fig. 6).
3.4.3. Calculation of THS
The THS was calculated for the semiquantitative estimation of the kidney injury based on the formula- THS = TD/2+TN+TIN/2. Based on the histological score, it was observed that THS was multi-folds increased at 14 days and reached a maximum at 18 days following ureter obstruction with respect to control as shown in Figure 7.
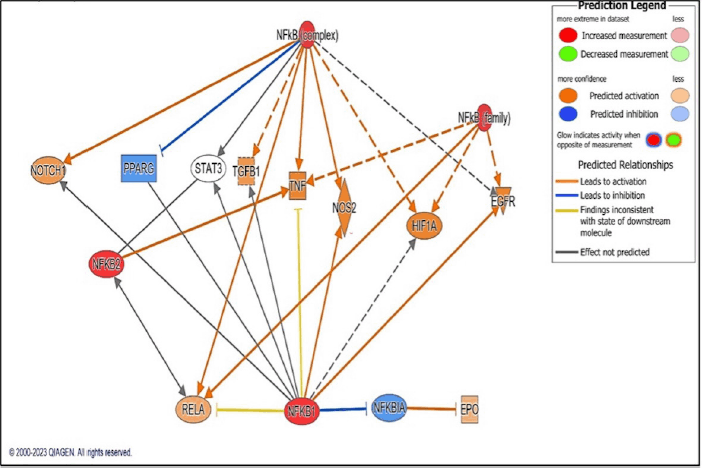 | Figure 4. Image depicting NF-?B as the master regulator of top 10 hub gene through core analysis approach of IPA. Strong red colour signify upregulation of genes whereas blue colour indicates downregulation of genes. [Click here to view] |
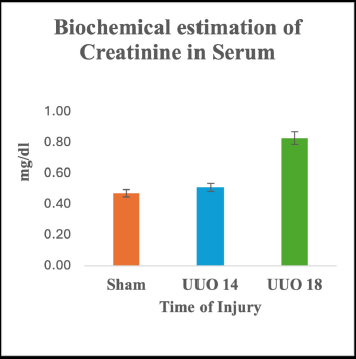 | Figure 5. Renal function test of UUO nephropathy in blood serum of rats- Changes in serum creatinine. Data represented as means +/- SD. *p < 0.05 w.r.t. to the control group. [Click here to view] |
3.4.4. Real time PCR analysis of TNF-α, TGF-β, STAT-3, and NF-κB gene with respect to CKD
In the UUO 14 group the fold change of TNF-α was inconspicuous in control versus right non-LK (1.4 w.r.t control; p = 0.091; 95% CI 0.087 to 1.024), while it was 2.1 times to that of control in the left LK (p < 0.05; 95% CI 0.369–2.269). However, in UUO 18 group the expression of TNF-α was 3 times to that of the control in the right kidney (p = 0.002; 95% CI 0.931–3.240) and 4.5 times in the left LK in comparison to the control (p = 0.001; 95% CI 1.743–5.418) (Fig. 8a).
A gradual decrease in the expression of STAT-3 gene in UUO 14 Group was observed. The fold change of STAT-3 in UUO 14 group was highest in the sham group, then in the right kidney, and least in the left kidney. However, the expression of STAT-3 gene was prominent in the left kidney in UUO 18 group, and it was almost triple, 2.8 times to that of the sham group (p = 0.01; 95% CI 0.549–3.213). However, the changes in the expression of the STAT-3 gene was found to be insignificant in sham group and right kidney (1.05 w.r.t sham group; p = 0.765; 95% CI 0.331–0.439) (Fig. 8b).
Next, the level of TGF-β gene was analyzed and a considerable upsurge in its expression was noticed in the right kidney of UUO 14 group which was double (2.61; p = 0.079; 95% CI 0.224–3.435) w.r.t. sham control kidney. However, its expression was found decreased in the left LK of the same group when compared with control (0.92; p = 0.764; 95% CI 0.485–0.643). On the contrary, in UUO 18 group, mRNA expression of TGF-β experienced a rise in the fold change of 1.3 in the right kidney (p = 0.343; 95% CI 0.480–1.269) and 4.1 (p = 0.001; 95% CI 1.701–4.663) in the left LK, (Fig. 8c).
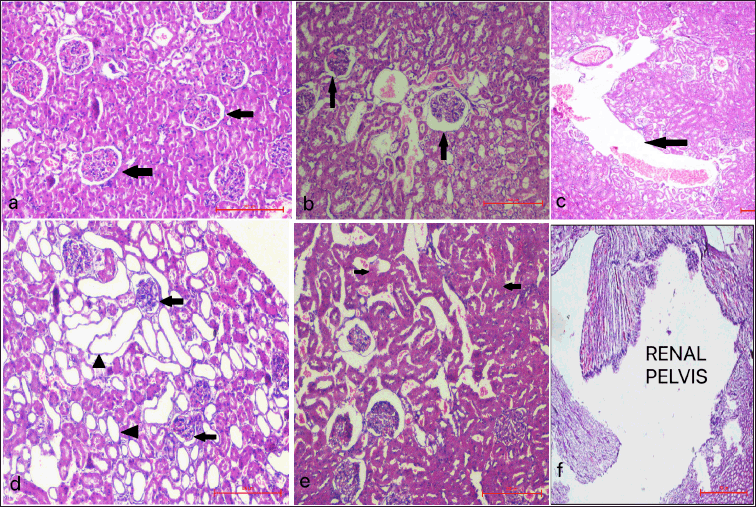 | Figure 6. Photomicrographs depicting histopathological changes (H&E staining) in rat kidney after UUO of Sham control group, UUO 14 and UUO 18 groups a) Sham group showing normal kidney architecture (black arrows- normal Bowman’s space, white arrows- normal renal tubules); b) UUO 14 group showing inflammation and dilatation of tubules (white arrows) and Bowman’s space (black arrows) of left LK; c) Widening of renal pelvis of left LK of UUO 14 group (black arrow); d) UUO 18 group showing significant dilatations of tubules (black arrow heads) and Bowman’s space (black arrows) of left LK; e) UUO 18 group showing mild interstitial fibrosis (black arrow heads) of left LK; f) Prominent widening of the renal pelvis of left LK UUO 18 group (scale bar = 100 μm). [Click here to view] |
 | Figure 7. Semiquantitative analysis of kidney injury by calculating THS based on degeneration, necrosis and inflammation of the tubules following ureter obstruction. Data represented as means +/- SD. *p < 0.05 w.r.t. to the control group. [Click here to view] |
NF-κB gene was found to be increased to more than double fold in the right kidney (2.56; p = 0.114; 95% CI 0.438–3.554) and almost seven times in the left kidney to that of the sham group in UUO 14 group (7.57; p < 0.001; 95% CI 3.997–9.138). In UUO 18 group also the expression of NF-κB gene in the right kidney was almost 6 times higher to that of the control. The left kidney also displayed a marked activation of NF-κB characterized by a prominent increase in its expression and it was found to be 19 times higher in comparison to sham group (19.13; p = 0.001; 95% CI 9.851–26.418) (Fig. 8d).
3.4.5. Immunohistochemical analysis
Expressions of TNF-α and TGF-β were further validated at the protein level and it was found that the immunofluorescence analysis for TNF-α and TGF-β were consistent with the results of PCR studies.
Results of immunohistochemical staining depicted that in Group II, the expression of TNF-α was increased in the LK as compared to that of the contralateral kidney. However, there was no perceivable expression in the sham control. In Group III, the expression was much more pronounced in the LK with respect to the contralateral kidney (Fig. 9).
Next, the tissue sections were evaluated for the expression of TGF-β and it was deciphered that there was not much variation in the protein expression was slightly higher in the LK as compared to the contralateral kidney in Group II. The sham control group again had a trivial expression of TGF-β as it was observed for TNF-α. However, in Group III, there was a significant increase in the expression in the interstitial and tubular cells (Fig. 10).
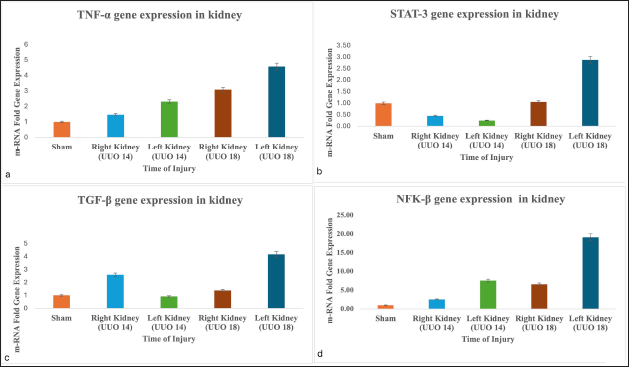 | Figure 8. Evaluation of a) TNF-α, b) STAT-3, c) TGF-β d) NFκB gene expression on UUO induced kidney injury in rats. The relative mRNA expression of the genes was analysed by quantitative real time PCR. (*p < 0.05 vs. control showed statistical significance). [Click here to view] |
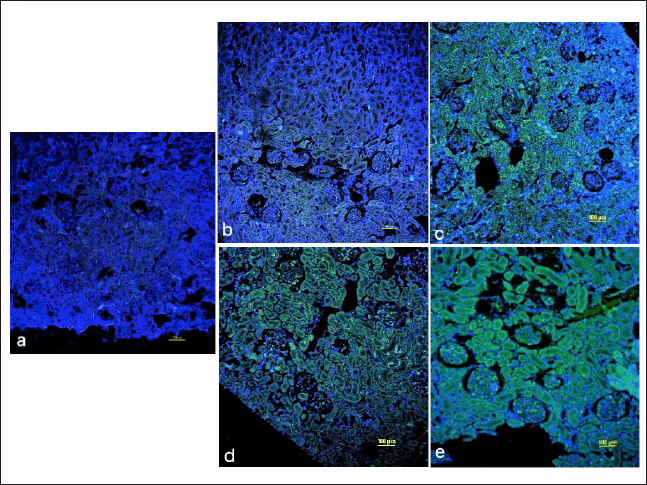 | Figure 9. Representative immune-histochemical stained images of rat kidney with FITC labelled TNF-α antibody (green colour) counterstained with DAPI (blue colour) of UUO Nephropathy of a) Sham group b) Right non LK of UUO 14 Group c) Left LK of UUO 14 Group d) Right non LK of UUO 18 Group e) Left LK of UUO 18 Group. Results of immunohistochemical staining depicted that in UUO 14 group, the expression of TNF-α was increased in left LK as compared to that of right non-LK. In UUO 18 group, the expression was much more pronounced in left LK with respect to right non-LK. However, there was no perceivable expression in the sham group. Images are observed at magnification of 10×. Scale bar: 100 µm. [Click here to view] |
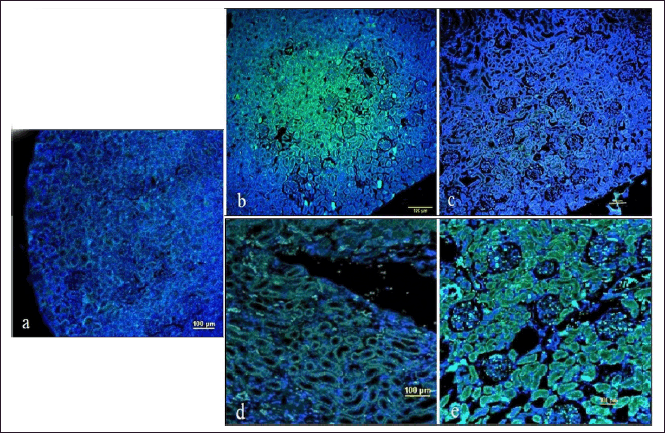 | Figure 10. Representative immune-histochemical stained images of rat kidney with FITC labelled TGF-β antibody (green colour) counterstained with DAPI (blue colour) of UUO Nephropathy of a) Sham group b) Right non-LK of UUO 14 group c) Left LK of UUO 14 group d) Right non-LK of UUO 18 group e) Left kidney of UUO 18 group. Examination of stained kidney sections of UUO14 group affirmatively manifested that the right contralateral kidney has higher protein expression of TGF-β as compared to the left LK. TGF-β expression in UUO 18 group followed the same pattern as was observed for TNF-α and manifested highest expression in the LK than in contralateral and least in the sham control group. Sham control displayed no noticeable expression of TGF-β as such. Images are observed at magnification of 10×. Scale bar: 100 µm. [Click here to view] |
4. DISCUSSION
To delineate the three central molecular processes, i.e., inflammation, apoptosis, and fibrosis, involved in CKD, the bioinformatic approach was utilized to look for the major set of genes possessing the potential in the disease pathophysiology [12,13]. The prediction analysis employing the bioinformatics tool Cytoscape was used for scoring and ranking genes in a network according to the three most influential topological parameters viz. Degree Centrality, BC and MNC [14]. The results filtered 10 essential overlapping genes in the top-ranked list (STAT3, TNFα, TGFβ, PPARγ, NOS2, EGFR, NOTCH1, HIF1A, EPO, and Rela). Three genes (STAT3, TNF, and TGF) with the highest score in accordance with the Cytoscape findings, were taken further for in vivo analysis. Additionally, NF-κB has been found to be the key mediator in regulating the expression of the above-obtained genes as observed through IPA.
For the experimental validation of the above-mentioned genes, UUO surgery was performed in rats on 14 and 18 days as the damage became apparent from day 14 and onwards. The extent of the damage was more pronounced on 18th day post ureter obstruction as compared to day 14th as depicted by the appearance of fibrosis on histopathological examination of the LK on day 18. This notion besides lending support gets reiterated by the fact that NF-kB exhibited enhanced expression indicating a significant function in UUO-induced CKD (p = 0.011; 95% CI 3.05–20.079). Experiments carried out in our laboratory prior to this study have shown the part played by NF-kB in folic acid-induced AKI, we now in the present frame of work, we display the role of NF-kB in UUO-induced CKD.
Other reports also lend strong corroboration to such a conclusion where in NF-kB is shown to be a key inflammatory ensuing molecules in humans as well as animal models of CKD. In one of the reports, it is demonstrated that chronic kidney failure gets attenuated by inhibiting the expression of NF-kB by geniposide injections [15]. Reports have also suggested that NF-kB is responsible for kidney injury in the UUO mouse model. They showed that telbivudine, drug with antiviral properties, targeted NF-kB signaling pathway and resulted in decreased inflammation and fibrosis [16]. Consistent with the previous reports, our work now proposes that NF-kB has a much more complicated role, being a major regulator in the pathology of CKD.
NF-kB activation takes a creative lead throughout the process of inflammation which serves as a trademark for CKD. NF-kB resides in the cytoplasm complexed with inhibitory-kB proteins. To get activated, it relies on the phosphorylation of the inhibitory protein, thereby its release and translocation into the nucleus. Following this, NF-kB act as a trigger for the activation of multiple inflammatory genes accountable for kidney diseases.
NF-kB is itself acted upon by several other cytokines for its activation. Amongst the several other stimuli, TNF-α is one of them, that is held responsible for eliciting the response of NF-kB by coupling to its receptor, thus, ensuing a domino effect on each other’s stimulation [17,18]. This engagement of NF-κB and TNF-α in a circular loop has been suggested to be the significant factor responsible for the mechanism of inflammation in various CKD [19]. Likewise similar increase was also seen in the expression of TNF-α parallel to that of NF-κB in the left LKs of animals in comparison to the sham control as well as right non-LK, signifying a strong association among them. Previous studies [20,21] have reported similar pattern of elevation in the gene expression of these cytokines thus ensuring their roles in inflammation during UUO-induced renal damage.
Studies on animal models have shown the role of TNF-α in the progression of CKD from AKI by the upregulation of M1 macrophages and advancing the stage to fibrosis. High expression of TNF-α has been evidenced in several experimental animal models for CKD including diabetic kidney disease [22]. A similar statistically significant rise in TNF-α expression (p = 0.05; 95% CI 0.002–4.329) was observed on 18th day with respect to 14th day in the ligated left kidney in our study (Fig. 8). Moreover, one of the members of TNF superfamily, i.e., TWEAK caused renal fibrosis of UUO mice through activation of NF-κB signaling. The reports also pointed to the observations wherein, TWEAK, one of the sole cytokines manifested to activate nonclassical NF-κB signaling in tubules of the kidney [23,24]. One more study, it is reported that isoliquiritigenin, a flavonoid, is anti-inflammatory and antifibrotic in its action. Following treatment with this flavonoid, UUO induced kidney injury remarkably improved by downregulating the levels of TNF-α, IL-6, and IL-1β gene expression and simultaneously suppressing the phosphorylation of NF-κB [25]. In yet another study it was suggested that following treatment with nifuroxazide, an antibacterial drug, the expression of TNF-α, TGF-β1 and IL-1 β was diminished and consequently resulted in reduced inflammation and fibrosis, which were liked with targeting NF-κB signaling [26]. Thus, it is strongly believed that this cyclic interplay of TNF and NF-?B must be playing a major role in UUO-induced renal tissue inflammation.
Eventually, following inflammation and apoptosis, fibrosis is the main concluding pathway of all sorts of CKD and literature reports evidence have shown that NF-?B/TNF-α are primarily responsible in provoking fibrosis by augmenting the expression of IL-6, and TGF-β1 in glomerular diseases [27,28]. It is very well documented in plenty of reports that NF-?B is opulently expressed during chronic inflammation and releases proinflammatory cytokine IL-6, which consequently results in the activation of STAT-3 gene [29]. It has been exhibited that NF-?B communicates directly with STAT3, thereby, aiding its recruitment to the promoter region of NF-?B and vice-versa and thus, are shown to symbiotically regulate various inflammatory and apoptotic genes [30]. Uncontrolled stimulation of Stat 3 gene has been depicted in various disorders specifically in cancer and autoimmune diseases, but its role has currently been manifested for its involvement in several kidney diseases [31]. Elevated expression of STAT-3 has been delineated in earlier studies in the tubular and interstitial region of the kidney in UUO rat model of kidney fibrosis [32]. In another subsequent research, activation of STAT-3 has been exhibited in mice model for kidney fibrosis generated by ureter obstruction. In their study, they treated the mice with S3I-201, following UUO and found a remarkable decline in the expression of proinflammatory cytokines, fibronectin, alpha smooth muscles actin, and type I collagen, resulting a decrease in the level of fibrosis [33]. Another study demonstrated the participation of IL-6 in the activation of NF-κB and STAT-3 in fibroblasts by the subsequent elicitation of TGF-β resulting in myofibroblast proliferation and extracellular matrix (ECM) deposition [34]. Therefore, turning on these pathways results in more serious renal injury in these cases.
The present work also witnessed NFκB coupled rise in the expression of TGF-β in UUO 18 group (p < 0.001; 95% CI 1.717–4.806) certifying the role of TGF-β in fibrosis as demonstrated in other studies [35]. TGF-β1 is considered as the principal profibrotic cytokine and myo?broblasts as the dominant cells responsible for generating fibrotic ECM. In some of the in-vitro studies also it was found that the epithelial cells attain the characteristics of mesenchymal cells phenotype when kidney epithelial cell lines were treated with TGF-β [36]. In yet another parallel study, increased expression of TGF-β was observed in IgA nephropathy patients indicating its involvement in fibrosis in the pathogenesis of CKD [37]. All these studies lend strong support to the present work.
However, TGF-β level in UUO 14 group was found to be highest in the non-LK in comparison to sham control and the left LK (Fig. 8). This can be elucidated on the basis that TGF-β has pleiotropic role and is not just involved in fibrosis, as mentioned above, it has a diverse roles like proliferation of cells, anti-inflammatory, cell differentiation and ECM deposition [38]. Another plausible reason for the decrease of TGF-β in the LK is compensatory hypertrophy. TGF-β is witnessed to play as one of the significant molecules responsible for ensuing compensatory hypertrophy in the contralateral kidney [39,40]. It has been very well documented that mesangial cells are one of the principal sources of TGF-β [41]. Obstructive nephropathy results in increased filtration of the glomerulus accompanied by increased production of mesangial cells in the contralateral kidney. This is further followed by accumulation of TGF-β which prompts tubular cells to encounter compensatory hypertrophy in the contralateral kidney. When an ample amount of TGF-β gets collected, it in turn prevents the amplification of mesangial cells consequently resulting in an impediment of compensatory hypertrophy of the contralateral kidney. However, when the stage of fibrosis is reached, TGF-β gets shifted in the LK. Several reports have suggested the relation of the contralateral kidney in compensatory hypertrophy in obstructive nephropathy animal models [42,43].
The bioinformatics observation of the present study, through IPA, further revealed the interaction of NF-?B and TGF-β in the advancement of kidney injury. NF-?B /TNF-α are primarily responsible in provoking kidney inflammation and fibrosis by augmenting the expression of IL-1β, and TGF-β1 in several kidney diseases. One of the studies has demonstrated that Artemisinin, an antimalarial drug has been shown to have anti-inflammatory and anti-fibrotic role by decreasing the expression of TGF-β and simultaneously suppressing NF-?B pathway in 5/6 nephrectomy rat model [44]. Ferulic acid, another pharmacological drug, significantly reduced the expression of NF-?B, TGF-β1and TNF-α in the diabetic nephropathy rat model [45]. It appears from these studies that TGF-β and NF-?B are getting decreased together, it can be assumed that both these genes work in co-ordination.
The present result is thus, in compliance with the other studies signifying the role of NF-?B and TGF-β in fibrosis. The present work also offers an additional advantage with respect to previous studies as the majority of the findings have analyzed the expression of the genes in the LK. However, the role of the participating genes in the unobstructed kidney has not formerly been analyzed. Thus, in the present piece of work, the expression level of predictive genes was considered in the contralateral as well as in the LK.
4.1. Limitations
There are a few limitations of the study
First, the inefficiency of UUO model to precisely analyse the variations in kidney function due to the presence of unobstructed kidney as the serum creatinine levels cannot be used as a parameter for assessment of CKD. Second, Rat UUO model experiences pronounced inflammatory component as compared to humans, hence the degree of gene expression may be variable and different in humans. Third, UUO model does not cast light on established disease as observed in clinical studies. Fourth, the present study used literature search for genes from NCBI only, other transcriptomic or proteomic dataset repositories can also be used. Fifth, clinical studies would be required to validate our results in humans.
5. CONCLUSION
NF-?B has been ascertained as a major participant in various forms of kidney diseases. Our study too revealed that NF-?B has a major influence on leading genes impacting CKD. For the first time, the study speculated, from the results drawn from integrated systems biology and in-vivo approach, that the top three genes (STAT-3, TNF-α, and TGF-β) work in coordination in disease progression and NF-kB being a major regulator of genes, participating in manifestation of CKD. The coupled increase in the expression of these genes proposed, that these might be significant contributors in the advancement of the disease. Our results suggested that NF-?B plays a pertinent role in the manifestation of CKD and thus can be restrained by therapeutic manoeuvres. The crosstalk amongst these molecular entities must be investigated for various kidney diseases specifically for CKD. With the evolution of modern techniques in the medical field, the present information will certainly enhance the knowledge of the underlying molecular etiology of CKD and eventually can be harnessed for potentially therapeutic targets.
6. ACKNOWLEDGMENT
Panjab University, Chandigarh, India is acknowledged for providing the resources for conducting this study.
7. LIST OF ABBREVIATIONS
BC, Betweenness centrality; BSA, Bovine serum albumin; CKD, Chronic kidney disease; DAPI, 6-diamidino-2-phenylindole; FITC, Fluorescein isothiocyanate; IPA, Ingenuity pathway analysis; LK, Left kidney; MCODE, Molecular complex detection; MNC, Maximum neighborhood centrality; NCBI, National centre for biotechnology information; NF-?B, Nuclear factor kappa-light-chain-enhancer of activated B cells; PBS, Phosphate buffered saline; PBST, Phosphate buffered saline with tween-20; PCR, Polymerase chain reaction; PIN, Protein interaction network; RK, Right kidney; ROS, Reactive oxygen species; STAT3, Signal transducer and activator of transcription 3; STRING, Search tool for the retrieval of interacting genes; TGFβ, Transforming growth factor beta; TNFα, Tumor necrosis factor alpha; UUO, Unilateral ureter obstruction.
8. AUTHOR CONTRIBUTIONS:
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
9. DATA AVAILABILITY:
All the data is available with the authors and shall be provided upon request.
10. PUBLISHER’S NOTE:
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
11. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
12. SUPPLEMENTARY MATERIAL:
The supplementary material can be accessed at the journal's website: Link here [https://jabonline.in/admin/php/uploadss/1298_pdf.pdf].
13. CONFLICTS OF INTEREST
The authors have no financial or proprietary interests in any material discussed in this article.
14. FUNDING
This work was funded by Indian Council of Medical Research (ICMR), New Delhi, India (ID No.- 2020-8718/CMB-BMS), and “DBT-Builder” grant vide No. BT/INF/22/SP41295/2020, dated 25/01/2021 sanctioned by Government of India, Ministry of Science and Technology, Department of Biotechnology, New Delhi, India.
15. ETHICS STATEMENT
The study protocol was approved by the Institutional Animal Ethics Committee of the Centre for Stem Cell, Tissue Engineering & Biomedical Excellence, Panjab University, Chandigarh, India (Approval No.: PU/45/99/CPCSEA/IAEC/2020/459).
REFERENCES
1. Cole A, Ong VH, Denton CP. Renal disease and systemic sclerosis: an update on scleroderma renal crisis. Clin Rev Allergy Immunol 2023;64(3):378–91; CrossRef
2. Evans M, Lewis RD, Morgan AR, Whyte MB, Hanif W, Bain SC, et al. A narrative review of chronic kidney disease in clinical practice: current challenges and future perspectives. Adv Ther 2022;39(1):33–43; CrossRef
3. Hojs R, Ekart R, Bevc S, Vodošek Hojs N. Chronic kidney disease and obesity. Nephron 2023;147(11):660–4; CrossRef
4. Niculae A, Gherghina ME, Peride I, Tiglis M, Nechita AM, Checherita IA. Pathway from acute kidney injury to chronic kidney disease: molecules involved in renal fibrosis. Int J Mol Sci 2023;24(18):14019; CrossRef
5. Choueiri R, Faddoul J, Ghorra C, Al Najjar J, Akiki BB, Boustany S, et al. A case report: 19-year-old male diagnosed with C1q nephropathy requiring renal replacement therapy. Explor Med 2022;3:386–92; CrossRef
6. Tuttle KR, Jones CR, Daratha KB, Koyama AK, Nicholas SB, Alicic RZ. Incidence of chronic kidney disease among adults with diabetes, 2015-2020. N Engl J Med 2022 Oct 13;387(15):1430–1; CrossRef
7. Havasi A, Borkan SC. Apoptosis and acute kidney injury. Kidney Int 2011;80:29–40; CrossRef
8. Tanaka S, Portilla D, Okusa MD. Role of perivascular cells in kidney homeostasis, inflammation, repair and fibrosis. Nat Rev Nephrol 2023;19(11):721–32; CrossRef
9. Song N, Thaiss F, Guo L. NFκB and kidney injury. Front Immunol 2019;10:815; CrossRef
10. Kumar D, Singla SK, Puri V, Puri S. The restrained expression of NF-kB in renal tissue ameliorates folic acid induced acute kidney injury in mice. PLoS One 2015;10(1):e115947; CrossRef
11. Gupta A, Puri S, Puri V. Bioinformatics unmasks the maneuverers of pain pathways in acute kidney injury. Sci Rep 2019;9:11872; CrossRef
12. Jin L, Yu B, Armando I, Han F. Mitochondrial DNA-mediated inflammation in acute kidney injury and chronic kidney disease. Oxid Med Cell Longev 2021;2021:9985603; CrossRef
13. Nicolic-Paterson DJ, Grynberg K, Ma FY. JUN amino terminal kinase in cell death and inflammation in acute and chronic kidney disease. Integr Med Nephrol Androl 2021;8:10; CrossRef
14. Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science 2002;296:910–3; CrossRef
15. Hu X, Zhang X, Jin G, Shi Z, Sun W, Chen F. Geniposide reduces development of streptozotocin-induced diabetic nephropathy via regulating nuclear factor-Kappa B signaling pathways. Fundam Clin Pharmacol 2017;31(1):54–63; CrossRef
16. Chen J, Li D. Telbivudine attenuates UUO-induced renal fibrosis via TGF-β/Smad and NF-κB signaling. Int Immunopharmacol 2018;55:1–8; CrossRef
17. Wertz IE. TNFR1-activated NF-kappa B signal transduction: regulation by the ubiquitin/proteasome system. Curr Opin Chem Biol 2014;23:71–7; CrossRef
18. Zhang H, Sun SC. NF-kappaB in inflammation and renal diseases. Cell Biosci 2015;5:63; CrossRef
19. Donnahoo KK, Shames BD, Harken AH, Meldrum DR. Review article: the role of tumor necrosis factor in renal ischemia-reperfusion injury. J Urol 1999;162(1):196–203; CrossRef
20. Lee WC, Jao HY, Hsu JD, Lee YR, Wu MJ, Kao YL, et al. Apple polyphenols reduce inflammation response of the kidneys in unilateral ureteral obstruction rats. J Funct Foods 2014;11:1–11; CrossRef
21. Chung AC, Huang XR, Zhou L, Heuchel R, Lai KN, Lan HY. Disruption of the Smad7 gene promotes renal fibrosis and inflammation in unilateral ureteral obstruction (UUO) in mice. Nephrol Dial Transplant 2009;24:1443–54; CrossRef
22. Cheng D, Liang R, Huang B, Hou J, Yin J, Zhao T, et al. Tumor necrosis factor-α blockade ameliorates diabetic nephropathy in rats. Clin Kidney J 2019;14:301–8; CrossRef
23. Sanz AB, Sanchez-Nino MD, Ortiz A. TWEAK a multifunctional cytokine in kidney injury. Kidney Int 2011;80:708–18; CrossRef
24. Ucero AC, Benito-Martin A, Fuentes-Calvo I, Santamaria B, Blanco J, Lopez-Novoa J, et al. TNF-related weak inducer of apoptosis (TWEAK) promotes kidney fibrosis and Ras dependent proliferation of cultured renal fibroblast. Biochim Biophys Acta 2013;1832:1744–55; CrossRef
25. Liao Y, Tan RZ, Li JC, Liu TT, Zhong X, Yan Y, et al. Isoliquiritigenin attenuates UUO-induced renal inflammation and fibrosis by inhibiting mincle/syk/NF-kappa B signaling pathway. Drug Des Devel Ther 2020;14:1455–68. CrossRef
26. Hassan NME, Said E, Shehatou GSG. Nifuroxazide suppresses UUO-induced renal fibrosis in rats via inhibiting STAT-3/NF-κB signaling, oxidative stress and inflammation. Life Sci 2021;272:119241; CrossRef
27. Sun SC, Chang JH, Jin J. Regulation of nuclear factor-kappaB in autoimmunity. Trends Immunol 2013;34:282–9; CrossRef
28. Lan T, Tang T, Li Y, Duan Y, Yuan Q, Liu W, et al. FTZ polysaccharides ameliorate kidney injury in diabetic mice by regulating gut-kidney axis. Phytomed Int J Phytother Phytopharmacol 2023;118:154935; CrossRef
29. Dai Y, Gu L, Yuan W. Podocyte specific deletion of signal transducer and activator of transcription 3 attenuates nephrotoxic serum induced glomerulonephritis. Kidney Int 2013;84(5):950–61; CrossRef
30. Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NF B. Genes Dev 2007;21:1396–408; CrossRef
31. Shawky AM, Almalki FA, Abdalla AN, Abdelazeem AH, Gouda AM. A comprehensive overview of globally approved JAK inhibitors. Pharmaceutics 2022;14(5):1001; CrossRef
32. Sarapultsev A, Gusev E, Komelkova M, Utepova I, Luo S, Hu D. JAK-STAT signaling in inflammation and stress-related diseases: implications for therapeutic interventions. Mol Biomed 2023;4(1):40; CrossRef
33. Pang M, Ma L, Gong R, Tolbert E, Mao H, Ponnusamy M, et al. A novel STAT3 inhibitor, S3I-201, attenuates renal interstitial fibroblast activation and interstitial fibrosis in obstructive nephropathy. Kidney Int 2010;78:257–68; CrossRef
34. Shi Y, Tao M, Ni J, Tang L, Liu F, Chen H, et al. Requirement of histone deacetylase 6 for interleukin-6 induced epithelial-mesenchymal transition, proliferation, and migration of peritoneal mesothelial cells. Front Pharmacol 2021;12:722638; CrossRef
35. Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. Targeted disruption of TGF-beta1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest 2003;112:1486–94; CrossRef
36. Wu W, Wang X, Yu X, Lan HY. Smad3 signatures in renal inflammation and fibrosis. Int J Biol Sci 2022;18:2795–806; CrossRef
37. Yang M, Liu JW, Zhang YT, Wu G. The role of renal macrophage, AIM, and TGF-β1 expression in renal fibrosis progression in IgAN patients. Front Immunol 2021;12:646650; CrossRef
38. Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol 2012;13:616–30; CrossRef
39. Fujita H, Omori S, Ishikura K, Hida M, Awazu M. ERK and p38 mediated high-glucoseinduced hypertrophy and TGF-beta expression in renal tubular cells. Am J Physiol Renal Physiol 2004;286:F120–6; CrossRef
40. Sinuani I, Averbukh Z, Gitelman I, Rapoport MJ, Sandbank J, Albeck M, et al. Mesangial cells initiate compensatory renal tubular hypertrophy via IL-10 induced TGF- _ secretion: effect of the immunomodulator AS101 on this process. Am J Physiol Renal Physiol 2006;291:F384–94; CrossRef
41. Kitamura M, Suto Y, Yokoo T, Shimizu F, Fine LG. Transforming growth factor-beta 1 is the predominant paracrine inhibitor of macrophage cytokine synthesis produced by glomerular mesangial cells. J Immunol 1996;156:2964–71; CrossRef
42. Zhou W, Wu WH, Si ZL, Liu HL, Wang H, Jiang H, et al. The gut microbe Bacteroides fragilis ameliorates renal fibrosis in mice. Nat Commun 2022 Oct 14;13(1):6081; CrossRef
43. Xiong Y, Chang Y, Hao J, Zhang C, Yang F, Wang Z, et al. Eplerenone attenuates fibrosis in the contralateral kidney of UUO rats by preventing macrophage-to-myofibroblast transition. Front Pharmacol 2021 Feb 24;12:620433; CrossRef
44. Wen Y, Pan MM, Lv LL, Tang TT, Zhou LT, Wang B, et al. Artemisinin attenuates tubulointerstitial inflammation and fibrosis via the NF-κB/NLRP3 pathway in rats with 5/6 subtotal nephrectomy. J Cell Biochem 2019;120:4291–300; CrossRef
45. Qi MY, Wang XT, Xu HL, Yang ZL, Cheng Y, Zhou B. Protective effect of ferulic acid on STZ-induced diabetic nephropathy in rats. Food Funct 2020;11:3706–18; CrossRef