1. INTRODUCTION
One of the challenges in medical practice is the search for active substances that can accelerate or enhance the natural healing process of fractures, skin wounds, tendons, and skeletal muscle tears. This is due to the fact that there are only a few pharmaceutical products that are specifically designed to promote healing.
Peptides are of significant interest as biologically active substances (BAS) for the aforementioned application. According to the authors [1–3], some peptide sequences have pronounced regenerative properties.
The authors [4] claim that peptides can bind to collagen in the extracellular matrix (ECM), which is exposed during inflammation. According to the data [5], some peptides have proven effective in specific clinical cases, in the early and late stages of wound healing (scarring), and even in nerve regeneration after spinal cord injury. Some peptides target bone tissue or fractures, binding to either tartrate-resistant acid phosphatase deposited by osteoclasts or hydroxyapatite deposited by osteoblasts [6].
Despite their effectiveness, peptides remain difficult to create (predict and design) because bioactivity is influenced by many characteristics, including structure, number of amino acids, sequence, molecular weight, total charge, hydrophobicity, lipophilicity, and so on [7]. At the same time, many new peptides have been developed, but only a few of them have regenerative properties [8]. The pharmaceutical industry faces a significant challenge in bringing peptides to market, as many of them use more environmentally friendly peptide synthesis methods that are more expensive than traditional approaches [9]. This presents a significant challenge to the study, design, and production of peptides. However, with the transition to large-scale production, the economic efficiency of peptide production can be dramatically improved.
It should be noted that biopeptides can vary in length, and it is likely that these small amino acid chains can interact with many non-target objects. Therefore, it is difficult to design peptides that can selectively bind to the desired target while avoiding non-target interactions [10].
Another obstacle to the introduction of peptides is their low biological stability. Like all biological materials, peptides are susceptible to enzymatic decomposition. Certain technologies have been developed to increase the stability of peptides [9]. In particular, two methods can be used simultaneously to obtain proteolysis-resistant peptides: 1. Molecular peptide transplantation: the introduction of a biologically active peptide fragment into another peptide; 2. The incorporation of a peptide fragment into a known stable cyclic peptide framework [11].
Peptide toxicity is associated with low stability and bioavailability, lack of selectivity for target cells, and so on. Therefore, these properties of peptides should be evaluated as early as possible, i.e., not at the design stage before synthesis.
In this regard, the study aimed to predict the toxicity and biological activity of a peptide with regenerative properties during the design stage, followed by synthesis and validation of the results in an in vitro experiment.
2. MATERIALS AND METHODS
2.1. Object of Research
The synthesized peptide with the amino acid sequence CTKSICTKKTLRTCPPIC (Pepmic Co., Ltd, China) was used as an object of research. The peptide was named CC-18.
2.2. Development of a Biologically Active Peptide
The development of a peptide with regenerative properties was based on an analysis of the scientific literature [12–14]. Due to the variety of terminology used, a search query was defined that included the following keywords: "peptide sequence", "peptides with regenerative properties", "amino acids", "biological activity", "total charge", "molecular weight", and so on. An additional manual search was performed for publications referred to by the authors of the most informative articles. Sources included MEDLINE, PubMed, EMBASE, Library, Scopus, Web of Science, and Google Scholar.
The base of cyclic peptides, Sybase, was also employed for the development of the peptide (https://www.sybase.ru) (Sybase CIS, USA). The Endogenous regulatory oligopeptides EROP-Moscow database (http://erop.inbi.ras.ru/index.html) (A.N. Bach Institute of Biochemistry of the Russian Academy of Sciences, Russia) was used for the peptide identification.
The prediction of the peptide biological activity was performed using the PeptideRanker server (http://distilldeep.ucd.ie/PeptideRanker), which is based on a novel N-to-1 neural network. The server predicts how likely it is that the peptide is bioactive for wound healing. PeptideRanker was trained with a threshold of 0.5, i.e., any peptide predicted above the 0.5 threshold is biologically active. Prediction of the biochemical and biophysical properties of the peptide that determine its bioactivity was performed using the APD database (https://aps.unmc.edu/home). Peptide properties were predicted using the Peptide Property Calculator (http://pepcalc.com) (Awesome Labs LLC, USA).
The ADMET platform was used to predict the bioavailability, biological activity, and toxicity of the designed peptide (https://admetmesh.scbdd.com/). First, the peptide sequence was converted into SMILES string using the PepSMI tool.
The peptide was synthesized at Pepmic Co., Ltd (Suzhou, China). The solid-phase Fmoc (SPPS) method was used to synthesize the peptide, followed by purification by high-performance liquid chromatography (HPLC). The SHIMADZU Inertsil ODS-SP chromatography column (SHIMADZU, Japan) was used for the chromatographic purification of the peptide. Trifluoroacetic acid (Sigma-Aldrich, USA), triisopropylsilane (Sigma-Aldrich, USA), 1,3-diisopropylcarbodiimide (Fluka, Germany), 1-hydroxybenzotriazole (NovaBiochem-Merck, Germany), N,N-dimethylformamide (DMF) (Vetec, Brazil), diisopropyl ether (Vetec, Brazil), and acetonitrile for HPLC (JT Baker, USA) were used for peptide synthesis. All solvents used in the HPLC system were manufactured by Tedia (Brazil).
2.3. Identification and Determination of Molecular Weight Distribution, Study of Biochemical and Biophysical Characteristics of the Peptide
The molecular weight distribution of the peptide was evaluated by mass spectrometry [15]. The peptide was identified by MALDI-TOF MS Ultraflex method (Bruker, Germany) [16]. The mass spectra were analyzed using the Mascot program and peptide Fingerprint option (Matrix Science, USA). The study and prediction of peptide properties were performed using PeptideRanker (http://distilldeep.ucd.ie/PeptideRanker), APD (https://aps.unmc.edu/home), and Peptide Property Calculator (http://pepcalc.com).
2.4. Determination of Toxicity
The acute toxicity of the peptide was predicted using the ADMET platform (https://admetmesh.scbdd.com/).
A primary culture of human lung fibroblasts (Fibr) was used to determine the cytotoxicity of the peptide. The cells were cultured in DMEM medium (10% fetal bovine serum, 1% L-glutamine, 1% sodium pyruvate, and 1% penicillin/streptomycin) (Capricorn Scientific, Germany). Fibroblast cells were cultured in a Galaxy CO170R CO2 incubator (New Brunswick, Canada) for 7 days. Cultivation conditions: 95% relative humidity, 37°C, 5% CO2. The medium was changed every 3 days. Cells were examined for the presence of mycoplasma before each experiment.
Cell viability was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay [17]. Cells were plated at 0.5*10^6 cells per well in 100 µl medium in a 96-well plate (Eppendorf, Germany). After reaching 70% confluence, CC-18 extract was added to the cells at different concentrations (1, 5, 10, and 50 µg/ml). The cells were incubated with the peptide for 2 days. On day 3, 10 µl of a solution of MTT with a final concentration of 0.5 mg/ml was added to the cells. The cells were then incubated for 4 hours in a Galaxy CO170R CO2 incubator (New Brunswick, Canada).
2.5. Determination of Regenerative Properties
As a model of regeneration, the in vitro cell migration approach or "wound healing assay" has been used, which consists of creating a "wound" with a spout and observing the rate of cell migration into the wound to fill the empty space. The main steps include creating a "wound" in the cell monolayer, taking images at the beginning and at regular intervals during cell migration to close the wound, and comparing images to quantify the rate of cell migration.
To study the effect of the peptide on fibroblast regeneration, cells were seeded at a density of 2 × 104 cells per well in a 24-well plate. Once the cells had reached 100% confluence, a 1,000 µl wound was inflicted on them with a spout. The cells were then washed with Hank's Balanced Salt Solution (Capricor Scientific, Germany) and a medium containing C18 at a concentration of 10 µg/ml was added. Images of cell migration were obtained using an inverted Axio Observer A2 microscope (Zeiss, Germany). Prior to wound repair, the rate of wound closure was determined by monitoring the cells every 12 hours. The experiment was performed in triplicate, and data were obtained using Image J 1.38 software and expressed as mean ± standard deviation (x ± s, n = 3) at ** p < 0.01 compared with control.
The wound closure rate (%) was calculated using the formula:
Wound closure rate (%) = (A0 − At)/A0 × 100% (1)
where A0 represents the area of the wound after, and At represents the same at the specified time.
2.6. Changes in the Expression of Migration Markers
The cells were plated in a 6-well plate at a density of 5 × 104 cells per well. On the subsequent day, the C18 peptide was introduced to the cells at the specified concentration. Following a 48-hour incubation period, the cells were harvested for RNA isolation via trypsinization. The RNA was extracted using the ExtractRNA reagent (Eurogene, Russia) in accordance with the manufacturer's instructions. Reverse transcription was conducted using the SuperScript IV kit (Invitrogen, USA), and quantitative PCR was performed using the 50X SYBR Green I kit with the primers listed in Table 1.
 | Table 1. Primers for assessing changes in the expression of migration markers. [Click here to view] |
3. RESULTS
3.1. Peptide Preparation
The object of the research was a synthesized peptide with a cyclic structure. The known Sybase peptide with the number [Nphe5]SFTI-1(100) was used as a base for peptide synthesis. The protein map of the [Nphe5]SFTI-1(100) peptide is presented in Figure 1. The synthesized Sybase is known to be resistant to the action of proteases. This property of the peptide allows it to be used as a basis for the synthesis of new peptides. The advantage of this approach is the simplicity of obtaining biologically active low molecular weight molecules of protein nature, the absence of stages of protein isolation from natural material and stages of protein hydrolysis.
As a result of molecular peptide transplantation of the TKKTLRT sequence into the proteolysis-resistant cyclic peptide [Nphe5]SFTI-1(100) and alignment according to the APD database, a new cyclic peptide with the amino acid sequence CTKSICTKKTLRTCPPIC was obtained (Fig. 2).
When searching the EROP-Moscow database for a peptide with the sequence CTKSICTKKTLRTCPPIC, this peptide was not found, indicating its distinctness (Fig. 3).
3.2. Prediction of the Peptide Biological Activity
The PeptideRanker server predicted the peptide's biological activity to be 0.668782 units. Peptides containing more than 0.5 units are considered bioactive (Fig. 4).
The physicochemical properties of the peptide were predicted from the APD database, resulting in the following characteristics: the molecular formula is C83H150N24O24S4, the molecular weight is 1996 Da, the total charge is +4, and the total hydrophobic ratio is 3. The peptide exhibits a hydrophobicity of 9%, as determined by the Wimley-White method, which assesses the energy required for the transfer of the peptide from water to a POPC surface in the absence of a residue. Its protein binding potential, as quantified by the Boman index, is 1.4 kcal/mol. The peptide sequence contains an equal number of C residues and is capable of forming a defensin-like beta structure stabilized by disulfide bonds.
 | Figure 1. Protein map of the [Nphe5]SFTI-1 peptide(100). [Click here to view] |
 | Figure 2. A new cyclic peptide with the CTKSICTKKTLRTCPPIC sequence and cyclization by bonds: C1-C3, C2-C4; C1-C4, C2-C3. [Click here to view] |
 | Figure 3. Search for the CC-18 peptide in the EROP-Moscow database. [Click here to view] |
 | Figure 4. The results of predicting the CC-18 peptide bioactivity using the PeptideRanker server. [Click here to view] |
The results obtained were confirmed using the peptide property calculator according to the PepCalc program (http://pepcalc.com) (Fig. 5).
In addition, the peptide was found to be highly soluble in water and its isoelectric point was determined to be 9.34.
3.3. Determination of the Molecular Weight Distribution of the Peptide and Study of the Biochemical and Biophysical Characteristics of the Peptide
A comprehensive three-phase analysis of the CC-18 peptide was conducted. The resulting chromatogram of the synthesized CC-18 peptide is presented in Figure 6. Figure 7 depicts the mass spectrum of the synthesized CC-18 peptide.
 | Figure 5. The results of predicting the CC-18 peptide properties on the PepCalc calculator. [Click here to view] |
The obtained chromatogram and mass spectrum demonstrate that the synthesized peptide exhibits properties consistent with those predicted.
The biophysical properties of the peptide are the determining factor in its bioavailability and efficacy. The biophysical and biochemical properties of the CC-18 peptide, as predicted by the APD database, are presented in Table 2.
3.4. Study of the Cytotoxicity of the Peptide
The ADMET platform was used to predict the IC50 for blocking the human ether-a-Go-Go potassium channel (HERG K+). The HERG K+ channel regulates the electrical activity of the heart, and the heartbeat, and is considered a molecular target responsible for the cardiotoxicity of a large number of drugs. Therefore, compounds that block HERG K+ channels may be toxic. The obtained predicted IC50 values are important for the assessment of cardiac toxicity of new biologically active compounds [18]. The in silico predicted IC50 value of the CC-18 peptide is 0.008 units, which indicates the absence of toxic effects on the heart (not hERG blockers with a value of 0–0.3 units).
In a study designed to assess the acute toxicity of the CC-18 peptide when administered orally to rats, no evidence of acute toxicity was observed. Therefore, the conditional value of the aforementioned indicator was 0.06 units, which is greater than 500 mg/kg. A threshold toxic dose for humans could not be determined. The data obtained from the ADMET platform indicate that the CC-18 peptide is not neurotoxic. The obtained value for neurotoxicity is 0.183 units, indicating that the peptides are not neurotoxic at values from 0.0 to 0.3 units. Similarly, the level of hematotoxicity of the peptide is 0.003 units, which indicates its absence, as it is not hematotoxic at a value from 0.0 to 0.3 units. The result of the prediction of toxicity of the peptide against Hek293 cells is 0.001 units, which corroborates the absence of toxicity (it is not cytotoxic at a value from 0 to 0.3 units).
 | Figure 6. Chromatogram of the CC-18 peptide. [Click here to view] |
Studies were conducted on the effect of C-18 peptide on the viability of fibroblast culture isolated from human lungs. The absence of a cytotoxic effect was found in the studied concentrations of 1–50 µg/ml. Thus, the viability of fibroblasts at concentrations of peptide extract 1–10 and 50 µg/ml of medium was 100 and 97% and did not change significantly.
3.5. Determination of Regenerative Properties
Studies were conducted to confirm the results of the prediction of the regenerative properties of the CC-18 peptide on fibroblast cell culture.
Figure 8 shows the area of the wound after its creation in the control and experimental (using the CC-18 peptide in the culture medium) cell groups.
Figure 9 shows the wound area after 48 hours in the control and experimental groups.
Figure 10 shows the percentage of wound closure (%) at 12, 24, and 48 hours after creation.
The rate of wound closure in the presence of the peptide was found to be significantly higher (p < 0.01) compared to the control. Consequently, at the 12-, 24-, and 48-hour marks of the experiment, the wound closure area exhibited a 13%, 17%, and 5% increase, respectively, in comparison to the control.
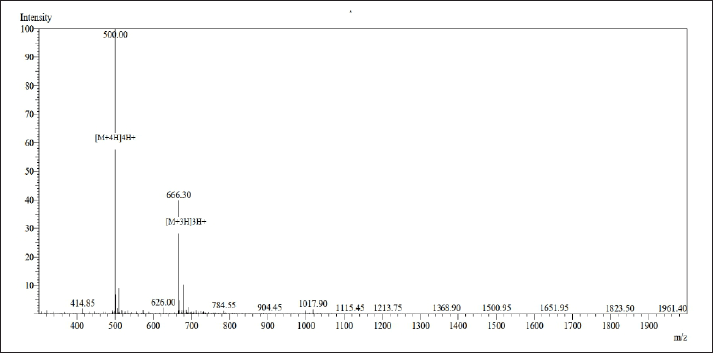 | Figure 7. Mass spectrum of the CC-18 peptide. [Click here to view] |
 | Table 2. Biophysical and biochemical characteristics of the CC-18 peptide. [Click here to view] |
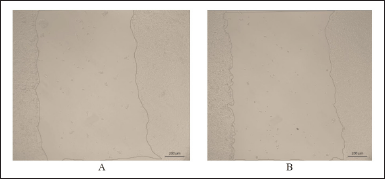 | Figure 8. The area of the wound after its creation using the CC-18 peptide in a culture medium: A -control, B –experimental. [Click here to view] |
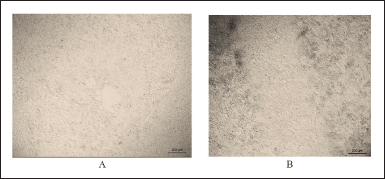 | Figure 9. Images of cell migration 48 hours after the wound was created: A—control, B—experimental. [Click here to view] |
 | Figure 10. The percentage of wound closure 48 hours after its creation: A—control, B—experimental. [Click here to view] |
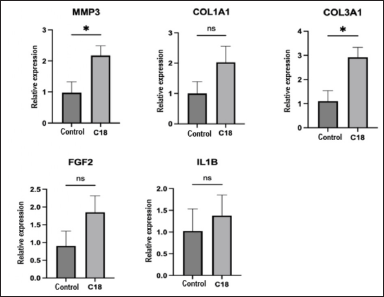 | Figure 11. Expression of genes associated with cell migration to the wound area. Accurately at *p < −0.05. [Click here to view] |
3.6. Studies of the Expression of Markers of Cell Migration to the Wound Area
The expression of genes associated with cell migration to the wound area, including GAPDH, MMP3, COL3A1, FGF2, IL1B, and COL1A1, was examined. The results are presented in Figure 11.
The levels of MMP3 and COL3A1 proteins exhibited a notable increase (p < 0.05) in the presence of CC-18 peptide, with a 133% and 183% rise, respectively, compared to the control. This confirms the beneficial impact of CC-18 peptide on the acceleration of the wound healing process.
4. DISCUSSION
Zhang's research has demonstrated that the amino acid sequence TKKTLRT exhibits regenerative properties and is capable of binding to type 1 collagen. This property serves to reduce the likelihood of proteolysis by collagenase and also stimulates tissue regeneration in the event of damage. A cyclic peptide with regenerative properties and resistance to proteolysis has been obtained. The peptide composition includes the lysylserine (KS) sequence, which is responsible for the peptide's regenerative properties. Additional research has demonstrated that the peptide containing the SK sequence exhibits notable regenerative properties. Thus, the cyclic peptide CARSKNKDC with an active sequence of SK and/or KS with cyclization through C1-C2 bonds restores damaged blood vessels during angiogenesis in inflammatory diseases, whose receptors are heparan sulfate proteoglycans [12]. Hussain et al. [13] came to similar conclusions when they investigated the cyclic peptide CARGGLKSC. The authors [13] demonstrated that the composition of the peptide contains the KS amino acid sequence, which contributes to the recovery of tissues infected with Staphylococcus aureus.
Peptides with the amino acid threonine (T) contribute to the restoration of nerve tissue and are used to identify nerves during surgery, in particular the Np41 peptide with the sequence NTQTLAKAPEHT. This peptide consists of two threonine (T) molecules [19].
The regenerative properties of the designed and synthesized peptide are additionally due to the presence of a sequence such as TL. Our conclusion is supported by Morgan's research. The authors showed that the peptide with the sequence RTLAFVRFK is used for the regeneration of blood vessels. The regenerative properties of this peptide are due to the presence of linked amino acids such as threonine (T) and leucine (L) [5].
According to Wang et al. [6], the TCLSYLKGLVTVG peptide with the threonylcysteine (TC) sequence, when interacting with the tartrate-resistant acid phosphatase receptor, restores bone tissue after fractures. A peptide with the leucylarginine (LR) sequence isolated from recombinant proteins is effective in treating inflammation of damaged tissue [4]. The developed peptide contains LR, TC, and KS sequences, suggesting its regenerative properties.
One of the mechanisms of action of regenerative peptides, as evidenced by data [20,21], maybe as follows: the peptide regulates inflammation by physically interacting with collagen fibers and serving as a structural component of collagen fibers, thereby affecting cellular functions such as proliferation, spreading, migration, and differentiation.
Some regenerative peptides have anti-fibrotic properties. In particular, the antifibrotic function of the aspartyl-cysteine-asparagine (DCN) peptide is related to its capacity to act as a natural inhibitor of transforming growth factor-β (TGF)-β, a growth factor responsible for scarring and fibrosis. The scar-inducing activity of TGF-β1 is mediated by connective tissue growth factors (CTGF/CCN2) and epidermal growth factor family receptors (ERBBs). DCN also neutralizes CCN2, ERBBs, and myostatin, which contribute to the development of scarring in multiple organs. Consequently, a single DCN molecule is capable of simultaneously neutralizing numerous fibrosis-inducing growth factors, as the binding sites of these growth factors are distributed across different regions of the DCN molecule [21]. Consequently, peptides are capable of inhibiting scarring and fibrosis due to their multi-purpose orientation.
The predicted toxicity indicators of the peptide allow for the indication of its safety in use. The data obtained on the physicochemical characteristics of the CC-18 peptide demonstrate that it is bioavailable, which is consistent with the empirical findings of other researchers [22,23]. These studies have shown that peptides with an average mass of up to 2 kDa can reach the cytoplasm of enterocytes and can be detected in the bloodstream after oral administration.
The peptide contains one NRing (a predictor of intestinal absorption), indicating high permeability through the intestinal walls and absorption into the blood.
Oral bioavailability is an important molecular descriptor in the development of BAS for absorption and metabolism during the first passage through the liver [24]. Absorption depends on the solubility and permeability of the compound across cell membranes, as well as its interaction with transporters and metabolizing enzymes in the gastrointestinal tract. The calculated molecular descriptor used to assess peptide absorption is the predicted water solubility (QPlogS). The predicted solubility of the CC-18 peptide was −2.1 log mol/l with an optimal value in the range of −4 to 0.5 log mol/l, indicating that the peptide is able to penetrate cell membranes and is bioavailable.
The synthesized peptide has a charge of 4, which is within the optimal charge range (from 0 to 4), which improves its bioavailability because it can attach to the cell due to the potential difference between the cell membrane and the peptide and then penetrate the cell.
The results of predicting the lipophilicity of the peptide are of particular interest since this property is the ability of the molecule to dissolve in lipid-like substances such as oils or non-polar solvents (toluene, cyclohexane, chloroform, and so on). Hydrophobic compounds prefer to be found in hydrophobic organelles/compartments such as lipid bilayers of the cell, while hydrophilic compounds prefer to dissolve in blood serum. The indicator logP ≤0 indicates high hydrophilicity and logP >0 indicates increased lipophilicity [25]. The last criterion is based on the distribution of molecules between the organic and aqueous phases, as in the n-octanol/water mixture, traditionally expressed as the negative logarithm of the distribution coefficient (logP organic water or simply logP). A study using monolayers of Caco-2 cells showed that optimal permeability of the compound is achieved at logP values of 4 to 5, while increasing the logP value outside this range decreases the permeability of the preparations [26].
The studied peptide has a logP of 2.9, confirming its ability to penetrate cell membranes.
The l log D7.4 indicator reflects the distribution of n-octanol in a buffer solution at pH = 7.4 and characterizes the lipophilicity/hydrophobicity balance of the test substance and influences the effective penetration through the cell membrane, absorption, distribution, metabolism, excretion, and toxicity [27]. For the studied peptide, it is 0.869 with an optimal value for medicinal substances ranging from 0.5 to 3 [28].
According to the ADMET platform, the pKa indicator shows the acidity/basicity of the test substance. Therefore, pKa reflects solubility, absorption, distribution, and elimination. A pKa value below the baseline (7.868) for the peptide indicates high absorption. Thus, the pKa of the tested peptide is 2,643 units, which is 66.4% lower than the base.
Fibroblasts are present in all tissues and adopt specialized phenotypes and activation states to perform important functions in development, wound healing, and maintenance of tissue architecture, as well as pathological functions such as tissue inflammation, fibrosis, and cancer reactions [29]. Fibroblast migration can accelerate the process of wound reepithelialization and promote wound closure during the healing process [30]. COL3A1 genes were identified in inflammatory processes and their regenerative function was confirmed using Funrich software [31]. In addition, Korsunsky et al. [32 ] demonstrated that the interacting COL3A1 and MMP9 genes are also involved in the PI3K/AKT signaling pathway, ECM receptor interaction, and other inflammatory signaling pathways (IL-17 signaling pathway, cytokine-cytokine receptor interaction, TNF signaling pathway, and chemokine signaling pathway). Numerous studies have shown that inflammatory pathways, including PI3K/AKT, IL-17, TNF, NF-kB, and MAPK pathways, are involved in the inflammatory process. These data suggest that COL3A1 and MMP9 play an important role in reducing the inflammatory process in tissues when damaged, which is consistent with the results of our studies.
A significant increase in the migration of COL3A1+ proteins interacting with fibroblasts was observed. The data obtained are consistent with studies [32] in which a decrease in the inflammatory process in the lungs, joints, and intestine was found in animal models on the background of migration of COL3A1 proteins.
In the studies of the authors [33,34], it was proved that COL3A1 expression levels increased at the early stages of inflammatory processes, in particular with damage to compounds, and decreased at later stages. Our results suggest that COL3A1 can be used as a diagnostic biomarkers of inflammation in tissue damage, which is consistent with studies [35], in which an increase in COL3A1 expression was found in joint injuries.
The MMP protein plays an important role in maintaining and restoring tissue homeostasis after tissue damage and performs a wide range of functions [36]. Our studies showed an increase in MMP3 expression, which is consistent with data [37] showing similar results in patients with healing wounds. In addition, the authors [37] analyzed the transcriptomics of migration genes in inflammation, in which the predominant localization of MMP3 was established—in the direction of the wound bed versus the wound edge or intact skin. Our results allow confirming the idea of the microenvironment during wound healing and the genes of proteins that may be crucial for accelerating healing, which can serve as the basis for new therapeutic approaches and the creation of food products for therapeutic and preventive purposes based on regenerative peptides for wound treatment.
In studies [38] on laboratory mice with excised wounds, the compensatory role of MMP3 expression was demonstrated with a decrease in MMP14 in adult skin fibroblasts (MMP14Sf-/-), leading to accumulation of type I collagen and increased MMP3 expression. MMP3 deficiency results in delayed wound closure in mice, but over time the wounds closed and epidermal integrity was restored. It should be noted that the density of myofibroblasts in MMP3-deficient wounds was lower than in the control on day 7 and higher on day 14. Furthermore, in vitro fibroblasts lacking MMP3 retained their ability to differentiate into myofibroblasts in response to mechanical stress. Thus, the results of our studies of increased MMP3 expression indicating accelerated wound closure are consistent with in vivo studies in mice with excised wounds in the authors' experiment [38].
It is possible that the data obtained on the increase of MMP3 expression is related to the activation of latent TGFß1. This growth factor has been identified as a regulator not only of myofibroblast resorption but also of myofibroblast formation during wound healing, among other processes [39,40].
Inactively expressed in a latent complex with a latent-associated peptide (LAP), TGFß1 is secreted into the extracellular space and bound by latent TGFß-binding protein 1 (LTBP1), where it remains available for activation [41]. By cleaving LTBP1 or LAP, MMP3 participates in the activation of TGFß1. MMP3 processes cell-bound LTBP1 [42]. Thus, TGFß1 activation and signaling occur under the action of MMP3, leading to accelerated wound healing.
5. CONCLUSION
A proteolysis-resistant CC-18 peptide with regenerative properties was designed and synthesized as a result of research using databases. The obtained results of prediction of toxicity, physicochemical properties, and bioavailability of the CC-18 peptide indicate its safety and efficacy in use and its ability to penetrate through the gastrointestinal tract into the bloodstream and then into cells. The synthesis makes it possible to obtain a peptide with the desired properties. The absence of toxicity of the ??-18 peptide was confirmed in an experiment on cell lines. The regenerative properties of the CC-18 peptide were confirmed in an in vitro experiment. However, to apply the developed and synthesized regenerative peptide in practice, it is necessary to confirm its effectiveness in preclinical and clinical trials.
6. ACKNOWLEDGMENTS
This study was not financially supported. No artificial intelligence was used (translation, revision, grammar check, and so on), while preparing this paper.
7. CONFLICTS OF INTEREST
The authors declare that there is no conflict of interest.
8. DATA AVAILABILITY
All supporting data are available through the corresponding author.
9. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Apone F, Barbulova A, Colucci MG. Plant and microalgae derived peptides are advantageously employed as bioactive compounds in cosmetics. Front Plant Sci 2019;10:756.
2. Järvinen TAH, Pemmari T. Systemically administered, targetspecific, multi-functional therapeutic recombinant proteins in regenerative medicine. Nanomaterials 2020;10:226.
3. Maldonado H, Savage BD, Barker HR, May U, Vähätupa M, Badiani RK, et al. Systemically administered wound-homing peptide accelerates wound healing by modulating syndecan-4 function. Nat Commun 2023;14:8069.
4. Katsumata K, Ishihara J, Mansurov A, Ishihara A, Raczy MM, Yuba E, et al. Targeting inflammatory sites through collagen affinity enhances the therapeutic efficacy of anti-inflammatory antibodies. Sci Adv 2019;5:eaay1971.
5. Morgan CE, Dombrowski AW, Rubert Perez CM, Bahnson ES, Tsihlis ND, Jiang W, et al. Tissue-factor targeted peptide amphiphile nanofibers as an injectable therapy to control hemorrhage. ACS Nano 2016;10:899–909.
6. Wang Y, Newman MR, Ackun-Farmmer M, Baranello MP, Sheu TA, Puzas JE, et al. Fracture-targeted delivery of beta-catenin agonists via peptide-functionalized nanoparticles augments fracture healing. ACS Nano 2017;11:9445–58.
7. Woolfson DN. A brief history of De Novo protein design: minimal, rational, and computational. J Mol Biol 2021;433(20):167160.
8. McDonald EF, Jones T, Plate L, Meiler J, Gulsevin A. Benchmarking alphafold2 on peptide structure prediction. Structure 2023;31:111–9.
9. Cooper BM, Iegre J, O' Donovan DH, Ölwegård Halvarsson M, Spring DR. Peptides as a platform for targeted therapeutics for cancer: peptide-drug conjugates (PDCs). Chem Soc Rev 2021;15;50(3):1480–94.
10. Burnside D, Schoenrock A, Moteshareie H, Hooshyar M, Basra P, Hajikarimlou M, et al. In silico engineering of synthetic binding proteins from random amino acid sequences. iScience 2018;11:375–87.
11. Jaradat DM. Solid-phase peptide cyclization with two disulfide bridges. Methods Mol Biol 2022;2371:19–29
12. Griffin DR, Archang MM, Kuan CH, Weaver WM, Weinstein JS, Feng AC, et al. Activating an adaptive immune response from a hydrogel scaffold imparts regenerative wound healing. Nat Mater 2021;20:560–9.
13. Hussain S, Joo J, Kang J, Kim B, Braun GB, She ZG, et al. Antibiotic-loaded nanoparticles targeted to the site of infection enhance antibacterial efficacy. Nat Biomed Eng 2018;2:95–103.
14. Zhang J, Ding L, Zhao Y, Sun W, Chen B, Lin H, et al. Collagen-targeting vascular endothelial growth factor improves cardiac performance after myocardial infarction. Circulation 2009;119:1776–84.
15. Hubler SL, Craciun G. Periodic patterns in distributions of peptide masses. Biosystems 2012;109(2):179–85.
16. Webster J, Oxley D. Protein identification by MALDI-TOF mass spectrometry. Methods Mol Biol (Clifton, N.J.) 2012;800:227–40.
17. Ghasemi M, Turnbull T, Sebastian S, Kempson I. The MTT assay: utility, limitations, pitfalls, and interpretation in bulk and single-cell analysis. Int J Mol Sci 2021;22(23):12827.
18. Hill AP, Sunde M, Campbell TJ, Vandenberg JI. Mechanism of block of the hERG K + channel by the scorpiontoxin CnErg1. Biophys J 2007;92(11):3915–29.
19. Whitney MA, Crisp JL, Nguyen LT, Friedman B, Gross LA, Steinbach P, et al. Fluorescent peptides highlight peripheral nerves during surgery in mice. Nat Biotechnol 2011;29:352–6.
20. Järvinen TA, Prince S. Decorin: a growth factor antagonist for tumor growth inhibition. Biomed Res Int 2015;2015:54765.
21. Järvinen TA?, Ruoslahti E. Generation of a multi-functional, target organ-specific, anti-fibrotic molecule by molecular engineering of the extracellular matrix protein, decorin. Br J Pharmacol 2019;176:16–25.
22. Graziani D, Ribeiro JVV, Cruz VS, Gomes RM, Araújo EG, Santos Júnior ACM, et al. Oxidonitrergic and antioxidant effects of a low molecular weight peptide fraction from hardened bean (Phaseolus vulgaris) on endothelium. Braz J Med Biol Res 2021;54:e10423.
23. Rodrigues WPS, Ribeiro JVV, da Silva CRB, de Campos ITN, Xavier CH, Dos Santos FCA, et al. In vivo effect of orally given polyvinyl alcohol/starch nanocomposites containing bioactive peptides from Phaseolus vulgaris beans. Colloids Surf B 2022;209:112213.
24. Waterbeemd HVD, Gi?ord E. ADMET in silico_modelling: towards prediction paradise? Nat Rev Drug Discov 2003;2(3):192–204.
25. Sawant PD, Luu D, Ye R, Buchta R. Drug release from hydroethanolic gels. Effect of drug’s lipophilicity (logP), polymer-drug interactions and solvent lipophilicity. Int J Pharm 2010;396:45–52.
26. Yang Y, Engkvist O, Llinas A, Chen H. Beyond size, ionization state, and lipophilicity: influence of molecular topology on absorption, distribution, metabolism, excretion, and toxicity for druglike compounds. J Med Chem 2012;55:3667.
27. Duan YJ, Fu L, Zhang XC, Long TZ, He YH, Liu ZQ, et al. Improved GNNs for Log?D7.4 prediction by transferring knowledge from low-fidelity data. J Chem Inf Model 2023;24:2345–59.
28. Chiu ML, Goulet DR, Teplyakov A, Gilliland GL. Antibody structure and function: the basis for engineering therapeutics. Antibodies 2019;8(4):55.
29. Koliaraki V, Prados A, Armaka M, Kollias G. The mesenchymal context in infammation, immunity and cancer. Nat Immunol 2020;21:974–82
30. Liu X, Wong SS, Taype CA, Kim J, Shentu TP, Espinoza CR, et al. Thy-1 interaction with Fas in lipid rafts regulates fibroblast apoptosis and lung injury resolution. Lab Invest 2017;97(3):256–67.
31. Li S, Wang H, Zhang Y, Qiao R, Xia P, Kong Z, et al. COL3A1 and MMP9 serve as potential diagnostic biomarkers of osteoarthritis and are associated with immune cell infiltration. Front Genet 2021;12:721258.
32. Korsunsky I, Wei K, Pohin M, Kim EY, Barone F, Major T, et al. Cross-tissue, single-cell stromal atlas identifies shared pathological fibroblast phenotypes in four chronic inflammatory diseases. Med 2022;3(7):481–518.
33. Tang S, Deng S, Guo J, Chen X, Zhang W, Cui Y. Deep coverage tissue and cellular proteomics revealed IL-1β can independently induce the secretion of TNF-associated proteins from human synoviocytes. J Immunol 2018;200:821–33.
34. Rai MF, Tycksen ED, Cai L, Yu J, Wright RW, Brophy RH. Distinct degenerative phenotype of articular cartilage from knees with meniscus tear compared to knees with osteoarthritis. Osteoarthr Cartil 2019;27:945–55.
35. Bollmann M, Pinno K, Ehnold LI, Märtens N, Märtson A, Pap T, et al. MMP-9 mediated Syndecan-4 shedding correlates with osteoarthritis severity. Osteoarthr Cartil 2021;29:280–9.
36. Caley MP, Martins VL, O’Toole EA. Metalloproteinases and wound healing. Adv Wound Care N Rochelle 2015;4:225–34.
37. Theocharidis G, Thomas BE, Sarkar D, Mumme HL, Pilcher WJ, Dwivedi B, et al. Single cell transcriptomic landscape of diabetic foot ulcers. Nat Commun 2022;13:181.
38. Kümper M, Zamek J, Steinkamp J, Pach E, Mauch C, Zigrino P. Role of MMP3 and fibroblast-MMP14 in skin homeostasis and repair. Eur J Cell Biol 2022;101(4):151276.
39. Hinz B, Lagares, D. Evasion of apoptosis by myofibroblasts: a hallmark of fibrotic diseases. Nat Rev Rheuma 2020;16:11–31.
40. Abarca-Buis RF, Mandujano-Tinoco EA, Cabrera-Wrooman A, Krotzsch E. The complexity of TGFβ/activin signaling in regeneration. J Cell Commun Signal 2021;15:7–23.
41. Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu Rev Immunol 2014;32:51–82.
42. Maeda S, Dean DD, Gomez R, Schwartz Z, Boyan BD. The first stage of transforming growth factor beta1 activation is release of the large latent complex from the extracellular matrix of growth plate chondrocytes by matrix vesicle stromelysin-1 (MMP3). Calcif Tissue Int 2002;70:54–65.