1. INTRODUCTION
Cancer is the leading cause of death and a significant impediment to living longer across the globe [1]. The World Health Organization 2019 estimate reveals that cancer is the first or second major cause of death in 112 countries [1,2]. The global burden of cancer incidence and death is on the rise; simply due to population increase and age, the worldwide burden of cancer has been anticipated to rise to 27.5 million new cancer cases and 16.3 million cancer deaths by 2040. In socioeconomically transitioning countries, the future burden will undoubtedly be more significant due to an increase in the incidence of cancer risk factors associated with unhealthy lifestyles such as smoking, consumption of alcohol and junk food, less physical activity, and low natality rates [1-5].
The primary liver cancer is the sixth most commonly diagnosed disease and the fourth major cause of cancer mortality globally. Furthermore, the prognosis of the disease is poor, with an estimated 5-year net survival rate of 19% [6,7]. Hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC) are the two most common histologic forms of the primary liver cancer. HCC comprises about 75% of the liver cancer cases, whereas ICC accounts for only about 12–15% of all the liver cancer cases throughout the world [6-9]. Hemangiocarcinoma accounts for 1% of all liver cancers, whereas secondary liver cancer or liver metastasis accounts for the rest of the liver cancer cases [10]. HCC develops in hepatocytes, usually due to oxidative stress, inflammation, and underlying liver disease, whereas ICC develops in the cholangiocytes that border the intrahepatic bile duct [8,11]. Despite breakthroughs in cancer treatment, liver cancer remains one of the most challenging malignancies to treat. Surgery, local destructive therapy, and liver transplantation have all been shown to be curative for people with the early HCC, but recurrence of HCC remains a hindrance in the treatment of the disease with an incidence rate of 70% or more. HCC is frequently discovered at an advanced stage, and many patients in these stages are ineligible for curative treatments. Furthermore, standard chemotherapy has low efficacy and provides less benefit in terms of survival [12]. Thus, several species of microbes, plants, animals, and marine animals are now proving to be promising sources for discovering new therapeutic candidates [13].
With the discovery and development of different natural substances such as cytotoxic podophyllotoxins, vincristine, vinca alkaloids, and vinblastine in the 1950s, the quest for anticancer agents from plant sources began [14]. The successful clinical use of cancer chemotherapeutic drugs derived from plants for nearly half a century has piqued interest for further research [14-16]. With advancements in technology, scientists have improved natural product drug discovery in an industrial setting. Several natural anticancer drugs have been launched in the market, and several drugs originating from various organisms are currently being tested in cancer-related clinical trials [14-17]. There are numerous routes involved in the development of cancer. Blocking one or more components of a specific pathway involved in cancer formation is a common strategy for developing anticancer drugs [9].
The molecular docking approach may be used to represent the atomic level interaction between a molecule and a protein, allowing us to define the molecule’s behavior in target protein binding sites as well as to elucidate key biochemical processes [18-20]. The molecules identified through docking are more likely to advance to the next stage of the drug development process. Nowadays, in vitro and in silico techniques are commonly utilized to examine the pharmacokinetic features of the chemical entities, and molecular modeling is widely used for optimizing leads in drug development.
The present study investigates chemicals and phytochemicals as potential drug inhibitors against liver cancer through in silico approach. To examine the potential of phytochemicals, different protein targets were selected. These targets included apoptotic proteins, transcription factors, serine/threonine protein kinases, growth factor receptors, mitogen-activated protein kinases (MAPK), and cell division protein kinase. We have performed a molecular docking study of ligands against all these proteins. Further, information from this study could be valuable in exploring a new compound, which could be very useful in developing a drug against HCC.
2. MATERIALS AND METHODS
2.1. Target Protein Retrieval
A total of 25 potent protein targets are involved in hepatocarcinoma, ranging from MAPK, nuclear receptor, apoptotic proteins, transcription factors, topoisomerases, epidermal growth factor receptor, cyclin dependent kinases (CDK), serine/threonine protein kinases, and various other proteins were selected for docking against the selected ligands [Table 1].
Table 1: List of protein targets for docking analysis.
| PDB ID | Protein description | Protein function | Protein | Resolution (A°) | References |
|---|---|---|---|---|---|
| 1CM8 | Proline-directed serine/threonine kinases | MAP kinases are dominant factors in the processes that govern embryogenesis, cell differentiation, proliferation, and cell death | MAP kinase | 2.4 | [21] |
| 1ERR | Nuclear transcription factors | Responsible for reproductive, skeletal, cardiovascular, and central nervous systems | ER- α | 2.6 | [22] |
| 1GFW | Cysteine proteases | Major executioner caspase during the demolition phase of apoptosis | Caspase 3 | 2.80 | [9] |
| 1JUN | Transcription factor | The factor plays a role in cellular processes, which includes cell proliferation, survival, and transformation of cell | AP-1 | Highest | [23] |
| 1NFK | Transcription factor | Plays a role in cell proliferation and differentiation, inflammatory and immune response, cell survival and apoptosis, cellular stress reactions, and tumorigenesis | NF-κB p50 subunit | 2.3 | [22] |
| 1P22 | Nuclear effector of canonical Wnt signaling in the nucleus | Tissue homeostasis, cell renewal, and regeneration | β-catenin | 2.95 | [23] |
| 1ZXM | Complex multifunctional enzymes | Modulation of topological state, chromosome segregation, and chromatin condensation | Human topoisomerase IIa | 1.87 | [24] |
| 2AR9 | Cysteine proteases | The caspase-9 function is vital for apoptosis during normal development of the central nervous system | Caspase 9 | 2.8 | [9] |
| 2CLS | Rho GTPase | Human RND1 expression is dysregulated in tumors and is involved in oncogenesis | Human RND1 GTPase in the active GTP bound state | 2.31 | [19] |
| 2E2B | Tyrosine-protein kinase | Cell growth and survival such as cytoskeleton remodeling in response to extracellular stimuli, cell motility and adhesion, receptor endocytosis, autophagy, DNA damage response, and apoptosis | c-Abl kinase domain in complex with INNO-406 | 2.20 | [20] |
| 2HY8 | Serine/Threonine protein kinases | Plays a role in cell cycle, cell motility, apoptosis, and cell survival | PAK1 | 2 | [22] |
| 2HZI | Tyrosine kinase | Responsible for cell differentiation, cell division, cell adhesion, and stress response | Proto-oncogene tyrosine-protein kinase ABL1 | 1.7 | [25] |
| 2OH4 | Belongs to PDGF supergene family | Signals for endothelial cell growth and survival (angiogenic) | Vascular EGFR-2 | 2.05 | [9] |
| 2OJI | Protein kinase | Proliferation, differentiation, transcription, and cellular motility | ERK2 | 2.60 | [22] |
| 3EZV | Serine/Threonine protein kinase | Role in the G1/S transition, the initiation of DNA synthesis, and the regulation of the exit from the S phase | CDK-2 | 1.99 | [24] |
| 3UPI | RNA-dependent RNA polymerase | HCV RNA replication | NS5B | 2.00 | [26] |
| 4AQ3 | BLC-2 family members regulate apoptosis | Family (such as BCL-2 and BCL-XL) inhibit apoptosis; meanwhile, others (such as BAX and BAK) promote cell death | BCL-2 | 2.40 | [22] |
| 4JSX | Serine/threonine protein kinase that belongs to Phosphatidylinositol-3-kinase related kinase superfamily | mTORC1 controls protein synthesis, cell growth, proliferation, autophagy, cell metabolism, and stress responses, whereas mTORC2 seems to regulate cell survival and polarity | MTOR | 3.50 | [22] |
| 4LQM | Family of receptor tyrosine kinases | Role in cell proliferation, survival, differentiation, and migration | EGFR | 2.50 | [9] |
| 4Y7R | Oncoprotein transcription factor | Cell growth, proliferation, loss of differentiation, and apoptosis | c-Myc | 1.898 | [23] |
| 4Y83 | Proto-oncogenic kinase, which belongs to the MAP3K family | COT contributed to G1 phase progression of T-lymphocytes, perhaps via regulation of IL-2 secretion | COT kinase | 2.89 | [27] |
| 5G4N | Tumour-suppressor protein | Role of p53 in the regulation of glycolysis and autophagy, the repair of genotoxic damage, cell survival and regulation of oxidative stress, invasion and motility, cellular senescence, angiogenesis, differentiation, and bone remodeling | p53 | 1.35 | [23] |
| 5LA9 | Heterodimeric transcription factor | Role in body’s response to low oxygen concentrations, or hypoxia | HIF-1α | 2.81 | [23] |
| 5CT7 | Kinase | Role in cellular proliferation, differentiation, and survival | BRAF in complex with RAF265 | 3.17 | [20] |
| 6HH1 | Receptor tyrosine kinase | Intracellular signaling and the mutated form of c-Kit plays a crucial role in the occurrence of some cancers | c-Kit with allosteric inhibitor 3G8 | 2.25 | [20] |
NF-κB: Nuclear factor Kappa-B, ER- α: Estrogen receptor-α, CDK-2: Cyclin-dependent kinase-2, PAK1: p21-activated kinase, NS5B: Non-structural protein 5B, BCL-2: B-cell lymphoma 2, MTOR: Mammalian target of rapamycin, EGFR: Endothelial growth factor receptor, COT: Cancer Osaka thyroid, HIF-1α: Hypoxia-inducible factor 1α, PDGF: Platelet-derived growth factor, and ERK2: Extracellular-signal-regulated kinase 2, HCV: Hepatitis B virus
PDB IDs for these proteins were obtained from protein data bank (PDB) [21], and their respective PDB format files were downloaded. To obtain processed protein structures free from any duplicate chains, these PDB files were opened using a 3D molecular visualization software, the BIOVIA Discovery Studio Visualizer (https://discover.3ds.com/discovery-studio-visualizer-download) and then the prebound ligand groups, heteroatoms and water molecules, and any undesired interference from the groups mentioned above were eliminated by their deletion.
2.2. Drug Preparation
For this study, a total of 1259 potential phytochemical drugs which might exhibit anticancer activity against HCC were retrieved from the Dr. Duke’s Phytochemical and Ethnobotanical Databases (http://phytochem.nal.usda.gov/). These databases aid in an extensive search for herbal and therapeutic plants and their chemical, bioactive, ethnobotanical characteristics using common, and scientific botanical names. The PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was used to obtain the 2D structure SDF files for each drug.
2.3. Energy Minimization of Docking Entities
The energy of processed proteins was then minimized using the Chimera software version 1.15 [22] through its Surface/Binding Analysis Tool for Dock prep, which facilitated the addition of required hydrogen atoms and charges acquired from Dunbrack Library at relevant positions in protein structure. The ligands finalized for docking analysis were initially energy minimized through the optimization algorithm by first loading their 2D structure SDF files on the PyRx – Virtual Screening Tool Software [23] and then converted to PDBQT format through its Open Babel tool. At this point, both the docking entities were ready for the next step.
2.4. Docking Methodology
In the molecular docking domain, to obtain a desired stabilized drug-receptor complex, it is essential to erase any unnecessary initial interactions of the two molecules with other prebound molecules to facilitate binding of the ligand at a competitive site on the receptor macromolecule. Before setting up the docking process, the energy minimized file of each target protein was first converted to a macromolecule. The docking operation was then launched using the Vina Wizard Control from the PyRx – Virtual Screening Tool Software by setting the Vina Search Space at its maximum as well as setting up the Vina Run exhaustiveness at value eight and finally continuing with the procedure that displays results in the form of binding affinity (kcal/mol) and root mean square deviation (RMSD) lower and upper bound values for each run up until the selected run value.
2.5. Visualization and Analysis of Docking Results
Out of all the RMSD values, only the highest scores were screened. Therefore, the ten topmost ligands displaying the highest binding affinity scores against each protein were selected for further analysis. The complex files of the drugs with their ligands were visualized in the form of a 2D drug-receptor complex in the BIOVIA Discovery Studio Visualizer. The structure of the desired ligand as per the binding affinity score and target-protein complex for each of the 25 proteins was visualized for Van der Waals, conventional H-bond, π-anion, π-cation, amide π-stacked, alkyl and π-alkyl, π-sigma, π-π T shaped, Carbon H-bond, π-Sulfur, π-π Stacked, Unfavorable Donor-Donor, and Unfavorable Acceptor-Acceptor interactions.
2.6. Bioavailability Radar and Pharmacokinetic (SWISS ADME) Profile Analysis
The canonical SMILES of each of the selected ligands with a high binding affinity against their targets were obtained from the simple search engine of the Indian Medicinal Plants, Phytochemistry and Therapeutics database [24]. Further using this data, the Pharmacokinetic analysis report was obtained from the SWISS ADME web tool (http://www.swissadme.ch/), a division of the web tools group provided by the Swiss Drug Design platform supported by the Swiss Institute Bioinformatics that act an as efficient computer-aided drug designing platform. The above-mentioned web tool platform evaluated the drug-likeness as per the bioavailability radar image and data obtained for each ligand as per Lipinski’s rule of five.
2.7. Computational Toxicity Prediction
Ligands following Lipinski’s rule of five were further evaluated to predict numerous toxicity parameters using the webtools ADMETlab 2.0 [25] and Protox II [26]. ADMETlab 2.0 aids in screening the drug molecules for Absorption, Distribution, Metabolism, Excretion, and Toxicological characteristics (ADMET), whereas Protox II, a virtual in silico laboratory, facilitated in mimicking its conventional counterpart and thereby delivered toxicity prediction of small molecules. Each ligand’s canonical smile was used to study its toxicity profile. Protox II helps determine the median lethal dose 50 (LD50) value in mg/kg along with toxicity class prediction. The toxicity classes are subdivided into six subtypes from Class I to VI, wherein Class I (LD50 ≤ 5) depicts an LD50 value that can be lethal if the drug is swallowed, and Class VI (LD50 > 5000) represents a non-toxic LD50 value. Furthermore, the tool also provides a toxicity model report which consists of variables such as Hepatotoxicity, Carcinogenicity, Immunotoxicity, Mutagenicity, and Cytotoxicity.
ADMET laboratory 2.0 precisely evaluates the toxic potential of the molecule through toxicity variables such as human ether-a-go-go gene-related blockers, human-hepatotoxicity, drug-Induced liver injury, ames test for mutagenicity, eye irritation, rat oral acute toxicity, FDAMDD (maximum recommended daily dose), skin sensitization, carcinogenicity, eye corrosion, respiratory toxicity, and environmental toxicity.
3. RESULTS AND DISCUSSION
Liver cancer is among the most prevalent malignant tumors in the world. The Global Cancer Statistics 2018 indicates that it ranks seventh and third in terms of global incidence and mortality, respectively, among all known cancer types. In all, 840,000 new cases and 780,000 deaths associated with HCC were reported in 2018 alone [27,28]. Chronic alcoholism, hepatitis B virus and hepatitis C virus infections, non-alcoholic fatty liver disease, and several other factors such as drinking water pollution, microcystins, and aflatoxin B1 also play a significant role in the incidence of HCC across the globe [29-32]. HCC is difficult to diagnose and cure; thus, using a phytochemical for developing a cure for this disease would be interesting and intriguing. In this study, we have investigated the binding of different phytochemicals to 25 protein targets associated with liver cancer using computational docking approaches.
A total of 1259 phytochemicals were chosen based on their ability to inhibit the proteins associated with HCC. These phytochemicals were retrieved from Dr Duke’s phytochemical and ethnobotanical databases. The phytochemicals chosen were organic compounds such as fatty acids, carbohydrates, benzene and its derivatives, chromones, polyphenols, and amino acids.
3.1. Molecular Docking
Molecular docking can investigate the interaction of a protein with ligands. It aims to find the potent drug by binding the correct ligand poses into the protein binding pockets [Table 2]. It elucidates whether the protein-ligand complex improves or inhibits the protein activity. It filters the best protein-ligand pair according to their binding scores so that the most potent drug candidate can be used for further testing. Molecular docking was carried out with the selected 25 proteins and the 1259 bioavailable ligands. A total of 250 ligands consisting of the top ten ligands of each protein ranked on the basis of their binding energy values are presented in Table 3. The higher binding affinity scores indicate that the ligands show a perfect fit into the protein cavity; the higher binding affinity can be attributed to the presence of hydrogen bonds in the complex. The π-cation bonds balance the negative atoms such as chlorine and fluorine in the drug, whereas the π-sigma bond introduces the charges responsible for the intercalation of the drug in the binding sites of the protein.
Table 2: Grid box parameters selected for target enzyme, based on the binding site residues.
| Target proteins | Active site residues | Grid box parameters | |
|---|---|---|---|
| 1CM8 | GLU-181, MET-182, MET-201, TYR-203, GLY-184, TRP-200, ARG-73, ARG-176, LYS-69, ARG-70, ARG152, ASP-153, VAL-186, VAL-187, ARG-192, ARG-180, HIS-231, ASP-230, PRO-110, MET-109, ILE-87, ALA-54, PHE-111, MET-112, LYS-56, LEU-170, ASP-171, VAL-41, ALA-40, ALA-160, ASN-158, GLY-157, LYS-155, LYS-118, VAL-33, THR-114, ASP-115 | 26.51*54.90*25.77 | 83.52*86.38*95.40 |
| 1ERR | MET-343, PHE-404, LEU-346, LEU-349, THR-347, ARG-394, LEU-391, ALA-350, ASP-351, GLU-353, LEU-354, LEU-539, LEU-536, VAL-376, HIS-377, LEU-378, GLY-457, SER-456, LEU-379, GLU-380, ALA-382, TRP-383, LEU-387, LEU-384, GLU-385, MET-388, LEU-391, ARG-394, LEU-525, GLY-521, MET-522, SER-518, ASN-519, ARG-515, HIS-524, LEU-428, ILE-424, PHE-425, MET-421, GLY-420 | 50.09*37.17*68.42 | 69.16*60.29*59.30 |
| 1GFW | GLU-125, GLY-122, HIS-121, SER-120, MET-61, CYS-163, ALA-162, TYR-204, SER-205, TRP-206, PHE-256, ARG-207, SER-65, SER-209 | 26.71*22.60*37.12 | 57.21*54.90*53.24 |
| 1JUN | GLY-274, CYS-273, CYS-273, GLY-274, GLY-275, ARG-276, ILE-277, ALA-278, LEU-280, GLU-281, ARG-276, ILE-277, LEU-280, GLU-281, ALA-278, GLY-275 | 2.40*1.04*1.63 | 23.87*67.83*22.32 |
| 1NFK | GLY-52, PRO-68, GLY-66, GLY-65, SER-78, SER-72, GLU-73, GLY-77 | 15.42*20.85*11.10 | 59*82*59 |
| 1P22 | ARG-431, ALA-39, GLY-38, GLY-432, GLY-408, ALA-391, ALA-392, LEU-351, | 17.92*32.00*-2.12 | 88*85*52 |
| 1ZXM | LYS-168, PHE-142, ALA-167, GLY-166, TYR-165, ILE-141, GLN-376, THR-147, GLY-164, ASN-163, SER-148, TYR-34, SER-149, ARG-162, ASN-150, GLY-161, ILE-125, LYS-378, THR-215, ALA-92, ARG-98, ASN-120, ASN-91, ASP-94, ASN-95, LYS-123, GLY-160, GLU-87, GLY-124 | 63.65*7.81*18.65 | 54.05*49.76*75.15 |
| 2AR9 | PRO-357, ASP-356, ARG-355, TRP-354, SER-353 | 15.73*41.00*0.96 | 101*129*147 |
| 2CLS | LEU-129, PHE-170, LYS-126, ALA-169, SER-168, MET-29, LYS-26, GLY-25, CYS-24, TYR-38, ASP-21, THR-68, GLY-70, SER-69, SER-71, THR-45, VAL-22, PRO-39, GLU-40, THR-41, PRO-44, TYR-42, VAL-43, | 14.88*-17.11*25.90 | 71*64*73 |
| 2E2B | ARG-362, ILE-360, PHE-359, HIS-361, LEU-354, VAL-289, ILE-293, GLU-286, MET-290, VAL-379, ASP-381, ALA-380, LEU-298, PHE-382, VAL-299, LEU-370, TYR-253, GLY-321, MET-318, THR-319, LEU-248, THR-315, ILE-313, GLU-316, LYS-271, PHE-317, VAL-256, ALA-269, VAL-270 | 13.64*71.86*39.60 | 59.28*90.41*91.77 |
| 2HY8 | GLY-349, GLY-350, ALA-348, LEU-347, SER-351, LEU-396, VAL-328, ASP-393, ASN-394, THR-406, ASP-407, GLU-315, ALA-297, TYR-346, GLU-345, MET-344, ILE-276, GLY-277, VAL-284, GLN-278, ARG-299, GLY-279, SER-281 | 11*79*14 | 53*58*44 |
| 2HZI | GLU-286, PHE-382, MET-290, ASP-381, ALA-380, ILE-313, VAL-299, THR-315, GLU-316, TYR-253, LYS-271, VAL-256, LEU-370, VAL-270, THR-315, ALA-269, GLU-316, GLY-249, ASN-322, LEU-248, GLY-321, TYR-320, THR-319 | 19.34*12.33*14.72 | 40*50*60 |
| 2OH4 | ASP-1056, ASP-1054, PRO-1055, LYS-869, ALA-842, LYS-1053, PRO-837, ARG-840, GLY-841, VAL-1058, ILE-1051, VAL-846, GLY-839, ARG-1049, ASP-1050, ARG-1059, LEU-838, LYS-866, LEU-1047, ALA-1048, ALA-864, GLY-1046, ASP-1062, GLU-883, PHE-1045, VAL-914, ILE-886, ASP-1044, PHE-916, LEU-887, GLU-915, GLY-920, LYS-918, CYS-1043, ASN-1031, ARG-1030, CYS-917, PHE-919, VAL-897, LEU-1033, HIS-1024, ILE-890, ILE-1042, VAL-896, LEU-1017 | 5.13*37.98*23.09 | 64*44*68 |
| 2OJI | ALA-33, GLY-32, GLU-31, GLY-30, ILE-29, GLY-35, VAL-37, LYS-52, LYS-112, GLU-69, ASP-165, ALA-50, ASN-152, ASP-109, THR-108, GLN-103, CYS-164, ASP-104, LEU-105, LEU-154, MET-106 | -4.33*8.39*46.93 | 64*44*68 |
| 3EZV | PHE-146, ASP-145, ALA-144, VAL-64, PHE-80, GLU-81, LYS-33, VAL-18, ALA-31, LEU-134, PHE-82, LEU-83, GLN-131, ILE-10, ASP-86, GLN-85, HIS-84 | 2.23*30.54*21.73 | 64*44*68 |
| 3UPI | ASP-318, ASP-319, GLY-317, ASN-316, SER-367, CYS-366, SER-368, ARG-386, SER-556, TYR-555, GLY-449, TYR-448, PHE-193, GLN-446, ILE-447, GLY-410, ASN-411, MET-414, PRO-197, TYR-415, ARG-200, LEU-384 | 59.11*-8.23*-24.60 | 48.42*65.43*56.95 |
| 4AQ3 | MET-74, ASP-70, PHE-71, TYR-67, ARG-66, ASP-62, PHE-63, ALA-59, TYR-161, LEU-160, PHE-112, VAL-92, LEU-96, GLU-95, THR-91, ALA-108, VAL-107, ARG-105, TRP-103, ASN-102, PHE-157 | -13.93*14.15*-10.15 | 40.6*34.74*39.13 |
| 4JSX | TRP-2239, VAL-2240, CYS-2243, LEU-2185, TYR-2225, ILE-2237, ASP-2244, ILE-2163, PRO-2169, LYS-2187, SER-2165, THR-2245, MET-2345, ILE-2356, SER-2342, ASP-2195, ASP-2357, GLU-2190 | 69.64*-12.59*-49.70 | 89.77*104.60*117.47 |
| 4LQM | GLN-791, THR-790, GLU-762, LEU-792, MET-793, ILE-789, LEU-788, ALA-743, ILE-744, LYS-745, VAL-726, LEU-718, THR-854, LEU-844, ASP-855, GLY-796, CYS-797, ARG-841, ASP-800 | 23.20*24.98*21.05 | 65.73*109.12*112.22 |
| 4Y7R | CYS-134, PHE-133, SER-175, ASP-92, SER-91, CYS-261, TYR-260, ILE-266, PHE-263, SER-50, SER-306, SER-49, ILE-264, ILE-305 | 13.02*23.79*8.34 | 53.34*54.94*37.99 |
| 4Y83 | LEU-134, LYS-133, TRP-132, GLU-217, ILE-144, GLY-213, SER-214, VAL-152, ARG-146, SER-257, GLY-210, ALA-165, LYS-167, ALA-209, VAL-260, ASN-258, GLU-208, MET-207, VAL-269, ASP-270, ALA-191 | 17.52*-34.78*-13.22 | 40.96*45.69*49.23 |
| 5G4N | VAL-157, GLU-221, CYS-220, LEU-257, THR-230, PRO-222, PRO-223, LEU-145, CYS-229, ASP-228, TRP-146, PRO-151, PHE-109, VAL-147, THR-150 | 130.5*91.75*-29.77 | 40.96*45.69*49.23 |
| 5LA9 | TRP-389, MET-299, THR-387, THR-325, PRO-402, ASP-315, ALA-301, ILE-327, ALA-385, ASP-254, PHE-366, ARG-252, HIS-313, TYR-303, TYR-310, TYR-329, ARG-383, HIS-374, VAL-376, LEU-343 | -24.02*23.32*16.59 | 54.03*47.37*39.90 |
| 5CT7 | GLY-534, GLU-533, ILE-463, PHE-583, CYS-532, TRP-531, ALA-481, VAL-471, GLN-530, VAL-482, PHE-595, THR-529, LYS-483, ILE-592, LEU-514, GLY-593, ASP-594, ILE-527, ILE-513, LEU-567, HIS-574, THR-508, LEU-505, ILE-572, VAL-504, GLU-501 | -15.73*10.38*0.88 | 64*44*68 |
| 6HH1 | LEU-783, CYS-788, ILE-789, HIS-790, ILE-808, ILE-653, LEU-647, LEU-799, CYS-809, VAL-654, ASP-810, VAL-643, LEU-644, CYS-673, PHE-811, GLU-671, GLU-640, TYR-670, ALA-621, LEU-595, VAL-603, LYS-623 | 57.54*38.47*14.68 | 54.58*50.44*57.58 |
Table 3: Top ten ligands showcasing highest binding affinity scores against each protein candidate.
| Protein | Ligand Chemical Name | PubChem ID | Binding affinity (-kcal/mol) |
|---|---|---|---|
| 1CM8 | Apiforol | 443638 | −11.9 |
| Adrenosterone | 223997 | −10.5 | |
| Alstonine | 441979 | −9.7 | |
| Shinjulactone A | 460537 | −9.7 | |
| Hippeastrine | 441594 | −-9.5 | |
| Kurchinicin | 9873539 | −9.4 | |
| 3’Methoxycoumestrol | 44151023 | −9.4 | |
| Coumestrol | 5281707 | −9.3 | |
| Carbamazepine | 2554 | −9.2 | |
| Uzarigenin | 92760 | −-9.2 | |
| 1ERR | Venalstonine | 426061 | −10.1 |
| Sorgolactone | 5281395 | −9.7 | |
| Butin | 92775 | −-9.5 | |
| Epicatechin | 255538 | −9.5 | |
| Naringenin | 439246 | −9.5 | |
| 7,3’,4’Trihydroxyflavone | 5322065 | −9.4 | |
| Enterolactone | 10685477 | −9.4 | |
| Garbanzol | 442410 | −-9.3 | |
| Gibberellin A51 | 443458 | −9.3 | |
| Apiforol | 443638 | −9.3 | |
| 1GFW | Pamoic acid | 8546 | −9.1 |
| Uzarigenin | 92760 | −8.9 | |
| Digitoxigenin | 4369270 | −8.9 | |
| 4Hydroxystrychnine | 211181 | −8.8 | |
| Jatrophatrione | 5281372 | −8.8 | |
| Strychnine | 441071 | −8.7 | |
| CHEMBL3612189 | 5245667 | −8.7 | |
| Pseudostrychnine | 21723446 | −8.6 | |
| Diterpene II (lactone) | 339816 | −8.5 | |
| Brucine | 442021 | −8.5 | |
| 1JUN | Psoralenol | 5320772 | −6.4 |
| Corylidin | 5316096 | −6.2 | |
| Isoarboreol | 21722929 | −6.2 | |
| Alectrol | 5281353 | −6.1 | |
| Gmelanone | 21722946 | −6.1 | |
| Pamoic acid | 8546 | −6 | |
| Jatrophatrione | 5281372 | −5.9 | |
| Indirubin | 5359405 | −5.9 | |
| Protopine | 4970 | −5.8 | |
| β-colubrine | 251890 | −5.8 | |
| 1NFK | Amarolide | 460539 | −8.5 |
| Corylidin | 5316096 | −8.4 | |
| 2Hydroxy3methoxystrychnine | 21680050 | −8.4 | |
| Pseudostrychnine | 21723446 | −8.2 | |
| Protopine | 4970 | −8.1 | |
| 4Hydroxystrychnine | 211181 | −8.1 | |
| β-colubrine | 251890 | −8.1 | |
| Pamoic acid | 8546 | −8 | |
| Stylopine | 440583 | −8 | |
| Sorgolactone | 5281395 | −8 | |
| 1P22 | Pseudostrychnine | 21723446 | −8.8 |
| Strychnine | 441071 | −8.6 | |
| Isoarboreol | 21722929 | −8.6 | |
| Uzarigenin | 92760 | −8.4 | |
| Digitoxigenin | 4369270 | −8.4 | |
| Gmelanone | 21722946 | −8.4 | |
| 4Hydroxystrychnine | 211181 | −8.3 | |
| CHEMBL3612189 | 5245667 | −8.3 | |
| 2Hydroxy3methoxystrychnine | 21680050 | −8.2 | |
| 10-Hydroxycamptothecin | 97226 | −8.2 | |
| 1ZXM | Gmelanone | 21722946 | −11.4 |
| (+) Teframidine | 436140 | −11.2 | |
| Stylopine | 440583 | −11.1 | |
| Seneciphylline | 5281750 | −11.1 | |
| Diterpene II (lactone) | 339816 | −10.9 | |
| Alectrol | 5281353 | −10.8 | |
| Ent7alpha, 12betadihydroxy16kauren19,6betaolide | 5088389 | −10.7 | |
| Fumaritine | 21627986 | −10.6 | |
| 19hydroxytabersonine | 443325 | −10.2 | |
| Gibberellin A51 | 443458 | −10.2 | |
| 2AR9 | 10-hydroxycamptothecin | 97226 | −9.4 |
| Berberine | 2353 | −9 | |
| Corytuberine | 160500 | −8.6 | |
| Cleomiscosin A | 442510 | −8.6 | |
| Berberastine | 442180 | −8.4 | |
| Allamcin | 5477870 | −8.3 | |
| 4’Hydroxychalcone 4’glucoside | 23144947 | −8.2 | |
| Protopine | 4970 | −8.1 | |
| Pseudoyohimbine | 251562 | −8 | |
| Yohimbine | 8969 | −7.9 | |
| 2CLS | Isoarboreol | 21722929 | −10.6 |
| Gmelanone | 21722946 | −10.4 | |
| Cleomiscosin A | 442510 | −10.3 | |
| Bavachromanol | 5321790 | −10.2 | |
| (+) Neoolivil | 9976812 | −9.8 | |
| Psoralenol | 5320772 | −9.7 | |
| 7,3’,4’Trihydroxyflavone | 5322065 | −9.7 | |
| Aureusidin | 5281220 | −9.6 | |
| Fisetin | 5281614 | −9.6 | |
| Dihydrofisetin | 246330 | −9.5 | |
| 2E2B | Coumestrol | 5281707 | −10.9 |
| CHEBI: 73029 | 363863 | −10.8 | |
| Berberine | 2353 | −10.8 | |
| Indirubin | 5359405 | −10.7 | |
| Anhydroglycinol | 442667 | −10.6 | |
| Benzo[a] phenoxazin-9-ylidene (dimethyl) azanium | 81507 | −10.5 | |
| Hematoxylin | 442514 | −10.5 | |
| 2hydroxychrysophanol | 442759 | −10.5 | |
| Jutamansinone | 759294 | −10.5 | |
| Equol | 91469 | −10.4 | |
| 2HY8 | Methoxystrychinine | 21723445 | −10.3 |
| β-colubrine | 251890 | −10.1 | |
| Strychnine | 441071 | −10.1 | |
| CHEMBL3612189 | 5245667 | −10.1 | |
| Pseudostrychnine | 21723446 | −10 | |
| 4Hydroxystrychnine | 211181 | −9.7 | |
| Brucine | 442021 | −9.7 | |
| Dehydroevodiamines | 9817839 | −9.7 | |
| Protopine | 4970 | −9.6 | |
| 10-hydroxycamptothecin | 97226 | −9.6 | |
| 2HZI | Indirubin | 5359405 | −11.8 |
| Benzo[a] phenoxazin-9-ylidene (dimethyl) azanium | 81507 | −11.4 | |
| CHEMBL3612189 | 5245667 | −11.4 | |
| Medicagol | 5319322 | −11.4 | |
| Paulownin | 3084131 | −11.3 | |
| Psoralenol | 5320772 | −11.2 | |
| Carbamazepine | 2554 | −11.1 | |
| CHEBI: 73029 | 363863 | −11.1 | |
| 6Methoxypulcherrimin | 44260092 | −11.1 | |
| Pulviatide | 90471471 | −11 | |
| 2OH4 | Fisetin | 5281614 | −9.7 |
| Butin | 92775 | −9.6 | |
| Orobol | 5281801 | −9.6 | |
| Pseudobaptigenin | 5281805 | −9.6 | |
| 7,3’,4’Trihydroxyflavone | 5322065 | −9.6 | |
| Santal | 9926336 | −9.6 | |
| Naringenin | 439246 | −9.5 | |
| Garbanzol | 442410 | −9.5 | |
| Kaempferol | 5280863 | −9.5 | |
| Genistein | 5280961 | −9.5 | |
| 2OJI | Pseudostrychnine | 21723446 | −10 |
| Shinjulactone A | 460537 | −9.9 | |
| CHEMBL3612189 | 5245667 | −9.9 | |
| Corylidin | 5316096 | −9.8 | |
| Strychnine | 441071 | −9.7 | |
| Ent7α,12βDihydroxy16kauren19,6βolide | 5088389 | −9.4 | |
| Molephantinin | 5281485 | −9.4 | |
| 4Hydroxystrychnine | 211181 | −9.3 | |
| β-colubrine | 251890 | −9.3 | |
| Precalyone | 324879 | −9.3 | |
| 3EZV | Dehydroevodiamines | 9817839 | −10.3 |
| Corylidin | 5316096 | −10.2 | |
| Rosmaquinone B | 46883407 | −9.9 | |
| CHEMBL3612189 | 5245667 | −9.8 | |
| Bavachromanol | 5321790 | −9.8 | |
| Pamoic acid | 8546 | −9.6 | |
| 4Hydroxystrychnine | 211181 | −9.6 | |
| Aureusidin | 5281220 | −9.6 | |
| Sulfuretin | 5281295 | −9.6 | |
| (E) 7Hydroxy3 (4hydroxybenzylidene) chroman4one | 44443280 | −9.6 | |
| 3UPI | Corylidin | 5316096 | −10.2 |
| Protopine | 4970 | −9.8 | |
| Shinjulactone A | 460537 | −9.6 | |
| Pamoic acid | 8546 | −9.5 | |
| Medicagol | 5319322 | −9.5 | |
| β-colubrine | 251890 | −9.4 | |
| 4Hydroxystrychnine | 211181 | −9.2 | |
| Riddelliine | 5281744 | −9.2 | |
| Granisetron | 5284566 | −9.2 | |
| AC1L8OTG | 430162 | −9.1 | |
| 4AQ3 | Protopine | 4970 | −8.7 |
| (+) Teframidine | 436140 | −8.6 | |
| Jatrophatrione | 5281372 | −8.4 | |
| Gmelanone | 21722946 | −8.3 | |
| Methoxystrychinine | 21723445 | −8.3 | |
| CHEMBL3612189 | 5245667 | −8.2 | |
| Centratherin | 44409502 | −8.2 | |
| Brucine | 442021 | −8.1 | |
| Isopinnatal | 3035834 | −8.1 | |
| 4’Hydroxychalcone 4’glucoside | 23144947 | −8.1 | |
| 4JSX | Isoarboreol | 21722929 | −10.9 |
| Digitoxigenin | 4369270 | −10.7 | |
| Uzarigenin | 92760 | −10.6 | |
| Psoralenol | 5320772 | −10.5 | |
| Stylopine | 440583 | −10.4 | |
| (-)-8-oxotetrahydropalmatine | 49769861 | −10.2 | |
| Pseudoyohimbine | 251562 | −9.9 | |
| Medicagol | 5319322 | −9.9 | |
| Dihydrobaicalein | 9816931 | −9.9 | |
| Gibberellin A35 | 21596345 | −9.9 | |
| 4LQM | Strychnine | 441071 | −9.7 |
| Pseudostrychnine | 21723446 | −9.7 | |
| Protopine | 4970 | −9.5 | |
| Sorgolactone | 5281395 | −9.4 | |
| Diosbulbin G | 21723242 | −9.4 | |
| β-colubrine | 251890 | −9.2 | |
| Cathenamine | 443361 | −9.2 | |
| CHEMBL3612189 | 5245667 | −9.2 | |
| 10-Hydroxycamptothecin | 97226 | −9.2 | |
| Uzarigenin | 92760 | −9.1 | |
| 4Y7R | Pamoic acid | 8546 | −10 |
| Pseudostrychnine | 21723446 | −9.8 | |
| Shinjulactone A | 460537 | −9.7 | |
| Granisetron | 5284566 | −9.7 | |
| 2Hydroxy3methoxystrychnine | 21680050 | −9.4 | |
| 4Hydroxystrychnine | 211181 | −9.2 | |
| (+) Adlumine | 442155 | −9.1 | |
| Amarolide | 460539 | −9.1 | |
| Methoxystrychinine | 21723445 | −9.1 | |
| Gmelanone | 21722946 | −9.1 | |
| 4Y83 | Pamoic acid | 8546 | −10.8 |
| Texalin | 473253 | −10.7 | |
| (E) 7Hydroxy3 (4hydroxybenzylidene) chroman4one | 44443280 | −10.7 | |
| Persicogenin | 320054 | −10.5 | |
| Aureusidin | 5281220 | −10.5 | |
| 7,3’,4’Trihydroxyflavone | 5322065 | −10.5 | |
| 10-Hydroxycamptothecin | 97226 | −10.5 | |
| Calycosin | 5280448 | −10.4 | |
| Naringenin | 439246 | −10.3 | |
| Apiforol | 443638 | −10.3 | |
| 5G4N | Medicagol | 5319322 | −8.8 |
| CHEMBL3612189 | 5245667 | −8.5 | |
| Coumestrol | 5281707 | −8.3 | |
| CHEBI: 73029 | 363863 | −8.2 | |
| Hippeastrine | 441594 | −8.1 | |
| Dehydroevodiamines | 9817839 | −8.1 | |
| Protopine | 4970 | −8 | |
| Pamoic acid | 8546 | −8 | |
| Stylopine | 440583 | −8 | |
| 6Hydroxyrubiadin | 5319801 | −8 | |
| 5LA9 | Pamoic acid | 8546 | −8.5 |
| Strychnine | 441071 | −8.4 | |
| Shinjulactone A | 460537 | −8.4 | |
| 4Hydroxystrychnine | 211181 | −8.3 | |
| β-colubrine | 251890 | −8.3 | |
| Corylidin | 5316096 | −8.3 | |
| Akuammiline | 5367018 | −8.3 | |
| 2Hydroxy3methoxystrychnine | 21680050 | −8.3 | |
| Viroallosecurinine | 908416 | −8.2 | |
| Methoxystrychinine | 21723445 | −8.2 | |
| 5CT7 | Pseudobaptigenin | 5281805 | −10.5 |
| Santal | 9926336 | −10.2 | |
| 1,9pyrazoloanthrone | 8515 | −10 | |
| Genistein | 5280961 | −10 | |
| Thalidomide | 5426 | −9.9 | |
| 2’hydroxydihydrodaidzein | 440047 | −9.9 | |
| 2’Hydroxydaidzein | 5280520 | −9.9 | |
| Baptigenin | 9965663 | −9.9 | |
| Chebi: 73029 | 363863 | −9.8 | |
| 3’,4’,7trihydroxyisoflavone | 5284648 | −9.8 | |
| 6HH1 | CHEMBL3612189 | 5245667 | −10.1 |
| Stylopine | 440583 | −10 | |
| Sorgolactone | 5281395 | −9.9 | |
| Enterolactone | 10685477 | −9.9 | |
| Cathenamine | 443361 | −9.8 | |
| Dehydroevodiamines | 9817839 | −9.8 | |
| Paulownin | 3084131 | −9.7 | |
| Corylidin | 5316096 | −9.6 | |
| Psoralenol | 5320772 | −9.6 | |
| Protopine | 4970 | −9.5 |
Hydrophobic ligand interaction in the receptor-binding pocket is enhanced by bonds such as π-alkyl and alkyl [32]. The highest number of hydrogen bonds was observed in the case of ligands that displayed higher binding affinity against the protein.
3.2. Evaluation of Pharmacokinetic and Pharmacological Properties
The Lipinski’s rule of five was used to determine the drug-likeness of the phytochemicals examined in the study Table B of the supplementary data. The rule states that the drug-like compound must have LogP values ranging between 0 and 5, hydrogen bond donors <5, hydrogen bonds acceptors <10, and the drug’s molecular weight should not be more than 500 Da [33]. Thus, the rule is based on the molecular weight, the presence of hydrogen donor-acceptor bonds, and the drug’s lipophilicity. These factors are used to evaluate the drug’s absorption, penetration, metabolism, and excretion. The interaction of the membrane with the drug is affected when the compound violates the rule of five; poor absorption and permeation of the drug are more expected when the rules are violated [34]. Lipinski’s rule of five was used to determine the drug-likeness of the 250 phytochemicals that have higher binding affinity against the selected proteins. Compounds that violated one or more of Lipinski’s rules were excluded from the study, and the remaining compounds were further investigated to test their toxicity. Furthermore, hydrogen bonds play a key role in drug designing as they are involved in drug permeation, metabolism, and absorption. The bond type and the number of bonds present in each protein-ligand complex are displayed in Table C of the supplementary data.
3.3. Bioavailability Radar and Toxicity Prediction
Following the molecular docking, pharmacokinetic, and pharmacological analysis, the selected compounds were, further, investigated for their toxicity profile. Bioavailability radar was utilized to determine the drug-likeness of the phytochemical against the respective protein. The tool determines the bioavailability radar of the given phytochemical based on six properties: saturation, size, solubility, flexibility, lipophilicity, and polarity. The saturation, that is, a fraction of carbons in the sp3 hybridization, should not be <0.25, and the molecular weight should range from 150 to 500 g/Mol. Solubility, that is, LogS should not be higher than 6, the flexibility should not be more than nine rotatable bonds, the lipophilicity, that is, XLOGP3 should be between −0.7 and +5.0, and the polarity, that is, TPSA must be between 20 and 130 Å2 [35].
Oral drug formulations are widely used and more convenient to administer than the other drugs; they also have fewer sterility restrictions and are cost-effective. However, most of the new compounds have low water solubility, making oral drug consumption less suitable as a model for drug administration, thus making it a significant problem for drug developers. The high lipophilicity of the compounds also impacts the drug’s therapeutic effect. These problems should be tackled to make the drug bioavailable. The compounds investigated against HCC were found to be orally bioavailable. Computational methods and approaches can be used to test a drug’s safety during the developmental process. These have proven to have more advantageous over in vitro and in vivo studies [36]. The toxicity analysis of the shortlisted molecules was carried out using the PROTOX -II webtool. The parameters used for evaluating the compounds were: Cytotoxicity, Carcinogenicity, Hepatotoxicity, Immunotoxicity, Mutagenicity, Predicted LD50, and Toxicity (Oral toxicity).
Considering the bioavailability radars (Table B of the Supplementary data) of the best 250 phytochemicals that displayed the highest binding affinity scores against their respective receptors, only 20 ligands were found to fit in the pink colored optimal range of the bioavailability radar [Figure 1]. In addition, they were also found to be inactive for hepatotoxicity while being either active or inactive for other toxicity parameters, as well as displayed an appreciable number of hydrogen bonds with the respective HCC protein targets [Table 4].
 | Figure 1: Bioavailability Radar plot of each shortlisted ligand. [Click here to view] |
Table 4: Toxicity profile of ligands.
| Ligands | Toxicity Class | LD50 (mg/kg) | Hepato- toxicity | Carcino- genicity | Immuno- toxicity | Muta- genicity | Cyto-toxicity | Conventional hydrogen bonds |
|---|---|---|---|---|---|---|---|---|
| Sorgolactone | 6 | 5105 | Inactive | Active | Active | Inactive | Inactive | 1 |
| Alectrol | 6 | 5105 | Inactive | Inactive | Active | Inactive | Inactive | 6 |
| Jatrophatrione | 5 | 4600 | Inactive | Active | Active | Inactive | Inactive | 2 |
| Amarolide | 5 | 3900 | Inactive | Inactive | Active | Inactive | Inactive | 2 |
| Psoralenol | 5 | 2875 | Inactive | Inactive | Active | Inactive | Inactive | 1 |
| Enterolactone | 4 | 2000 | Inactive | Inactive | Active | Inactive | Inactive | 2 |
| Adrenosterone | 4 | 1870 | Inactive | Active | Active | Inactive | Inactive | 1 |
| Kurchinicin | 4 | 1680 | Inactive | Active | Active | Inactive | Active | 2 |
| Isoarboreol | 3 | 1500 | Inactive | Inactive | Active | Inactive | Inactive | 3 |
| Gmelanone | 4 | 1500 | Inactive | Active | Active | Inactive | Inactive | 4 |
| Protopine | 4 | 940 | Inactive | Active | Active | Active | Inactive | 1 |
| Stylopine | 4 | 940 | Inactive | Active | Active | Inactive | Active | 2 |
| Corylidin | 4 | 832 | Inactive | Active | Active | Active | Inactive | 3 |
| Shinjulactone A | 4 | 590 | Inactive | Inactive | Active | Inactive | Active | 5 |
| Hippeastrine | 3 | 230 | Inactive | Active | Active | Inactive | Inactive | 1 |
| Ac1l9c1h (synonyms: alstonine; senegasaponins b) | 3 | 215 | Inactive | Inactive | Active | Inactive | Inactive | 1 |
| Brucine | 3 | 150 | Inactive | Active | Inactive | Inactive | Inactive | 2 |
| Beta-colubrine | 3 | 150 | Inactive | Inactive | Active | Inactive | Inactive | 1 |
| 2 Hydroxy-3-methoxystrychnine | 3 | 150 | Inactive | Inactive | Active | Inactive | Active | 2 |
| Uzarigenin | 2 | 26 | Inactive | Inactive | Active | Inactive | Inactive | 3 |
Out of all the ligands displayed in Table 4, Enterolactone, Adrenosterone, Kurchinicin, Gmelanone, Protopine, Stylopine, Corylidin, and Shinjulactone A belonged to toxicity class IV (300 < LD50 ≤ 2000), indicating their harmfulness if swallowed. On the other hand, only three ligands, namely, Jatrophatrione, Amarolide, and Psoralenol, fell in the toxicity Class V (2000 < LD50 ≤ 5000), indicating their possibility of being toxic if swallowed. Finally, two phytochemical drugs, Sorgolactone and Alectrol, were observed to have the same LD50 value of 5105 mg/kg body weight. Such LD50 value >5000 mg/kg body weight indicates a Class VI toxicity level that is nontoxic. Based on these toxicity parameters, the phytochemicals Sorgolactone and Alectrol were considered as ideal drug targets against their respective HCC protein targets.
3.4. Selection of Drug Candidate for 6HH1
Based on the molecular docking, bioavailability radar, and toxicity profile analysis of the ligands, Sorgolactone was an ideal drug candidate against the tyrosine kinase receptor. Sorgolactone belongs to the group of strigolactones (SLs), which consist of terpenoid-derived endogenous plant hormones [37]. SLs have been known for three main physiological processes in plants: first, they aid in the germination of parasitic microbes which reside in the roots of the host plants of the genus Striga; second, they help in plant identification by the mutual association forming fungi; and finally, these phytochemicals are well known to prevent branching especially at plants’ the stem terminal region. Sorgolactone plays a key role as a plant growth regulator involved in developing several parts of the plant such as stems, leaves, flowers, and the ripening of fruits.
Sorgolactone exhibited binding energy of –9.9 kcal/mol against the tyrosine kinase receptor 6HH1. Figure 2 depicts seven different types of bonds formed by sorgolactone with the kinase receptor: the conventional hydrogen bond, the alkyl, the π-alkyl, the π-anion, the π-cation, the π-sigma, and the π-sulfur bond. The 2D representation of the protein-ligand complex revealed that the ligand formed one conventional hydrogen bond on the 6HH1 chain A amino acid ASP-810; seven alkyl bonds were formed on LYS-623, VAL-603, CYS-788, LEU-644, and VAL-643; two π-alkyl bonds were formed with VAL-654 and LEU-644; one π-cation bond was formed with LYS-623, whereas two π-anion bonds were formed with ASP-810; one π-sulfur bond was created with the receptor amino acid CYS-809, and one π-sigma bond was formed with VAL-643.
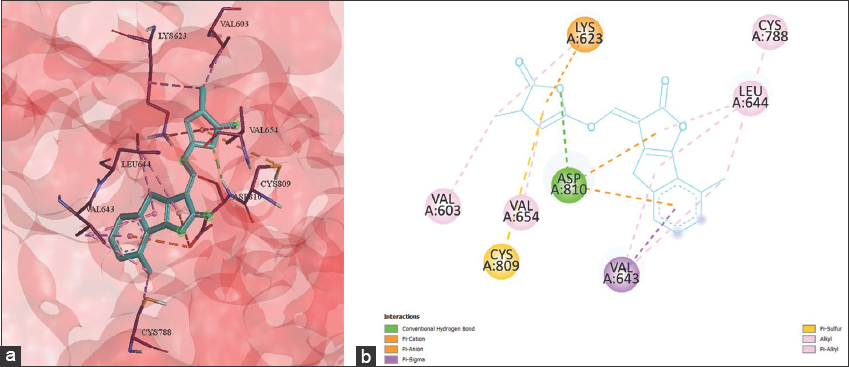 | Figure 2: (a) 3D and (b) 2D representation of Sorgolactone interacting with the binding pocket of TYR kinase receptor 6HH1. [Click here to view] |
3.5. Selection of a Drug Candidate for 1ZXM
Alectrol was selected as an appropriate candidate against the HCC protein target 1ZXM due to its high binding affinity, its accordance with Lipinski’s rule of five, and its non-toxic nature as compared to other drug candidates. Alectrol belongs to the class of strigolactone, a class of cartenoid-derived molecules known for controlling plants’ growth, development, adaptation, and abiotic stress regulation [38]. The binding affinity score of Alectrol against the protein molecule 1ZXM, a complex multifunctional enzyme Human topoisomerase IIa, was observed to be –10.8 kcal/mol and displays altogether ten bonds with the protein target. Figure 3 exhibits six conventional hydrogen bonds formed by electron on the receptor chain A amino acids, namely, GLY-164, ARG-162, SER-148, and SER-149, whereas two hydrogen bond linkages were formed with ASN-150. The drug was also observed to form a Carbon hydrogen bond with the amino acid LYS-168; a π-alkyl bond was formed with PHE-142, and one alkyl bond was created with the amino acids ILE-141 and ILE-125 of target protein 1ZXM.
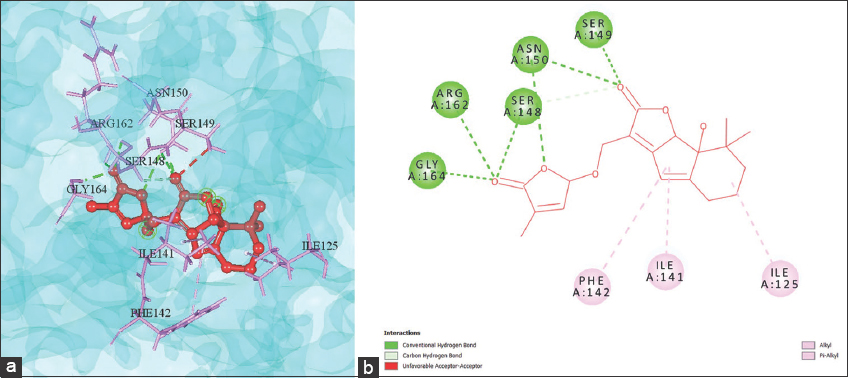 | Figure 3: (a) 3D and (b) 2D representation of Alectrol interacting with the binding pocket of the complex multifunctional enzyme 1ZXM. [Click here to view] |
4. CONCLUSION
Cancer has been a long thriving mortality threat to humanity for decades. Out of all the carcinoma types encountered to date, primary liver cancer continues to be the most frequently diagnosed disease and ranks fourth in terms of cancer mortality worldwide. Considering the threat posed by this carcinoma type, the search for effective drug candidates has been on the rise. As opposed to the synthetic drugs available for cancer treatment, naturally occurring chemicals known as phytochemicals are being investigated for their possible applications as anticancer agents. In our study, 1259 phytochemicals were docked against 25 HCC targets resulting in 250 probable drug candidates. With further screening using pharmacokinetic and pharmacological parameters and toxicity profiling of each possible drug candidate, only two non-toxic phytochemicals were identified as the most appropriate drug candidates against their respective carcinoma receptors.
Both the phytochemicals, Sorgolactone and Alectrol, belong to the class of SLs, which are widely known as plant hormones capable of restricting branching in plants [39]. Although our current study findings indicate that Sorgolactone and Alectrol have been identified as potential drug targets against the HCC protein targets 6HH1 and 1ZXM, respectively, both the phytochemicals were found to be active for immunotoxicity. In contrast, the former drug candidate was active for carcinogenicity [Table 4]. A probable solution to tackle this problem can be the introduction of a slight structural modification that can improve the chances of these drug candidates becoming less toxic. A review in the past has reported strigolactone analogues as potential anticancer agents that can affect the cancerous cells viability rates, promote apoptosis, and thereby lead to cell death at micromolar concentrations [40]. Another recent study conducted by Prandi et al. [41] reported the antiproliferative effects of SLs analogues on both solid and non-solid cancer cells lines such as prostate, colon, lung, melanoma, osteosarcoma, and leukemic cell line while being non-toxic to normal cell lines. In addition, the findings of the study report that SLs analogues have had an anti-proliferative effect on HCC-HepG2 cell line. Despite these studies, there has been no report till date towards the in silico and in vivo identification of Sorgolactone and Alectrol, both belonging to SLs, and their candidacy as potential drug target. Therefore, the results of this study make an attempt to shed light on the potential of these two phytochemicals as probable drugs for therapeutic treatment against HCC.
5. AUTHOR CONTRIBUTIONS
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
6. FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
7. CONFLICTS OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflicts of interest.
8. ETHICS APPROVAL
This study does not involve experiments on animals or human subjects.
9. DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin 2021;71:209-49. [CrossRef]
2. Global Health Estimates:Life Expectancy and Leading Causes of Death and Disability. Cause-specific Mortality, 2009-2019. Available from:https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates/ghe-leading-causes-of-death [Last accessed on 2020 Apr]
3. Omran AR. The epidemiologic transition:A theory of the epidemiology of population change. Milbank Q 2005;83:731-57. [CrossRef]
4. Gersten O, Wilmoth JR. The cancer transition in Japan since 1951. Demogr Res 2002;271-306. [CrossRef]
5. American Cancer Society. American Cancer Society. Cancer Facts and Figures 2021. Atlanta:American Cancer Society;2021. 1-72. Available from:https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html [Last accessed on 2020 Apr]
6. Dasgupta P, Henshaw C, Youlden DR, Clark PJ, Aitken JF, Baade PD. Global trends in incidence rates of primary adult liver cancers:A systematic review and meta-analysis. Front Oncol 2020;10:1-17. [CrossRef]
7. Howlader N, Noone AM, Krapcho M, Miller D, Brest A. SEER Cancer Statistics Review (CSR) 1975-2017. Available from:https://seer.cancer.gov/archive/csr/1975_2017 [Last accessed on 2020 Apr]
8. Petrick JL, McGlynn KA. The changing epidemiology of primary liver cancer. Curr Epidemiol Rep 2019;6:104-11. [CrossRef]
9. Suganya V, Anuradha V. In silico molecular docking of astaxanthin and sorafenib with different apoptotic proteins involved in hepatocellular carcinoma. Biocatal Agric Biotechnol 2019;19:101076. [CrossRef]
10. Liver Cancer Types. Available from:https://www.cancercenter.com/cancer-types/liver-cancer/types [Last accessed on 2022 Apr 20].
11. Ambade A, Mandrekar P. Oxidative stress and inflammation:Essential partners in alcoholic liver disease. Int J Hepatol 2012;2012:853175. [CrossRef]
12. Liu CY, Chen KF, Chen PJ. Treatment of liver cance. Cold Spring Harb Perspect Med 2015;5:a021535. [CrossRef]
13. Da Rocha AB, Lopes RM, Schwartsmann G. Natural products in anticancer therapy. Curr Opin Pharmacol 2001;1:364-9. [CrossRef]
14. Cragg GM Newman DJ. Plants as a source of anti-cancer agents. J Ethnopharmacol 2005;100:72-9. [CrossRef]
15. Pan L, Chai H, Kinghorn AD. The continuing search for antitumor agents from higher plants. Phytochem Lett 2010;3:1-8. [CrossRef]
16. Pan L, Chai HB, Kingorn AD. Discovery of new anticancer agents from higher plants. Front Biosci (Schol Ed) 2012;4:142-56. [CrossRef]
17. Sarkar B, Ullah MA, Islam SS, Rahman MH, Araf Y. Analysis of plant-derived phytochemicals as anti-cancer agents targeting cyclin dependent kinase-2, human topoisomerase IIa and vascular endothelial growth factor receptor-2. J Recept Signal Transduct Res 2021;41:217-33. [CrossRef]
18. McConkey BJ, Sobolev V, Edelman M. The performance of current methods in ligand-protein docking. Curr Sci 2002;83:845-56.
19. Rashdan HR, El-Naggar M, Abdelmonsef AH. Synthesis, molecular docking studies and in silico admet screening of new heterocycles linked thiazole conjugates as potent anti-hepatic cancer agents. Molecules 2021;26:1705. [CrossRef]
20. Sasidharan SP, Vasumathi AV. In vitro pharmacological, in vivo toxicological and in silico molecular docking analysis of glycopentalone, a novel compound from Glycosmis pentaphylla (Retz.) Correa. Med Chem Res 2017;26:1697-707. [CrossRef]
21. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat YN, Weissig H, et al. The protein data bank. Nuclei Acids Res 2000;28:235-42. [CrossRef]
22. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605-12. [CrossRef]
23. Dallakyan S, Olson A. Small-molecule library screening by docking with pyrx. Glob Food Secur Gov 2015;1263:1-11. [CrossRef]
24. Mohanraj K, Karthikeyan BS, Vivek-Ananth RP, Chand RP, Aparna SR, Mangalapandi P, et al. IMPPAT:A curated database of Indian medicinal plants, phytochemistry and therapeutics. Sci Rep 2018;8:4329. [CrossRef]
25. Xiong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, et al. ADMETlab 2.0:An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 2021;49:W5-14. [CrossRef]
26. Banerjee P, Eckert AO, Schrey AK, Preissner R. ProTox-II:A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res 2018;46:W257-63. [CrossRef]
27. Lin L, Yan L, Liu Y, Qu C, Ni J, Li H. The burden and trends of primary liver cancer caused by specific etiologies from 1990 to 2017 at the global, regional, National, age, and sex level results from the global burden of disease study 2017. Liver Cancer 2020;9:563-82. [CrossRef]
28. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018:GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [CrossRef]
29. El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012;142:1264-73. [CrossRef]
30. Whitworth AL, Mann NH, Larkum AW. Serum microcystins level positively linked with risk of hepatocellular carcinoma:A case-control study in Southwest China. Ultrasound Obs Gynecol 2006;50:776-80.
31. Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology 2010;51:1972-8. [CrossRef]
32. Varma AK, Patil R, Das S, Stanley A, Yadav L, Sudhakar A. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS One 2010;5:e12029. [CrossRef]
33. Munir A, Azam S, Mehmood A. Structure-based pharmacophore modeling, virtual screening and molecular docking for the treatment of ESR1 mutations in breast cancer. Drug Des Open Access 2016;5:1000137. [CrossRef]
34. Pollastri MP. Overview on the rule of five. Curr Protoc Pharmacol 2010;9:9-12. [CrossRef]
35. Bojarska J, Remko M, Breza M, Madura ID, Kaczmarek K, Zabrocki J, et al. A supramolecular approach to structure-based design with a focus on synthons hierarchy in ornithine-derived ligands:Review, synthesis, experimental and in silico studies. Molecules 2020;25:1135. [CrossRef]
36. Waring MJ. Lipophilicity in drug discovery. Expert Opin Drug Discov 2010;5:235-48. [CrossRef]
37. Bürger M Chory J. In-silico analysis of the strigolactone ligand-receptor system. Plant Direct 2020;4:e00263. [CrossRef]
38. Bhoi A, Yadu B, Chandra J, Keshavkant S. Contribution of strigolactone in plant physiology, hormonal interaction and abiotic stresses. Planta 2021;254:28. [CrossRef]
39. Bouwmeester H, Li C, Thiombiano B, Rahimi M, Dong L. Adaptation of the parasitic plant lifecycle:Germination is controlled by essential host signaling molecules. Plant Physiol 2021;185:1292-308. [CrossRef]
40. Hasan MN, Razvi S, Kuerban A, Balamash KS, Al-Bishri WM, Abulnaja KO, et al. Strigolactones a novel class of phytohormones as anti-cancer agents. J Pestic Sci 2018;43:168-72 [CrossRef]
41. Prandi C, Kapulnik Y, Koltai H. Strigolactones:Phytohormones with promising biomedical applications. Eur J Org Chem 2021;2021:4019-26. [CrossRef]