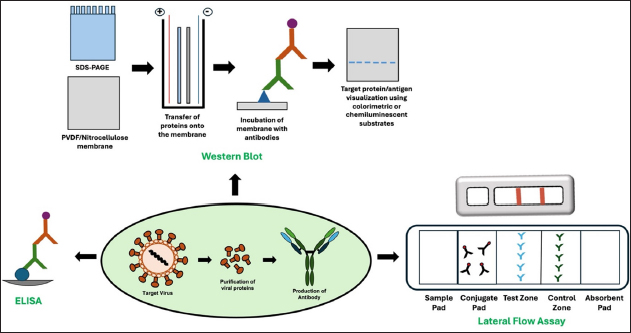
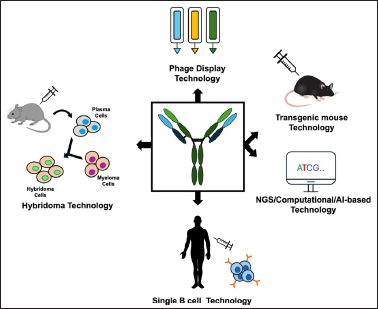
5. DEVELOPMENT OF MONOCLONAL ANTIBODIES FOR THE DETECTION OF VIRUSES OTHER THAN SARS-CoV-2
While the importance of monoclonal antibodies in diagnostics became prominently evident during the COVID-19 pandemic, it is noteworthy that even before this global health crisis, monoclonal antibodies were extensively utilized for detecting various viral diseases. Several research groups have reported the development and applications of monoclonal antibodies for detecting viruses other than SARS-CoV-2 [Table 4]. During the selection of targets for antibody development in diagnostic applications, a critical factor is choosing proteins abundant in the virus particle, such as spike and nucleocapsid proteins in the case of SARS-CoV-2, which are also highly immunogenic. These proteins facilitate the production of high-affinity antibodies essential for sensitive detection in immunodiagnostic assays.
Table 4: Studies reporting the development of monoclonal antibodies for detecting viruses other than SARS-CoV-2.
| S. No. | Virus | Immunogen | Method Used for Antibody Characterization | Detection Limit | References |
|---|
| 1. | Porcine Circovirus 3 | Capsid protein | Indirect ELISA, Western blot, IFA, Dot blot, EB-ELISA | Not reported | [22] |
| 2. | Varicella-zoster virus (VZV) | Glycoprotein E (gE) | IFA, immunoperoxidase monolayer assay, ELISA, Western Blot, LFA | 30 ng/mL of purified gE using LFA | [61] |
| 3. | Pseudorabies virus | Inactivated whole virus protein | Indirect ELISA, IFA, Western Blot | Not reported | [24] |
| 4. | Classical swine fever virus | Envelope protein E2 | Indirect ELISA, Western blot, competition ELISA | Not reported | [71] |
| 5. | Cache Valley virus (CVV) | Nucleoprotein | ELISA, Western blot, IFA, MAC-ELISA | Not reported | [23] |
| 6. | Hepatitis E virus (HEV) | HEV ORF3 protein | Indirect ELISA, Western blot, Competitive ELISA | Not reported | [70] |
| 7. | Zika virus | Envelope protein | ELISA, LFA | 33 μg/mL of recombinant E protein and 6.3×106 PFU/mL of ZIKV using LFA | [62] |
| 8. | H3 influenza A virus | Inactive H3 virus | Sandwich ELISA, Western Blot | >104 virus dilution using Sandwich ELISA; Amount not reported | [64] |
| 9. | Chikungunya Virus | Envelope 2 protein | ELISA, Immunoblotting | 0.7 µg/mL inactivated CHIKV using indirect ELISA | [68] |
| 10. | MERS-CoV | Spike protein | Sandwich ELISA | 5.89 ng/mL of MERS-CoV S protein using Sandwich ELISA | [65] |
| 11. | Zaire Ebola virus | Zaire Ebola virus glycoprotein | SDS-PAGE, Western blot, Dot blot, Indirect, IFA, sandwich ELISA | 3.6 ng/mL rGPdTM with Sandwich ELISA | [30] |
| 12. | Dengue Virus (DENV-4) | NS1 | Western Blot, Sandwich ELISA | 32.5 ng/mL of NS1 protein with Sandwich ELISA | [66] |
| 13. | Zucchini yellow mosaic virus | ZYMV virion | ACP-ELISA, dot-ELISA, tissue dot-ELISA, DAS-ELISA and IC-RT-PCR | ACP-ELISA (1:163840*), dot-ELISA (1:2560*), DAS-ELISA (1:327680*), IC-RT-PCR (1:1310720*)
* Dilution of ZYMV-infected crude extracts | [72] |
| 14. | Yellow fever virus | Recombinant Envelope protein and YF vaccine virus 17D | Indirect ELISA, Western blot, IFA, and sandwich ELISA | 1 x 103 FFU/mL of YFV 17D or 2ng/well of recombinant envelop protein using sandwich ELISA | [67] |
| 15. | MERS-CoV | Truncated Nucleocapsid Protein | Immunoprecipitation assay, Sandwich ELISA, LFA | 0.5 ng of recombinant protein and 3 x 104 copies of MERS-CoV virions using LFA | [63] |
| 16. | Chikungunya Virus | Capsid protein | ELISA, Western blot, IFA, and IHC | Not reported | [40] |
| 17. | Tembusu Virus | Envelope protein | Western Blot, Sandwich ELISA | Not reported | [69] |
| 18. | SARS-CoV | Spike protein subunit S1 | SDS-PAGE, Western Blot, Indirect ELISA, Sandwich ELISA | 0.019 µg/mL of S1 protein using Sandwich ELISA | [38] |
| 19. | Seneca Valley virus | Binary ethylenimine (BEI) inactivated SVV | Dot blot, IHC, competitive ELISA | Not reported | [41] |
Since LFAs can facilitate quick diagnosis and containment efforts, several groups have focused on developing LFA-based tests for detecting viruses other than SARS-CoV-2 as well. Wang et al., developed an LFA-based test for the detection of Varicella-zoster virus (VZV), which causes varicella and herpes zoster [61]. They developed mouse monoclonal antibodies against glycoprotein E present on the surface of VZV in high copy numbers, using hybridoma technology. Upon completing the antibody characterization, the antibodies 2F2 (as capture antibody) and 118H2 (as detector antibody) were selected for LFA. The test allowed the detection of 30 ng/mL VZV gE antigen without any cross-reactivity against Enterovirus 71 or Herpes Simplex Virus 1 and 2. This study emphasizes the potential of monoclonal antibodies for developing specific detection tests with no cross-reactivity to related antigens.
Similarly, Li et. al., developed an LFA-based test using mouse monoclonal antibodies against the envelope (E) protein of the Zika Virus (ZIKV), which is a structural protein present on the surface of the virus [62]. Out of 4 antibodies, antibody 9E-1 was found to be highly specific to ZIKV and had a sub-nanomolar affinity. It was used to develop an LFA-based test along with an antibody B1, and the test was able to detect 33 μg/mL of recombinant E protein and 6.3×106 PFU/mL of ZIKV in culture supernatant. Yamaoka et. al., developed an LFA-based test to detect Middle East respiratory syndrome coronavirus (MERS-CoV) [63]. Using hybridoma technology, they developed mouse monoclonal antibodies specific to nucleocapsid protein (122-413 amino acids). Seven antibodies specific to the target protein were tested using immunoprecipitation, which allowed the successful capture of the antigen from cell lysates. Further, sandwich ELISA was used to test all possible combinations of 7 antibodies as capture and detection reagents. Antibody pair 46/20 was found to exhibit maximum reactivity using antigen capture ELISA. It allowed the detection of 0.0625 ng recombinant antigen or 1.5 x 105 copies of MERS-CoV virions in a 0.1 mL sample. The same pair allowed the detection of 0.5 ng of purified protein and 3 x 106 copies of MERS-CoV virions in a 0.1 mL sample using colloidal gold-based LFA.
Another commonly explored format for developing monoclonal antibody-based diagnostics is sandwich ELISA, which typically offers high sensitivity and quantitative measurement of the target. However, it also necessitates specialized equipment (such as plate readers) and technical proficiency, in contrast to the rapid, user-friendly nature of LFAs, despite their lower sensitivity and limited quantification ability. Several groups have reported the development of sandwich ELISA-based tests to detect viruses. Luo et al., developed a sandwich ELISA-based method for detecting the avian influenza virus (AIV), which can lead to the emergence of zoonotic infections [64]. They developed mouse monoclonal antibodies using hybridoma technology. The specificity of antibody 9F12 was determined using hemagglutination inhibition (HI) against different viruses and was found to be specific to the H3 subtype with no cross-reactivity to other viruses. 9F12 also showed activity in Western blot and IFA and allowed specific detection of H3 strain in clinical specimens using sandwich ELISA. Lee et al. developed a sandwich ELISA to detect MERS-CoV [65]. They developed mouse monoclonal antibodies against the MERS-CoV spike protein. The best antibody pair allowed detection of MERS-CoV S protein with LOD of 5.89 ng/mL using sandwich ELISA. Zai et. al., developed mouse monoclonal antibodies against recombinant glycoprotein of Zaire Ebola virus (rGPdTM; glycoprotein without the transmembrane domain) using hybridoma technology [30]. Two monoclonal antibodies, 6E3 and 3F21, were characterized using multiple assays. In dot blot analysis, 3F21 demonstrated higher sensitivity compared to 6E3, a finding further supported by BLI results indicating that 3F21 exhibited higher affinity. Using these two antibodies in sandwich ELISA allowed the detection of 3.6 ng/mL rGPdTM. Gelanew and Hunsperger produced antibodies against the NS1 protein of the Dengue virus-4 serotype (DENV-4), which is detected by commercially available NS1 antigen tests with limited sensitivity [66]. Three selected antibodies were characterized using Sandwich ELISA, and the best antibody pair, 8A6F2 (capture) and 6D4B10 (detector), exhibited the LOD of 32.5 ng/mL of NS1 protein and allowed specific detection of DENV-4 in cell culture supernatants with no reactivity from other DENV serotypes. Adungo et. al., reported the development of 8 mouse monoclonal antibodies for detecting yellow fever virus (YFV), which spreads through mosquito bites [67]. They employed recombinant envelop protein as well as YF vaccine virus 17D for immunization in mice and successfully isolated 4 antibodies against each target. All the antibodies were found to be highly specific to YFV with no cross-reactivity to related DENV and Japanese Encephalitis virus. All the antibodies also showed reactivity in the IFA. Furthermore, using two antibodies, namely 4C9 (for capture) and 3F4 (for detection), enabled the detection of 1 x 103 focus forming units/mL of YFV 17D or 2 ng/well of recombinant envelope protein using sandwich ELISA. This study highlights the utility of monoclonal antibodies for developing cost-effective, highly sensitive, and specific diagnostic tests for arboviruses like YFV that are endemic in regions like Africa, where routine testing measures may be lacking. J. Kim et al., developed mouse monoclonal antibodies against the envelope protein E2 of Chikungunya Virus (CHIKV), which is another virus that spreads through mosquito bites [68]. Four antibodies were characterized using ELISA and Western blot, out of which, two antibodies, 9-1 and 21-1 efficiently recognized CHIKV-E2 protein, and the 9-1 antibody showed no cross-reactivity against other related viruses, such as ZIKV, JEV, and DENV. Further, the antibody 9-1 allowed the detection of as low as 0.7 µg/mL inactivated CHIKV using ELISA and can be explored for the development of new CHIKV diagnostic techniques. Goh et al. worked on a different CHIKV target and developed 11 mouse monoclonal antibodies against the CHIKV capsid protein using hybridoma technology and demonstrated their applications in multiple assays [40]. All antibodies were found to show reactivity against CHIKV using ELISA, Western blot, and immunofluorescence performed using cells expressing recombinant capsid protein or infected with CHIKV. One antibody, 5.5G9, also allowed target protein detection in scattered macrophage-like cells using IHC, demonstrating the versatility of monoclonal antibody-based assays for diagnostics. H. Chen et. al., reported a sandwich ELISA-based method for detecting Tembusu Virus (TMUV), which causes infection in waterfowls [69]. Using hybridoma technology, they developed 3 mouse monoclonal antibodies against the TMUV envelope protein. The antibodies were characterized using Western Blot and were highly specific to TMUV. Antibody 12B1 was used as a capture antibody with 2D2 for detection to develop TMUV-specific ELISA. The assay was compared to RT-PCR and was found to be 99.1% specific and 93.1% sensitive compared to RT-PCR, further underscoring that monoclonal antibody-based tests are a viable alternative to nucleic-acid-based tests. Sunwoo et al., reported the development of sandwich ELISA for the detection of SARS-CoV, responsible for the first outbreak of SARS in 2002 [38]. They developed mouse monoclonal antibodies using hybridoma technology against the spike protein subunit S1 of the SARS-CoV. Based on the antibody titers, three clones, P135.3F3, P1368D12, and F26G18, were selected for further characterization. They also generated bispecific antibodies by fusion of F26G18 and P136.8D12 hybridoma clones with anti-HRPO hybridoma YP4 to generate quadromas. The use of F26G18 as a coating antibody and its biotinylated version as a detector antibody, sandwich ELISA, allowed the detection of 0.037 µg/mL S1 antigen. Furthermore, the LOD improved to 0.019 µg/mL when they used bi-specific monoclonal antibody F157 (F26G18 x YP4) as the detector antibody, indicating that bispecific antibodies can be explored for enhancing the detection limits of monoclonal antibodies. Competition ELISA has also been explored for the detection of viruses. B. Zhang et al., developed a competitive ELISA for the detection of hepatitis E caused by genotype 1 of the hepatitis E virus (HEV) [70]. They developed 7 mouse monoclonal antibodies against recombinant genotype 1 HEV ORF3 protein using hybridoma technology, out of which 2 antibodies 3C11 and 1D2 were specific to the human HEV SAR-55 strain. Out of the two, one antibody, 1D2 was, showed higher reactivity in competitive ELISA, which can be helpful for large-scale serological testing and clinical diagnosis of HEV infections.
Apart from viruses affecting human health, monoclonal antibody-based tests have been developed to detect viruses responsible for diseases in animals as well. J. Wang et. al., developed four mouse monoclonal antibodies using hybridoma technology against the capsid protein of PCV3, which is a significant cause of disorders such as multi-organ inflammation, nephrotic syndrome, reproductive disorders, and dermatitis in swine [22]. These antibodies were characterized using indirect ELISA, Western Blot, IFA, and Dot Blot, and antibody 7E3 was found to have the highest binding affinity to the target protein. Finally, the B cell epitope of the 7E3 antibody was determined, and an epitope-blocking ELISA (EB-ELISA) was designed to detect PCV3 antibodies in sera, which showed high specificity and sensitivity. This study exemplifies using monoclonal antibodies for monitoring and managing PCV3 infections in swine farms. Similarly, J. Zhang et. al., developed mouse monoclonal antibodies against envelope protein E2 protein using hybridoma technology to detect classical swine fever virus (CSFV), which is a cause of concern in pig breeding industries. After thorough characterization, four antibodies were found to be specific to the sub-genotype 2.1 strain, and out of these, two antibodies, MM1 and MM5, were found to recognize critical epitopes on the E2 protein that were present in 90.9% of the genotype sequences available in GeneBank. Such antibodies can be promising reagents for developing assays like indirect and competition ELISA for detecting CSFV [71]. Guo et. al., also developed monoclonal antibodies using hybridoma technology against the glycoprotein E (gE) of the pseudorabies virus (PRV) that causes porcine pseudorabies (PR) [24]. One monoclonal antibody (1H5) was characterized using indirect ELISA, IFA, and Western Blot and was found to bind to a small epitope conserved in the gE of almost all PRV strains. It can be used to develop antigen detection tests [24]. Skinner et. al., developed mouse monoclonal antibodies against CVV, which is a mosquito-borne virus that causes disease in livestock and humans [23]. Antibodies were developed against inactivated CVV using hybridoma technology, and twelve hybridoma clones were found to show significant reactivity against CVV. Four mAbs, CVV14/15/17, and 18, were found to be highly specific for the detection test of anti-viral antibodies in human sera using IgM-antibody capture ELISA (MAC-ELISA), with MAb CVV14 exhibiting the highest specificity. Antibodies like CVV14 can be used as detector antibodies to develop promising serodiagnostic tools against CVV. Yang et al., developed an ELISA-based method for detecting the Seneca Valley virus (SVV), which has been linked to disease in pigs [41]. Using hybridoma technology, mouse monoclonal antibodies were produced against binary ethylenimine (BEI)-inactivated SVV. Five antibodies were characterized using Dot blot and were found to be specific to SVV. Furthermore, antibody F61SVV-9 exhibited the strongest competition with monospecific polyclonal sera in cELISA and resulted in 100% specificity, indicating that monoclonal antibodies can also be explored for serodiagnosis of viruses using cELISA [41]. The applications of monoclonal antibodies also extend to the accurate and sensitive detection of plant viruses, which is crucial for disease management and control in agriculture. Z. Chen et. al., reported the development of 3 mouse monoclonal antibodies for the detection of Zucchini yellow mosaic virus (ZYMV) using hybridoma technology [72]. Using ZYMV virion for immunization, three hybridoma clones, 16A11, 5A7, and 3B8, were developed and characterized using multiple immunoassays.
REFERENCES
1. Park KS. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens Bioelectron. 2018;102:179-88. [CrossRef]
2. Omar RF, Boissinot M, Huletsky A, Bergeron MG. Tackling infectious diseases with rapid molecular diagnosis and innovative prevention. Infect Dis Rep. 2024;16:216-27. [CrossRef]
3. Harsh, Tripathi P. Medical viruses: diagnostic techniques. Virol J. 2023;20:143. [CrossRef]
4. Louten J. Detection and diagnosis of viral infections. Essential Human Virology. Elsevier;2016:111-32. [CrossRef]
5. Verma V, Abhishek A. Production of SARS-CoV-2 nucleocapsid protein in Escherichia coli and its characterization. J Appl Biol Biotechnol. 2023;11(3):250-5. [CrossRef]
6. Quinteros DA, Bermúdez JM, Ravetti S, Cid A, Allemandi DA, Palma SD. Therapeutic use of monoclonal antibodies: general aspects and challenges for drug delivery. Nanostructures for Drug Delivery. Elsevier; 2017:807-33. [CrossRef]
7. Kuzman D, Bunc M, Ravnik M, Reiter F, Žagar L, Bon?ina M. Long-term stability predictions of therapeutic monoclonal antibodies in solution using Arrhenius-based kinetics. Sci Rep. 2021;11:20534. [CrossRef]
8. Kyosei Y, Yamura S, Namba M, Yoshimura T, Watabe S, Ito E. Antigen tests for COVID-19. Biophys Physicobiology. 2021;18:28-39. [CrossRef]
9. Mitra S, Tomar PC. Hybridoma technology; advancements, clinical significance, and future aspects. J Genet Eng Biotechnol. 2021;19:159. [CrossRef]
10. Lipman NS, Jackson LR, Trudel LJ, Weis-Garcia F. Monoclonal versus polyclonal antibodies: distinguishing characteristics, applications, and information resources. ILAR J. 2005;46:258-68. [CrossRef]
11. Morioka K, Fukai K, Sakamoto K, Yoshida K, Kanno T. Evaluation of monoclonal antibody-based sandwich direct ELISA (MSD-ELISA) for antigen detection of foot-and-mouth disease virus using clinical samples. PLoS ONE. 2014;9:e94143. [CrossRef]
12. Leow CH, Jones M, Cheng Q, Mahler S, McCarthy J. Production and characterization of specific monoclonal antibodies binding the Plasmodium falciparum diagnostic biomarker, histidine-rich protein 2. Malar J. 2014;13:277. [CrossRef]
13. Siddiqui MZ. Monoclonal antibodies as diagnostics; an appraisal. Indian J Pharm Sci. 2010;72:12-7. [CrossRef]
14. Law JW-F, Ab Mutalib N-S, Chan K-G, Lee L-H. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol. 2015;5. [CrossRef]
15. Byrne B, Stack E, Gilmartin N, O’Kennedy R. Antibody-based sensors: principles, problems and potential for detection of pathogens and associated toxins. Sensors. 2009;9:4407-45. [CrossRef]
16. Konstantinou GN. Enzyme-Linked Immunosorbent Assay (ELISA). In: Lin J, Alcocer M, editors. Food Allergens. New York, NY: Springer; 2017:79-94. [CrossRef]
17. Gan SD, Patel KR. Enzyme immunoassay and enzyme-linked immunosorbent assay. J Invest Dermatol. 2013;133:1-3. [CrossRef]
18. Engvall E. The ELISA, enzyme-linked immunosorbent assay. Clin Chem. 2010;56:319-20. [CrossRef]
19. Aydin S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides. 2015;72:4-15. [CrossRef]
20. Kim JW, Cho AH, Shin HG, Jang SH, Cho SY, Lee YR, et al. Development and characterization of phage display-derived monoclonal antibodies to the S2 domain of spike proteins of wild-type SARS-CoV-2 and multiple variants. Viruses. 2023;15:174. [CrossRef]
21. Terry JS, Anderson LBR, Scherman MS, McAlister CE, Perera R, Schountz T, et al. Development of a SARS-CoV-2 nucleocapsid specific monoclonal antibody. Virology. 2021;558:28-37. [CrossRef]
22. Wang J, Lei B, Zhang W, Li L, Ji J, Liu M, et al. Preparation of monoclonal antibodies against the capsid protein and development of an epitope-blocking enzyme-linked immunosorbent assay for detection of the antibody against porcine circovirus 3. Animals. 2024;14:235. [CrossRef]
23. Skinner B, Mikula S, Davis BS, Powers JA, Hughes HR, Calvert AE. Monoclonal antibodies to Cache Valley virus for serological diagnosis. Ainsworth SR, editor. PLoS Negl Trop Dis. 2022;16:e0010156. [CrossRef]
24. Guo Z, Zhang S, Xu H, Li W, Li C, Zhao J, et al. Preparation and identification of a monoclonal antibody against the pseudorabies virus gE glycoprotein through a novel strategy. Vet Sci. 2023;10:133. [CrossRef]
25. Ono E, Lafer MM, Weckx LY, Granato C, Moraes-Pinto MID. A simple and cheaper in house varicella zoster virus antibody indirect ELISA. Rev Inst Med Trop São Paulo. 2004;46:165-8. [CrossRef]
26. Alhajj M, Zubair M, Farhana A. Enzyme Linked Immunosorbent Assay. StatPearls. Treasure Island (FL): StatPearls Publishing; 2024. http://www.ncbi.nlm.nih.gov/books/NBK555922/
27. Verma V, Kaur C, Grover P, Gupta A, Chaudhary VK. Biotin-tagged proteins: reagents for efficient ELISA-based serodiagnosis and phage display-based affinity selection. PLOS ONE. 2018;13:e0191315. [CrossRef]
28. Shrivastava N, Kumar JS, Yadav P, Sharma S, Shete AM, Jain R, et al. Development and evaluation of indirect antibody ELISA assay for early diagnosis and surveillance of Crimean-Congo hemorrhagic fever infection in humans. Virus Res. 2022;313:198717. [CrossRef]
29. Bold D, Roman-Sosa G, Gaudreault NN, Zayat B, Pogranichniy RM, Richt JA. Development of an indirect ELISA for the detection of SARS-CoV-2 antibodies in cats. Front Vet Sci. 2022;9:864884. [CrossRef]
30. Zai J, Yi K, Xie L, Zhu J, Feng X, Li Y. Dual monoclonal antibody-based sandwich ELISA for detection of in vitro packaged Ebola virus. Diagn Pathol. 2018;13:96. [CrossRef]
31. Choudhary N, Roy A, Govindarajulu A, Nakhla MK, Levy L, Brlansky RH. Production of monoclonal antibodies for detection of Citrus leprosis virus C in enzyme-linked immuno-assays and immunocapture reverse transcription-polymerase chain reaction. J Virol Methods. 2014;206:144-9. [CrossRef]
32. Tabatabaei MS, Ahmed M. Enzyme-Linked Immunosorbent Assay (ELISA). In: Christian SL, editor. Cancer Cell Biology. New York, NY: Springer US; 2022:115-34. [CrossRef]
33. Sozzi E, Moreno A, Lelli D, Perulli S, Prosperi A, Brocchi E, et al. Development and validation of a monoclonal antibody-based competitive ELISA for detection of antibodies against porcine epidemic diarrhoea virus (PEDV). Res Vet Sci. 2018;121:106-10. [CrossRef]
34. He D, Sun M, Jiang X, Zhang S, Wei F, Wu B, et al. Development of an indirect competitive ELISA method based on ORF2 detecting the antibodies of novel goose astrovirus. J Virol Methods. 2023;311:114643. [CrossRef]
35. Hnasko TS, Hnasko RM. The Western Blot. In: Hnasko R, editor. ELISA. New York, NY: Springer New York; 2015:87-96. [CrossRef]
36. Morgan MS, Yan K, Le TT, Johnston RA, Amarilla AA, Muller DA, et al. Monoclonal antibodies specific for SARS-CoV-2 spike protein suitable for multiple applications for current variants of concern. Viruses. 2022;15:139. [CrossRef]
37. Mishra N, Teyra J, Boytz R, Miersch S, Merritt TN, Cardarelli L, et al. Development of monoclonal antibodies to detect for SARS-CoV-2 proteins. J Mol Biol. 2022;434:167583. [CrossRef]
38. Sunwoo HH, Palaniyappan A, Ganguly A, Bhatnagar PK, Das D, El-Kadi AOS, et al. Quantitative and sensitive detection of the SARS-CoV spike protein using bispecific monoclonal antibody-based enzyme-linked immunoassay. J Virol Methods. 2013;187:72-8. [CrossRef]
39. Magaki S, Hojat SA, Wei B, So A, Yong WH. An Introduction to the Performance of Immunohistochemistry. In: Yong WH, editor. Biobanking. New York, NY: Springer New York; 2019:289-98. [CrossRef]
40. Goh LYH, Hobson-Peters J, Prow NA, Gardner J, Bielefeldt-Ohmann H, Suhrbier A, et al. Monoclonal antibodies specific for the capsid protein of chikungunya virus suitable for multiple applications. J Gen Virol. 2015;96:507-12. [CrossRef]
41. Yang M, Van Bruggen R, Xu W. Generation and diagnostic application of monoclonal antibodies against Seneca Valley virus. J Vet Diagn Invest. 2012;24:42-50. [CrossRef]
42. Kohn J. An immunochromatographic technique. Immunology. 1968;15:863-5.
43. Salcedo N, Reddy A, Gomez AR, Bosch I, Herrera BB. Monoclonal antibody pairs against SARS-CoV-2 for rapid antigen test development. Brindley PJ, editor. PLoS Negl Trop Dis. 2022;16:e0010311. [CrossRef]
44. Xie C, Ding H, Ding J, Xue Y, Lu S, Lv H. Preparation of highly specific monoclonal antibodies against SARS?CoV?2 nucleocapsid protein and the preliminary development of antigen detection test strips. J Med Virol. 2022;94:1633-40. [CrossRef]
45. Richardson S, Kohn MA, Bollyky J, Parsonnet J. Validity of at-home rapid antigen lateral flow assay and artificial intelligence read to detect SARS-CoV-2. Diagn Microbiol Infect Dis. 2022;104:115763. [CrossRef]
46. Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495-7. [CrossRef]
47. Holzlöhner P, Hanack K. Generation of murine monoclonal antibodies by hybridoma technology. J Vis Exp JoVE. 2017. [CrossRef]
48. Parray HA, Shukla S, Samal S, Shrivastava T, Ahmed S, Sharma C, et al. Hybridoma technology a versatile method for isolation of monoclonal antibodies, its applicability across species, limitations, advancement and future perspectives. Int Immunopharmacol. 2020;85:106639. [CrossRef]
49. Mariotti S, Chiantore MV, Teloni R, Iacobino A, Capocefalo A, Michelini Z, et al. New monoclonal antibodies specific for different epitopes of the spike protein of SARS-CoV-2 and its major variants: additional tools for a more specific COVID-19 diagnosis. Biomedicines. 2023;11:610. [CrossRef]
50. Yamaoka Y, Miyakawa K, Jeremiah SS, Funabashi R, Okudela K, Kikuchi S, et al. Highly specific monoclonal antibodies and epitope identification against SARS-CoV-2 nucleocapsid protein for antigen detection tests. Cell Rep Med. 2021;2:100311. [CrossRef]
51. Shukra AM, Sridevi NV, Dev Chandran, Kapil Maithal. Production of recombinant antibodies using bacteriophages. Eur J Microbiol Immunol. 2014;4:91-8. [CrossRef]
52. Pedrioli A, Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021;42:1143-58. [CrossRef]
53. Verma V. Leveraging monoclonal antibodies as therapeutics to address antimicrobial resistance in bacteria. J Appl Biol Biotechnol. 2023;11:53-60. [CrossRef]
54. Prashar P, Swain S, Adhikari N, Aryan P, Singh A, Kwatra M, et al. A novel high-throughput single B-cell cloning platform for isolation and characterization of high-affinity and potent SARS-CoV-2 neutralizing antibodies. Antiviral Res. 2022;203:105349. [CrossRef]
55. Lonberg N. Human antibodies from transgenic animals. Nat Biotechnol. 2005;23:1117-25. [CrossRef]
56. Erasmus MF, Ferrara F, D’Angelo S, Spector L, Leal-Lopes C, Teixeira AA, et al. Insights into next generation sequencing guided antibody selection strategies. Sci Rep. 2023;13. [CrossRef]
57. Yang W, Yoon A, Lee S, Kim S, Han J, Chung J. Next-generation sequencing enables the discovery of more diverse positive clones from a phage-displayed antibody library. Exp Mol Med. 2017;49:e308. [CrossRef]
58. Cheng J, Liang T, Xie X-Q, Feng Z, Meng L. A new era of antibody discovery: an in-depth review of AI-driven approaches. Drug Discov Today. 2024;29:103984. [CrossRef]
59. Kim J, McFee M, Fang Q, Abdin O, Kim PM. Computational and artificial intelligence-based methods for antibody development. Trends Pharmacol Sci. 2023;44:175-89. [CrossRef]
60. Kim H-Y, Lee J-H, Kim MJ, Park SC, Choi M, Lee W, et al. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens Bioelectron. 2021;175:112868. [CrossRef]
61. Wang A, Niu Y, Zhao J, Liu H, Ding P, Chen Y, et al. Rapid detection of varicella-zoster virus based on an immunochromatographic strip. Virology. 2023;586:35-42. [CrossRef]
62. Li C-J, Huang P-H, Chen H-W, Chang S-C. Development and characterization of mouse monoclonal antibodies targeting to distinct epitopes of Zika virus envelope protein for specific detection of Zika virus. Appl Microbiol Biotechnol. 2021;105:4663-73. [CrossRef]
63. Yamaoka Y, Matsuyama S, Fukushi S, Matsunaga S, Matsushima Y, Kuroyama H, et al. Development of monoclonal antibody and diagnostic test for middle east respiratory syndrome coronavirus using cell-free synthesized nucleocapsid antigen. Front Microbiol. 2016;7. [CrossRef]
64. Luo S, Deng X, Xie Z, Huang J, Zhang M, Li M, et al. Production and identification of monoclonal antibodies and development of a sandwich ELISA for detection of the H3-subtype avian influenza virus antigen. AMB Express. 2020;10:49. [CrossRef]
65. Lee K, Ko HL, Lee E-Y, Park H-J, Kim YS, Kim Y-S, et al. Development of a diagnostic system for detection of specific antibodies and antigens against Middle East respiratory syndrome coronavirus: indirect and sandwich ELISA for MERS-CoV. Microbiol Immunol. 2018;62:574-84. [CrossRef]
66. Gelanew T, Hunsperger E. Development and characterization of serotype-specific monoclonal antibodies against the dengue virus-4 (DENV-4) non-structural protein (NS1). Virol J. 2018;15:30. [CrossRef]
67. Adungo F, Yu F, Kamau D, Inoue S, Hayasaka D, Posadas-Herrera G, et al. Development and characterization of monoclonal antibodies to yellow fever virus and application in antigen detection and IgM capture enzyme-linked immunosorbent assay. Clin Vaccine Immunol. 2016;23:689-97. [CrossRef]
68. Kim J, Yang J, Kim YB, Lee H-J, Kim S, Poo H. Development of a specific CHIKV-E2 monoclonal antibody for chikungunya diagnosis. Virol Sin. 2019;34:563-71. [CrossRef]
69. Chen H, Ou Q, Tang Y, Gao X, Wu L, Xue C, et al. Development and evaluation of a DAS-ELISA for rapid detection of tembusu virus using monoclonal antibodies against the envelope protein. PLoS ONE. 2014;9:e96366. [CrossRef]
70. Zhang B, Fan J, Luo Y, Lv H, Zhao Q, Fan M, et al. Development of a competitive ELISA for detecting antibodies against genotype 1 hepatitis E virus. Appl Microbiol Biotechnol. 2021;105:8505-16. [CrossRef]
71. Zhang J, Guo Z, Zhao Y, Yang Y, Huang P, Wang N, et al. Identification of novel monoclonal antibodies specific for the conserved epitopes in the E2 protein of genotype 2 classical swine fever virus: implication for differential diagnosis. Acta Virol. 2023;67:12124. [CrossRef]
72. Chen Z, Zhang M, Zhou X, Wu J. Development and detection application of monoclonal antibodies against Zucchini yellow mosaic virus. J Integr Agric. 2017;16:115-24. [CrossRef]
73. Zhang J-H, Shan L-L, Liang F, Du C-Y, Li J-J. Strategies and Considerations for Improving Recombinant Antibody Production and Quality in Chinese Hamster Ovary Cells. Front Bioeng Biotechnol. 2022;10:856049. [CrossRef]
74. Cardoso AR, Alves JF, Frasco MF, Piloto AM, Serrano V, Mateus D, et al. An ultra-sensitive electrochemical biosensor using the Spike protein for capturing antibodies against SARS-CoV-2 in point-of-care. Mater Today Bio. 2022;16:100354. [CrossRef]
75. Pantaleo G, Correia B, Fenwick C, Joo VS, Perez L. Antibodies to combat viral infections: development strategies and progress. Nat Rev Drug Discov. 2022;21:676-96. [CrossRef]
76. Saeed AFUH, Wang R, Ling S, Wang S. Antibody engineering for pursuing a healthier future. Front Microbiol. 2017;8. [CrossRef]
77. Moraes JZ, Hamaguchi B, Braggion C, Speciale ER, Cesar FBV, Soares GDFDS, et al. Hybridoma technology: is it still useful? Curr Res Immunol. 2021;2:32-40. [CrossRef]
78. Smith SA, Crowe JE. Use of human hybridoma technology to isolate human monoclonal antibodies. Microbiol Spectr. 2015;3:AID-0027-2014. [CrossRef]
79. Kumar R, Andrabi R, Tiwari A, Prakash SS, Wig N, Dutta D, et al. A novel strategy for efficient production of anti-V3 human scFvs against HIV-1 clade C. BMC Biotechnol. 2012;12:87. [CrossRef]
80. Shi W, Liao Y, Willis SN, Taubenheim N, Inouye M, Tarlinton DM, et al. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. 2015;16:663-73. [CrossRef]
81. Jat PS, Noble MD, Ataliotis P, Tanaka Y, Yannoutsos N, Larsen L, et al. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc Natl Acad Sci U S A. 1991;88:5096-100. [CrossRef]
82. Luckenbach G-A. Some recent aspect on hybridoma technology. In: Mayr WR, editor. Advances in Forensic Haemogenetics. Berlin, Heidelberg: Springer Berlin Heidelberg; 1988:267-67. [CrossRef]
83. Winter G, Milstein C. Man-made antibodies. Nature. 1991;349:293-9. [CrossRef]
84. Hammers CM, Stanley JR. Antibody phage display: technique and applications. J Invest Dermatol. 2014;134:1-5. [CrossRef]