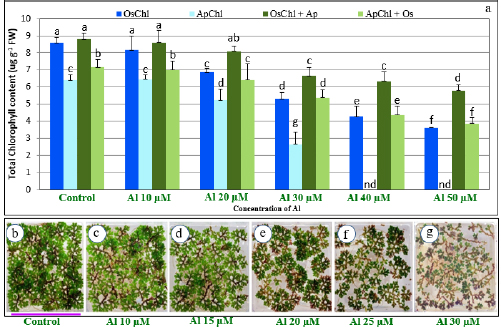
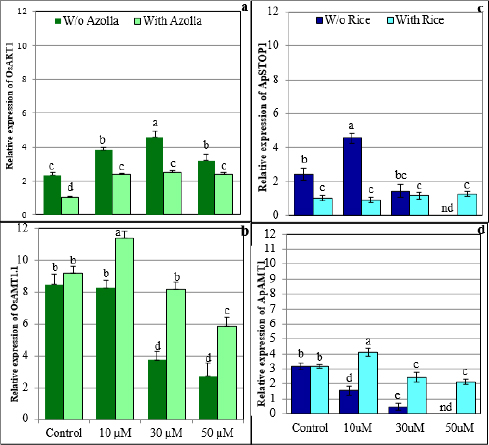
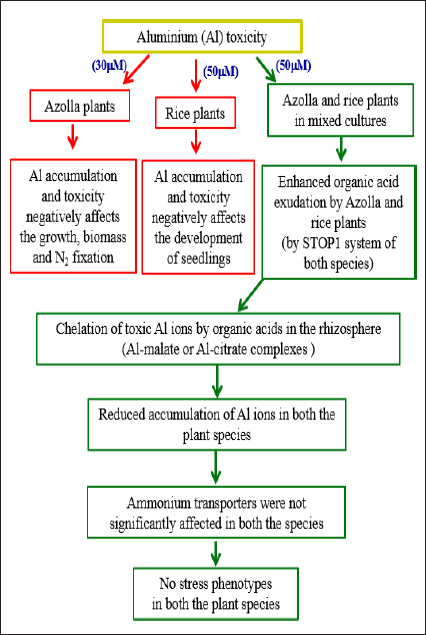
1. INTRODUCTION
Acidic soil is one of the most common issues faced by farmers in agricultural fields, where major crops are grown [1]. Acidic soil environments are stressful as well as toxic to plants [2]. When soil and surface water are acidified to a pH below 5.0, monomeric aluminum hydroxides and Al-containing minerals are dissolved to form polynuclear aluminum species and trivalent aluminum (Al3+) that are readily absorbed by plants [3,4]. However, soluble Al like aquo/hydroxo-Al complexes have deleterious effects on plant growth and survival, primarily by hindering the uptake of nutrients from the soil [5,6]. In plant tissues, Al content ranges from 0.2 to 1000 mg/kg, and variations in Al accumulation are regulated by different factors such as plant species, soil type, and soil nutrients. It has been reported that excess Al is found in those plants that grow in acidic soils mainly [7]. Al exists in soil in the form of various ions, among which Al3+ is the most toxic [6,8]. Al also forms insoluble oxides and hydroxides at pH 5.0 to 6.2, as well as polynuclear dissolved species below pH 5.0, that determine Al toxicity [5,9].
In Arabidopsis, Al3+ ions are recognized and regulated by several genes, transcription factors, and transporters [10]. The principal functional tool for plant tolerance to Al3+ includes Al3+-induced stimulation of transporters that accelerates the discharge of organic acids from the roots. When the amount of Al3+ increases in the soil, it is either chelated by organic acids that are excreted from the roots into the rhizosphere or Al enters the plant cells through the roots via absorption [11]. Multidrug and Toxic compound Extrusion [MATE] and aluminum activated Malate Transporters [ALMT1] are key transporters involved in the release of citrate and malate, respectively, in most plants. These transporters are regulated by the STOP1 system [12-14]. Rice plants displayed a high level of Al-tolerance when compared with other crop plants [15]. In particular, sensitive to aluminum rhizotoxicity 1 [STAR1] and sensitive to aluminum rhizotoxicity 2 [STAR2], which encode domains of a bacterial-type ATP-binding cassette [ABC] transporter, and Al-resistance transcription factor 1 [ART1], a homologue of STOP1, are key genes involved in Al tolerance. These genes are mainly expressed in roots and regulate multiple genes for Al tolerance in rice plants [16]. The Azolla plant is a heterosporous aquatic fern characterized by high rates of multiplication and biological nitrogen fixation potential due to the presence of cyanobacterial symbionts. The cyanobacterial symbiont in Azolla can fix a significant amount of nitrogen [0.15–0.17 mg Nh-1g-1 of dry biomass] compared with Rhizobia, which fixes only 0.08 mg Nh-1g-1 of dry biomass in Glycine max [17]. In addition, studies have found that under stress conditions, Azolla can also increase the nitrogen use efficiency (NUE) of rice plants, thereby increasing grain yield [18]. The peripheral zone of cavities in each dorsal frond of Azolla is filled with a mucilaginous fibrillar network in which N2-fixing cyanobacteria reside and release ammonium for utilization by their host [19,20]. Azolla plants can survive in various pH ranges [3.5 to 10] [21] and can also accumulate different metal ions [22-24]. Furthermore, Azolla is a plant frequently intercropped with rice to take advantage of nitrogen fixed by the Azolla-Anabaena symbiosis [25]. Apart from these, no other major advantages were recorded for intercropping rice with Azolla. Therefore, this study focuses on the effects of Al on the growth and development of Azolla and rice plants under acidic aquatic environmental conditions. Since soil acidity is increasing due to global warming and other factors [26], we selected Al toxicity studies in the rice-Azolla intercropping system. Likewise, the effects of Al stress on the growth and development of Azolla pinnata plants have not been studied. We also investigated the effects of Al stress on the expression profiles of ammonium transporters [AMTs] and STOP1 genes in both species. STOP1 regulates Al tolerance levels by controlling organic acid exudation, whereas AMT1 controls ammonium uptake in both plant species [12, 27].
Several intercropping methods have achieved remarkable results in rice improvement programs. Recent reports clearly demonstrated that the intercropping of rice with different plant species uncovered several advantages, like increased yield, reduced heavy metal accumulation in the plant, confined N fertilizer application, etc. [28,29]. In particular, intercropping of water spinach and rice plants can control pests, increase yields [30], reduce cadmium [28] and arsenic accumulation [31], and increase silicon absorption [29]. Reduced pest attack and increased yield parameters were also recorded in the rice-Pontederia cordata intercropping methods [32]. Interestingly, intercropping upland rice with forage grasses produces a higher yield, biomass, and N content [33]. In addition, polyculture of rice and water mimosa can help reduce the application of N fertilizer and reduce the loss of N from the agriculture field [34]. Studies that utilized fresh Azolla pinnata or its compost powder in rice fields, with or without inorganic nitrogen application, reported increased yield and N uptake of rice plants, improved nutrient content of soil, as well as reductions in weeds and ammonia volatilization [35-40]. Intriguingly, the impact of Al stress on the growth and development of rice plants is well-researched [41-46], but the effects induced by Al on Azolla and rice plants under mixed growth conditions have not been studied yet. Hence, the aim of this study was to characterize the potential of Azolla pinnata as an Al toxicity mitigator in the rice-Azolla pinnata intercropping system. We therefore investigated Azolla pinnata and IR64 rice seeds in individual cultures as well as in mixed cultures to study the effect of Al toxicity in an acidic medium on both plants in relation to phenotype, microscopic, physiological, and biochemical responses. We further studied the expression profiles of the STOP1 and AMT1 genes of both plants in both cultures. Lastly, aluminum uptake by both species was determined and discussed in relation to the application of Azolla pinnata in rice fields for alleviating Al toxicity in acid soil.
REFERENCES
1. Jones DL, Kochian LV. Aluminum inhibition of the inositol 1, 4, 5-trisphosphate signal transduction pathway in wheat roots: a role in aluminum toxicity? Plant Cell. 1995;7(11):1913-22. [CrossRef]
2. Bian M, Zhou M, Sun D, Li C. Molecular approaches unravel the mechanism of acid soil tolerance in plants. Crop J. 2013;1(2):91-104. [CrossRef]
3. Furrer G, Trusch B, Müller C. The formation of polynuclear Al13 under simulated natural conditions. Geochimica et cosmochimica acta. 1992;56(10):3831-8. [CrossRef]
4. Kinraide T. Reconsidering the rhizotoxicity of hydroxyl, sulphate, and fluoride complexes of aluminum. J Exp Bot. 1997;48(5):1115-24. [CrossRef]
5. Neenu S, Karthika KS. Aluminum toxicity in soil and plants. Harit Dhara. 2019;2(1):15-9.
6. Mossor-Pietraszewska T. Effect of aluminum on plant growth and metabolism. Acta Biochim Pol. 2001;48(3):673-86. [CrossRef]
7. Gensemer RW, Playle RC. The bioavailability and toxicity of aluminum in aquatic environments. Crit Rev Environ Sci Technol. 1999;29(4):315-450. [CrossRef]
8. Bojórquez-Quintal E, Escalante-Magaña C, Echevarría-Machado I, Martínez-Estévez M. Aluminum, a friend or foe of higher plants in acid soils. Front Plant Sci. 2017;8:1767. [CrossRef]
9. Kobayashi Y, Ohyama Y, Kobayashi Y, Ito H, Iuchi S, Fujita M, et al. STOP2 activates transcription of several genes for Al-and low pH-tolerance that are regulated by STOP1 in Arabidopsis. Mol Plant. 2014;7(2):311-22. [CrossRef]
10. Nguyen NT, Nakabayashi K, Thompson J, Fujita K. Role of exudation of organic acids and phosphate in aluminum tolerance of four tropical woody species. Tree Physiol. 2003;23(15):1041-50. [CrossRef]
11. Kobayashi Y, Kobayashi Y, Watanabe T, Shaff JE, Ohta H, Kochian LV et al. Molecular and physiological analysis of Al3+ and H+ rhizotoxicities at moderately acidic conditions. Plant Physiol. 2013;163(1):180-192. [CrossRef]
12. Kundu A, Das S, Basu S, Kobayashi Y, Kobayashi Y, Koyama H, Ganesan M. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for Aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 2019;21(1):35-44. [CrossRef]
13. Kundu A, Ganesan M. GhMATE1 expression regulates Aluminum tolerance of cotton and overexpression of GhMATE1 enhances acid soil tolerance of Arabidopsis. Curr Plant Biol. 2020;24:100160. [CrossRef]
14. Minh VQ, Vu PT, Giao NT. Soil properties characterization and constraints for rice cultivation in Vinh Long Province, Vietnam. J Applied Biol Biotechnol. 2024;12:98-105. [CrossRef]
15. Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell. 2009;21(10):3339-49. [CrossRef]
16. Arora M, Kaur A. Azolla pinnata, Aspergillus terreus and Eisenia fetida for enhancing agronomic value of paddy straw. Sci Rep. 2019;9(1):1341. [CrossRef]
17. Brouwer P, Schluepmann H, Nierop KG, Elderson J, Bijl PK, van der Meer I, et al. Growing Azolla to produce sustainable protein feed: the effect of differing species and CO2 concentrations on biomass productivity and chemical composition. J Sci Food Agri. 2018; 98(12):4759-68. [CrossRef]
18. Manna AB, Singh PK. Rice yields as influenced by Azolla N2 fixation and urea N-fertilization. Plant Soil. 1989;114(1):63-68. [CrossRef]
19. Singh PK. Effect of Azolla on the yield of paddy with and without application of N fertilizer. Curr Sci. 1977;46(18):642-644.
20. Xu H, Zhu B, Liu J, Li D, Yang Y, Zhang K, et al. Azolla planting reduces methane emission and nitrogen fertilizer application in double rice cropping system in southern China. Agron Sustainable Dev. 2017;37(4):1-9. [CrossRef]
21. Wagner GM. Azolla: a review of its biology and utilization. Botanic Rev. 1997;63(1):1-26. [CrossRef]
22. Naz M, Raza MA, Bouzroud S, Ali E, Bukhari SAH, Tariq M, Fan X. Sustainability Aspects of Nano-Remediation and Nano-Phytoremediation. In: Tonelli FMP, Bhat RA, Dar GH, editors. Nanotechnology For Environmental Pollution Decontamination, New York: Apple Academic Press;2023:509-25.
23. Roberts AE, Boylen CW, Nierzwicki-Bauer SA. Effects of lead accumulation on the Azolla caroliniana-Anabaena association. Ecotoxicol Environ Saf. 2014;102:100-4. [CrossRef]
24. Valderrama A, Carvajal DE, Peñailillo P, Tapia J. Accumulation capacity of cadmium and copper and their effects on photosynthetic performance in Azolla filiculoides Lam. under induced rhizofiltration. Gayana Botánica. 2016;73(2):283-91. https://revistas.udec.cl/index.php/gayana_botanica/article/view/4103
25. Akhtar M, Sarwar N, Ashraf A, Ejaz A, Ali S, Rizwan M. Beneficial role of Azolla sp. in paddy soils and their use as bioremediators in polluted aqueous environments: implications and future perspectives. Arch Agron Soil Sci. 2021;67(9):1242-55. [CrossRef]
26. Hajiboland R, Panda CK, Lastochkina O, Gavassi MA, Habermann G, Pereira JF. Aluminum toxicity in plants: Present and future. J. Plant Growth Regul. 2023; 42(7):3967-99. [CrossRef]
27. TsutsuiT, Yamaji N, Ma JF. Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 2011;156:925-31. [CrossRef]
28. Kang Z, Zhang W, Qin J, Li S, Yang X, Wei X, Li H. Yield advantage and cadmium decreasing of rice in intercropping with water spinach under moisture management. Ecotoxicol Environ Saf. 2020;190:110102. [CrossRef]
29. Ning C, Wang L, Liu R, Pan T, Cai Y, Tian J, et al. Plant-mediated rhizospheric interactions in rice and water spinach intercropping enhance Si uptake by rice. Plant Soil. 2022;477:183-99. [CrossRef]
30. Liang K, Yang T, Zhang S, Zhang JE, Luo M, Fu L, Zhao B. Effects of intercropping rice and water spinach on net yields and pest control: An experiment in southern China. Int J Agric Sustain. 2016;14(4):448-65. [CrossRef]
31. Huang SY, Zhuo C, Du XY, Li HS. Remediation of arsenic-contaminated paddy soil by intercropping aquatic vegetables and rice. Int J Phytoremediation. 2021;23(10):1021-9. [CrossRef]
32. Xiang H, Lan N, Wang F, Zhao B, Wei H, Zhang J. Reduced pests, improved grain quality and greater total income: benefits of intercropping rice with Pontederia cordata. J Sci Food Agric. 2021;101(14):5907-17. [CrossRef]
33. Crusciol CA, Momesso L, Portugal JR, Costa CH, Bossolani JW, Costa NR, et al. Upland rice intercropped with forage grasses in an integrated crop-livestock system: optimizing nitrogen management and food production. Field Crops Res. 2021;261:108008. [CrossRef]
34. Hei Z, Xiang H, Zhang J, Liang K, Zhong J, Li M, Lu Y. Rice intercropping with water mimosa (Neptunia oleracea Lour.) can facilitate soil N utilization and alleviate apparent N loss. Agricul Ecosys Environ. 2021;313:107378. [CrossRef]
35. Setiawati MR, Damayani M, Herdiyantoro D, Suryatmana P, Anggraini D, Khumairah FH. The application dosage of Azolla pinnata in fresh and powder form as organic fertilizer on soil chemical properties, growth and yield of rice plant. In: AIP Conference Proceedings. 2018;(Vol. 1927, No. 1). AIP Publishing.
36. Singh AL, Singh PK. Comparative studies on different methods of Azolla utilization in rice culture. J Agri Sci. 1986;107(2):273-8. [CrossRef]
37. Bhuvaneshwari K, Singh PK. Response of nitrogen-fixing water fern Azolla biofertilization to rice crop. 3 Biotech. 2015;5:523-9. [CrossRef]
38. Kandel S, Malla R, Adhikary BH, Vista SP. Effect of Azolla application on rice production at mid-hills condition of Nepal. Trop. Agroeco. 2020;1(2):103-6. 10.26480/taec.02.2020.103.106
39. Seleiman MF, Elshayb OM, Nada AM, El-leithy SA, Baz L, Alhammad BA, Mahdi AH. Azolla compost as an approach for enhancing growth, productivity and nutrient uptake of Oryza sativa L. Agronomy. 2022;12(2):416. [CrossRef]
40. Marzouk SH, Tindwa HJ, Amuri NA, Semoka JM. An overview of underutilized benefits derived from Azolla as a promising biofertilizer in lowland rice production. Heliyon. 2023; 9(1):e13040. [CrossRef]
41. Ma JF, Shen RF, Zhao Z, Wissuwa M, Takeuchi Y, Ebitani T, Yano M. Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol. 2002;43:652-9. [CrossRef]
42. Roy B, Bhadra S. Hydroponic screening for selection of aluminum tolerant rice (Oryza sativa L.) genotypes at seedlings stage using different indices. Cereal Res. Comm. 2014; 42(3):463-73. [CrossRef]
43. Awasthi JP, Saha B, Regon P, Sahoo S, Chowra U, Pradhan A, et al. Morpho-physiological analysis of tolerance to aluminum toxicity in rice varieties of North East India. PLOS ONE. 2017;12(4):e0176357. [CrossRef]
44. Wang M, Qiao J, Yu C, Chen H, Sun C, Huang L, Li C, Geisler M, Qian Q, Jiang DA, Qi Y. The auxin influx carrier, OsAUX3, regulates rice root development and responses to aluminum stress. Plant Cell Environ. 2019;42(4):1125-38. [CrossRef]
45. Tyagi W, Yumnam JS, Sen D, Rai M. Root transcriptome reveals efficient cell signaling and energy conservation key to aluminum toxicity tolerance in acidic soil adapted rice genotype. Sci Rep. 2020; 10(1):4580. [CrossRef]
46. Pradhan AK, Shandilya ZM, Sarma P, Bora RK, Regon P, Vemireddy LN, Tanti B. Concurrent effect of aluminum toxicity and phosphorus deficiency in the root growth of aluminum tolerant and sensitive rice cultivars. Acta Physiologiae Plantarum. 2023; 45(2):33. [CrossRef]
47. Naito S, Hirai MY, Chino M, Komeda Y. Expression of a soybean (Glycine max L. Merr.) seed storage protein gene in transgenic Arabidopsis thaliana and its response to nutritional stress and to abscisic acid mutations. Plant Physiol. 1994;104:497-503. [CrossRef]
48. Matsumoto H, Motoda H. Aluminum toxicity recovery processes in root apices. Possible association with oxidative stress. Plant Sci. 2012;185:1-8. [CrossRef]
49. Duan B, Lu Y, Yin C, Junttila O, Li C. Physiological responses to drought and shade in two contrasting Picea asperata populations. Physiol Plant. 2005;124(4):476-84. [CrossRef]
50. Parida AK, Das AB, Mohanty P. Defense potentials to NaCl in a mangrove, Bruguiera parviflora: differential changes of isoforms of some antioxidative enzymes. J Plant Physiol. 2004;161(5):531-42. [CrossRef]
51. Yoshida K, Kaothien P, Matsui T, Kawaoka A, Shinmyo A. Molecular biology and application of plant peroxidase genes. Appl Microbiol Biotechnol. 2003; 60(6):665-70. [CrossRef]
52. Ritchie RJ. Universal chlorophyll equations for estimating chlorophylls a, b, c, and d and total chlorophylls in natural assemblages of photosynthetic organisms using acetone, methanol, or ethanol solvents. Photosynthetica. 2008;46(1):115-6. [CrossRef]
53. Hampp R, Goller M, Fullgraf H. Determination of compartmented metabolite pools by a combination of rapid fractionation of oat mesophyll protoplasts and enzymic cycling. Plant Physiol. 1984;75(4):1017-21. [CrossRef]
54. Freschet GT, Pagès L, Iversen CM, Comas LH, Rewald B, Roumet C et al. A starting guide to root ecology: strengthening ecological concepts and standardising root classification, sampling, processing and trait measurements. New Phytol. 2021;232(3):973-1122. [CrossRef]
55. Teramoto S, Takayasu S, Kitomi Y, Arai-Sanoh Y, Tanabata T, Uga Y. High-throughput three-dimensional visualization of root system architecture of rice using X-ray computed tomography. Plant Methods. 2020;16(1):1-14. http://orcid.org/10.21203/rs.2.18397/v2
56. Kösesakal T, Yildiz M. Growth performance and biochemical profile of Azolla pinnata and Azolla caroliniana grown under greenhouse conditions. Arch Biol Sci. 2019;71(3):475-82. http://orcid.org/0000-0003-1818-3813
57. Tran TL, Miranda AF, Abeynayake SW, Mouradov A. Differential production of phenolics, lipids, carbohydrates and proteins in stressed and unstressed aquatic plants, Azolla filiculoides and Azolla pinnata. Biology. 2020;9(10):342. [CrossRef]
58. Li W, Du J, Feng H et al. Function of NHX-type transporters in improving rice tolerance to aluminum stress and soil acidity. Planta. 2020;251:1-13. [CrossRef]
59. Miftahudin M, Roslim DI, Fendiyanto MH, Satrio RD, Zulkifli A, Umaiyah EI, et al. OsGERLP: A novel aluminum tolerance rice gene isolated from a local cultivar in Indonesia. Plant Physiol Biochem. 2021;162:86-99. [CrossRef]
60. Phukunkamkaew S, Tisarum R, Pipatsitee P, Samphumphuang T, Maksup S, Cha-Um S. Morpho-physiological responses of indica rice (Oryza sativa sub. indica) to aluminum toxicity at seedling stage. Environ Sci Pollut Res. 2021;28:29321-31. [CrossRef]
61. Esterbauer H. Cytotoxicity and genotoxicity of lipid-oxidation products. Am J Clin Nutr. 1993;57(5):779S-86S. [CrossRef]
62. Kong W, Liu F, Zhang C, Zhang J, Feng H. Non-destructive determination of Malondialdehyde (MDA) distribution in oilseed rape leaves by laboratory scale NIR hyperspectral imaging. Sci Rep. 2016;6(1):1-8. [CrossRef]
63. Zhang X, Chan T, Mak M. Morphodynamic signatures of MDA-MB-231 single cells and cell doublets undergoing invasion in confined microenvironments. Sci Rep. 2021;11(1):1-10. [CrossRef]
64. Tognolli M, Penel C, Greppin H, Simon P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene. 2002;288:129-38. [CrossRef]
65. Wu Y, Yang Z, How J, Xu H, Chen L, Li K. Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol Biol. 2017;95:157-68. [CrossRef]
66. Apostolova EL, Dobrikova AG, Ivanova PI, Petkanchin IB, Taneva SG. Relationship between the organization of the PSII supercomplex and the functions of the photosynthetic apparatus. J Photochem Photobiol B. 2006;83(2):114-22. [CrossRef]
67. Azura AE, Shamshuddin J, Fauziah CI. Root elongation, root surface area and organic acid by rice seedling under Al 3+ and/or H+ stress. Am J Agric Biol Sci. 2011;6(93):324-31. [CrossRef]
68. Barker AV, Pilbeam DJ (Eds.). Handbook of plant nutrition. 2nd ed: CRC Press; 2007.
69. Zeng F, Chen S, Miao Y, Wu F, Zhang G. Changes of organic acid exudation and rhizosphere pH in rice plants under chromium stress. Environ Pollut. 2008;155(2):284-9. [CrossRef]
70. Sadhukhan A, Enomoto T, Kobayashi Y, Watanabe T, Iuchi S, Kobayashi M et al. Sensitive to proton rhizotoxicity1 regulates salt and drought tolerance of Arabidopsis thaliana through transcriptional regulation of CIPK23. Plant Cell Physiol. 2019;60(9):2113-26. [CrossRef]
71. Fang XZ, Fang SQ, Ye ZQ, Liu D, Zhao KL, Jin CW. NRT1. 1 dual-affinity nitrate transport/signalling and its roles in plant abiotic stress resistance. Front Plant Sci. 2021;12:715694. [CrossRef]
72. Ye JY, Tian WH, Zhou M, Zhu QY, Du WX, Zhu YX, et al. STOP1 activates NRT1.1-mediated nitrate uptake to create a favorable rhizospheric pH for plant adaptation to acidity. Plant Cell. 2021;33(12):3658-74. [CrossRef]
73. Bu Y, Takano T, Liu S. The role of ammonium transporter (AMT) against salt stress in plants. Plant Signal Behav. 2019;14(8):1625696. [CrossRef]
74. Straub T, Ludewig U, Neuhäuser B. The kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell. 2017;29(2):409-22. [CrossRef]
75. Luo L, Zhu M, Jia L, Xie Y, Wang Z, Xuan W. Ammonium transporters cooperatively regulate rice crown root formation responding to ammonium nitrogen. J Exp Bot. 2022;73(11):3671-85. [CrossRef]
76. Kishchenko O, Stepanenko A, Straub T, Zhou Y, Neuhäuser B, Borisjuk N. Ammonium uptake, mediated by ammonium transporters, mitigates manganese toxicity in Duckweed, Spirodela polyrhiza. Plants. 2023;12(1):208. [CrossRef]
77. Liu K, Sakuraba Y, Ohtsuki N, Yang M, Ueda Y Yanagisawa S. CRISPR/Cas9-mediated elimination of OsHHO3, a transcriptional repressor of three AMMONIUM TRANSPORTER1 genes, improves nitrogen use efficiency in rice. Plant Biotechnol. J. 2023;21(11):2169. [CrossRef]
78. Wu XX, Yuan DP, Chen H, Kumar V, Kang SM, Jia B, et al. Ammonium transporter 1 increases rice resistance to sheath blight by promoting nitrogen assimilation and ethylene signalling. Plant Biotechnol J. 2022;20(6):1085-97. [CrossRef]
79. Singh VP, Tripathi DK, Kumar D, Chauhan DK. Influence of exogenous silicon addition on aluminum tolerance in rice seedlings. Biol Trace Elem Res. 2011;144:1260-74. [CrossRef]
80. van Kempen MM. Azolla on top of the world: an ecophysiological study of floating fairy moss and its potential role in ecosystem services related to climate change (Doctoral dissertation, SI:[Sn]); 2013.
81. Jing CH, Zhao XQ, Shen RF. Molecular mechanisms of plant adaptation to acid soils: a review. Pedosphere. 2023;33(1):14-22. [CrossRef]
82. Das S, Ganesan M. Aluminum induced malate transporter (ALMT1) is regulating the Aluminum stress tolerance responses of mungbean seedlings. Plant Gene. 2022;32:100388. [CrossRef]