1. INTRODUCTION
In shrimp, fish, and crustacean farming, several factors influence the disease’s occurrence, including aquatic breed sources, environmental factors (water quality and agricultural management), and bacterial quantity or virulence. In the diseases caused by bacterial virulence, Vibrio spp., which belongs to the Gram-negative marine bacteria family, is one of the most serious [1]. Thus, in aquatic farming, bacteria, especially Vibrio spp., are strictly controlled by chemists, herbs, or biofactors.
ToxR is a Vibrio spp. virulence regulator gene, coding a protein in a group of transmembrane ones conserved throughout the bacteria. Several Vibrio spp. species use toxR to help them resist bile, and some strains choose toxR to cause infection [2] The protein helps bacteria survive and colonize by forming biofilms. However, in farm systems, the abuse of antibiotic usage in aquaculture leads to the emergence and increase of antibiotic-resistant bacteria [3]. Therefore, finding a biological therapy to replace antibiotics for bacterial disease control is urgently required.
One potential alternative therapy is using phages in farming, in which the bacterial resistance of phage expansion is nearly 10 times lower than that of antibiotics [4]. The phages still develop in harsh environments and constantly increase their concentration density until that of the infected host incredibly decreases [5]. Phages do not harm the environment’s natural and beneficial microbial flora [6]. In aquaculture, Vibrio spp. can form biofilms, which decrease the effectiveness of antibiotics. Nevertheless, some studies have indicated that phages can resist bacterial biofilms [7,8].
Bacteriophages are viruses that parasitize bacteria and are categorized into two groups based on their life cycle: virulent phages that exhibit a lytic cycle and temperate ones that initiate either the lytic or lysogenic cycle [9]. During infection with bacterial cells, the reproduction of virulent phages always leads to host death, while in the lysogenic cycle of temperate phages, the main mechanism is the integration of phage nucleic acid into the host genome, which does not directly cause bacterial death [10]. Nevertheless, integration and excision from host chromosomes can also affect bacterial characteristics [9]. It has been widely observed that temperate phages increase bacterial virulence, the most important factor causing infection. However, there is also some research indicating that phages can reduce viral virulence [11].
There have been many studies on isolating and applying lytic bacteriophages capable of inhibiting Vibrio spp. reported [12-14]. However, up to date, there are still very few studies on the effect of phage on the virulence genes of Vibrio spp. For the above reason, the study was conducted with the objective of isolating Vibrio spp. and their phages from shrimp and shrimp pond samples in some provinces of Viet Nam, the first step in determining the ability to infect bacteria by phages: directly reducing the number of bacteria or reducing the virulence of bacteria.
2. MATERIALS AND METHODS
2.1. Isolation and Identification of Vibrio spp.
Vibrio spp. Were isolated from 35 samples of pond water, button mud, and cultured shrimp from Kien Giang, Soc Trang, and Bac Lieu provinces. Ten cultured shrimp samples were collected for every five samples in Soc Trang and Kien Giang. Similarly, 10 samples of pond button mud were collected from Kien Giang and Bac Lieu, and 10 sick shrimp samples were collected from Bac Lieu and Soc Trang. The last five samples of pond siphon water were collected from Bac Lieu. Samples were diluted and inoculated on a thiosulfate-citrate-bile-salt-sucrose (TCBS) medium. The plates were incubated at room temperature for 24 h. Colonies specific to Vibrio spp. (large, blue, and yellow) were selected for domestication using the streak-plate culture method.
Bacterial cells were extracted through centrifugation at 12,000 rpm for 10 min at room temperature. Then, 800 μL of lysis buffer was added to 800 μL of chloroform: isoamyl (24:1 v/v), mixed well, and incubated at 28°C for 10 min. Then, the solution was centrifuged as described above to collect the supernatant. The mixture containing 700 μL of acquired and purified bacterial DNA supernatant and 700 μL of a 95% ethanol solution was mixed well and centrifuged at 12,000 rpm for 20 min to collect the precipitate. The precipitate was then washed with 500 μL of a 70% ethanol solution and centrifuged at 12,000 rpm for 10 min. Subsequently, the samples were vacuum dried at 45°C for 10 min. Afterward, DNA was dissolved in 100 μL of 0.1× TE solution and stored at 4°C. DNA quality was checked by 1% agarose gel electrophoresis.
Vibrio spp. and Vibrio parahaemolyticus were identified using polymerase chain reaction (PCR). The ToxR PCR contained 14 μL H2O, 2 μL DNA, 8 μL My TaqMix 2×, 0.5 μL F-primer: GTCTTCTGACGCAATCGTTG, and 0.5 μL R-primer: ATACGAGTGGTTGCTGTCATG [15]. The amplifier was carried out at 94°C for 10 min, 20 cycles: –94°C (60 s), 63°C (90 s), 72°C (90 s), and 72°C (10 min) [16]. Then, products were examined by 2% agarose gel electrophoresis with 1× TBE (10 mM Tris; 5 mM borate; 0.1 mM ethylenediaminetetraacetic acid) to find the bank at the position of 368 bp. The 16S rRNA PCR was carried out similarly to those of ToxR, except for the F-primer: CAGGCCTAACACATGCAAGTC, R-primer: GCATCTGAGTGTCAGTATCTGTCC [17], carried out at 95°C for 10 min, 30 cycles –95°C (60 s), 55°C (60 s), 72°C (90 s), and 72°C (5 min) [16] and find the bank at the position of 700 bp. Then, the nucleotides were sequenced and compared to the Genbank using the basic local alignment search tool.
2.2. Isolation of Vibrio spp. Phage
Vibrio spp. Phage were isolated from 144 samples of pond water, healthy shrimp, and diseased shrimp from Kien Giang, Soc Trang, and Bac Lieu provinces. Vibrio spp. phages were isolated using the double-agar plaque assay [18]. Ten microlitters of the crude phage mixture and 100 μL of the isolated V. parahaemolyticus (isolated in the experiment) culture were aliquoted into 5 mL of a sterile soft nutrient medium (0.4% agar TSA), kept warm at 55°C, mixed, and spread on a hard nutrient medium (1.7% agar TSA). Plates were incubated at 35°C for 24 h to monitor plaque formation. Lastly, individual plaques were collected with sterile tips, added to 1 mL of a sterile SM buffer, and stored at 4°C.
2.3. Determination of the Host Range of Isolated Phages
The host range of the Vibrio spp. phages was determined by the double agar drop method [18]. Ten μL of each phage were dropped on a soft nutrient medium (TSA 0.4% agar) with Vibrio spp. Next, the plaques were classified into 3 categories of clarity: clear, turbid, and no reaction [19].
2.4. Comparison of the toxR gene of pre- and post-phage-infecting
A mixture of phages and bacteria, with an MOI of 100, was incubated for 24 h. The bacterial samples were then subjected to DNA extraction through PCR with a toxR primer. Next, the PCR products were electrophoresed using the Bio-Rad electrophoresis kit with PC and a 2% agarose gel × 1× TBE buffer. The product was then stained with SafeView, and the gel was photographed under UV light using a Bio-Rad Gel Doc system. Subsequently, the tape images were analyzed using Quantity One software. Following this, the GeneRulerTM 100-bp DNA ladder was used with a standard scale to estimate the size of the DNA fragment. The electrophoretic PCR product was then sent for Sanger sequencing. Afterward, the bacterial toxR gene sequences were compared by BioEdit 6.0 and Multalin software.
2.5. Determination of Pre- and Post-phage Infecting Vibrio spp. V Concentration
A mixture of phages and bacteria, with an MOI of 100, was incubated for 24 h. Concentration of Vibrio spp. V before and after phage infection was determined by the spreading plate method on TCBS [20].
2.6. Statistical Analysis
Experiments were conducted with triplet replicates, the SD value was calculated by Excel software (Microsoft Corp., Redmond, WA, US), and statistics were analyzed by Statgraphics Centurion XV software (Statgraphics Technologies Inc., The Plains, Virginia, US).
3. RESULTS
3.1. Isolated Vibrio spp.
Thirty-two Vibrio spp. were isolated on a TCBS medium from the 35 collected samples with typical characteristics of Vibrio spp. colonies after 24 h of incubation as round, regular, blue-yellow colonies and 2 mm or more in diameter. To determine which strains belonged to Vibrio, a PCR technique targeting Vibrio spp. was used.
Through the use of a PCR with specific primers for Vibrio spp. [Figure 1], 31 bacterial strains all gave positive results. The strain at the 700 bp position shows that these bacterial strains were identified as Vibrio spp.
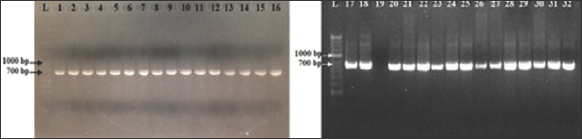 | Figure 1: Polymerase chain reaction amplification of 16S rRNA gene region of 32 isolated strains. [Click here to view] |
The PCR results with primer pairs specific for the V. parahaemolyticus strain [Figure 2] showed that 10 samples were positive. The strains appeared at a position of 368 bp, in which Well 1 was a positive control of V. parahaemolyticus ATCC 17802. It means that nine suspected bacterial strains were identified as V. parahaemolyticus.
 | Figure 2: Polymerase chain reaction amplification of the toxR gene region of 9 isolated strains. L: Ladder 100 bp; 1: Positive control Vibrio parahaemolyticus ATCC 17802; 2: B32; 3: V4.1; 4: B42.2; 5: B42.3; 6: V4.2; 7: V4.3; 8: V4.6; 9: V4.7; 10: V4; 11: Vibrio spp. Computed tomography (negative control). [Click here to view] |
Thus, this result indicated that 31 strains of Vibrio spp. were isolated, including 9 strains of V. parahaemolyticus. The prevalence of Vibrio spp. and V. parahaemolyticus in shrimp cultures was very high. The prevalence of Vibrio spp. and V. parahaemolyticus in samples collected from shrimp culture in the 3 provinces, Bac Lieu, Soc Trang, and Kien Giang, is presented in Table 1.
Table 1: Vibrio spp. isolation from shrimp culture in areas.
| Sampling location | Sample types | Samples | Vibrio spp. | Vibrio parahaemolyticus |
|---|---|---|---|---|
| Kien giang | Bottom mud | 5 | 4 | 3 |
| Pond water | 5 | 2 | 0 | |
| Bac lieu | Bottom mud | 5 | 7 | 0 |
| Diseased shrimp | 5 | 1 | 1 | |
| Soc trang | Pond water | 5 | 0 | 6 |
| Pond siphon water | 5 | 2 | 0 | |
| Diseased shrimp | 5 | 5 | 0 |
3.2. Isolated Vibrio spp. Bacteriophage
Bacteriophages were found in environments inhabited by bacterial hosts [Figure 3]. The abundance of phage populations in aquatic systems was estimated to be 104–108 virions/1 mL [21]. Indeed, 32 phages capable of infecting Vibrio spp. were isolated from 144 shrimp and shrimp pond samples in Soc Trang, Bac Lieu, and Kien Giang provinces. The list and origins of the phages are detailed in Table 2.
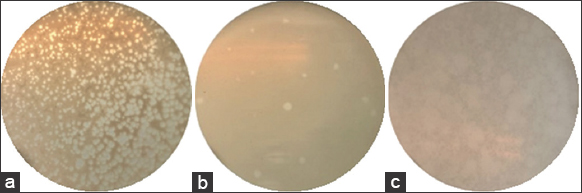 | Figure 3: Plaque morphology of isolated phages on the double-layer agar plates. (a) phage ST2/Vibrio spp. B2.2; (b) phage ST8/Vibrio spp. VP5.1; (c) phage BL4/Vibrio parahaemolyticus B42.3. [Click here to view] |
Table 2: Vibrio spp. phage isolation from shrimp culture in areas.
| Sampling location | Sample types | Samples | Vibrio spp. phages |
|---|---|---|---|
| Shrimp pond water | 16 | 1 | |
| Healthy shrimp | 16 | 14 | |
| Diseased shrimp | 16 | 1 | |
| Shrimp pond water | 16 | 6 | |
| Healthy shrimp | 16 | 0 | |
| Diseased shrimp | 16 | 0 | |
| Shrimp pond water | 16 | 2 | |
| Healthy shrimp | 16 | 8 | |
| Diseased shrimp | 16 | 0 |
When conducting a host range survey of 32 isolated phages on 6 strains of Vibrio spp. [Table 3], there were many characteristic morphological patterns of the plaque, as shown in Figure 4. These morphologies were divided into the four groups below: Clear plaques representing toxic or lytic phages, turbid plaques, plaques resistant to host bacteria, and no plaque (the phage was not able to infect the bacteria selected as the host range) [22].
Table 3: Host range of bacteriophages isolated from Vibrio spp.
| Vibrio spp./Phages | 17802 | VPV4.1 | VPB42.2 | VPB42.3 | VP5.1 | B2.2 |
|---|---|---|---|---|---|---|
| BL1 | ||||||
| BL2 | ||||||
| BL3 | ||||||
| BL4 | ||||||
| BL5 | ||||||
| BL6 | ||||||
| BL7 | ||||||
| BL8 | ||||||
| BL9 | ||||||
| BL10 | ||||||
| BL11 | ||||||
| BL12 | ||||||
| BL13 | ||||||
| BL14 | ||||||
| BL15 | ||||||
| BL16 | ||||||
| KG1 | ||||||
| KG2 | ||||||
| KG3 | ||||||
| KG4 | ||||||
| KG5 | ||||||
| KG6 | ||||||
| ST1 | ||||||
| ST2 | ¤ | ¤ | ¤ | |||
| ST3 | ¤ | ¤ | ¤ | ¤ | ¤ | |
| ST4 | ¤ | |||||
| ST5 | ¤ | ¤ | ||||
| ST6 | ¤ | |||||
| ST7 | ¤ | |||||
| ST8 | ¤ | ¤ | ||||
| ST9 | ||||||
| ST10 |
Host range of 33 phage strains isolated on 7 strains of Vibrio spp. Plaque morphology was classified into 3 categories: Clear (orange with symbol), turbid (orange), bacterial phage resistance (grey), and no reaction (no color).
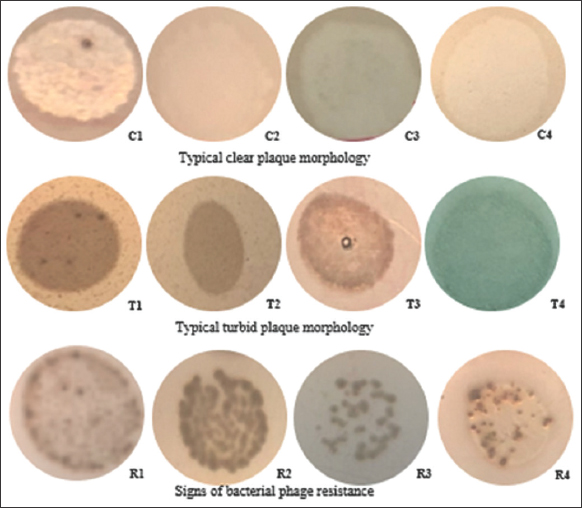 | Figure 4: Diversity of phage plaque morphology. The results were accessed by double agar drop method. [Click here to view] |
3.3. Bacterial toxR gene Sequence with and Without Phage Infection
The results indicated that the toxR nucleotide sequence of the bacterium infected by the phage KG6 remained unchanged as compared to that of the control, which was not infected by the bacteriophage [Figure 5]. However, in the case of infection by phages ST9 and ST10, the gene nucleotide sequence differed by 4 positions from that of the original bacterium. Phage ST9 caused the replacement of C with T, G with A, A with G, and T with A at 22, 93, 139, and 179 positions in the DNA sequence, respectively. Similarly, the replacement of C with T, G with A, A with G, and T with A was caused by phage ST10 at the specific positions of 25, 93, 142, and 181, respectively [Figure 6]. The Q values ranged from 26 to 62.
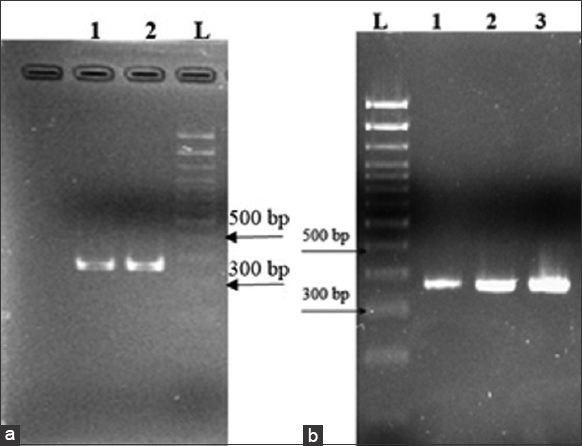 | Figure 5: ToxR primer pairs for Vibrio spp. V in polymerase chain reaction electrophoresis. (a) (1) Vibrio spp. V (no phage infection), (2) Vibrio spp. V infected by KG6 phage; (b) (1) Vibrio spp. V (no phage infection), (2) Vibrio spp. V infected by ST9 phage, (3) Vibrio spp. V infected by ST10 phage. [Click here to view] |
 | Figure 6: Results of alignment of the Vibrio spp. V toxR gene sequence with and without phage infection (D1_ toxR: toxR sequence of Vibrio spp. V (control); D3_ toxR: sequence of Vibrio spp. V infected by phage KG6, U2: Vibrio spp. V infected by phage ST10, and U1: Vibrio spp. V infected by phage ST9. [Click here to view] |
Then, the influence of phages on the bacterial concentration was carried out. Figure 7 showed that the concentration of phage ST9 and phage ST10 infected Vibrio spp. V and that of the control was not a statistically significant difference (P > 0.05). However, after being infected by phage KG6, Vibrio spp. V concentration significantly decreased as compared to that of the initial bacterium (control) (P < 0.05).
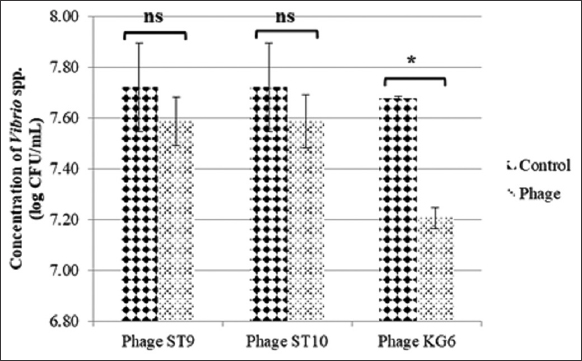 | Figure 7: Concentration of Vibrio spp. V with (Phage) and without (Control) phage infection. [Click here to view] |
4. DISCUSSION
The situation of antibiotic resistance of bacteria in the current period is becoming more common and dangerous [23]. Faced with this situation, bacteriophage therapy has been focused on again. In order to use bacteriophage in therapy, there are two priority groups: Virulent phages (strictly lytic phages) have only a lytic cycle, in which the bacteriophages use the host cell to create new particles and lyse the host cell to release new phages. The second group is temperate phages (capable of integration into the bacterial genome during the lysogenic cycle) and therefore plays a more important role in the evolution of bacteria and bacterial pathogenicity [9].
Previous studies have demonstrated temperate phages’ ability to increase host bacterial virulence [9]. In addition, many studies have identified the ability to reduce the bacterial virulence of temperate phages, specifically P2, which is a prophage that can attach to the plasmid of Klebsiella pneumoniae. When infecting mice with K. pneumoniae, the mutant strain without P2 caused a mortality rate of 100%, while the bacterial strain carrying P2 had a mortality rate of only 70%, suggesting that the presence of P2 reduces the virulence of bacteria [24]. Similarly, for temperate phage PHB09, when the prophage integrated into the pilin gene of Bordetella bronchiseptica, it significantly reduced the virulence of the strain Bb01 in mice, most likely due to disruption of pilin gene expression [25]. Furthermore, prophages can also change the gene sequence of bacteria through integration and excision. For instance, integration of prophage A118 leads to the inactivation of comK, an important gene of Listeria monocytogenes for the infection of animal cells, thereby inhibiting the infection process [26]. Temperate phages are abundant in nature; nearly half of sequenced bacteria are lysogen [27], so temperate phages are a potential group in phage therapy research, although the effect of pro-phages on host bacteria still has many issues that need to be studied.
The bacterial toxR gene was sequenced with and without phage infection, and phage inhibition was measured via bacterial concentration (The toxR gene was discovered as the gene regulating the cholera toxin operon and many other genes in Vibrio cholerae [28]. For V. parahaemolyticus and Vibrio alginolyticus, toxR is required in biofilm formation [29,30]). The results showed that the phage ST9 and phage ST10 phages did not directly reduce the bacterial concentration, but they could change the toxR DNA sequence and virulence regulator gene in Vibrio spp. Preliminary results show that when infecting bacteria, some phages could change the nucleotide sequence of the toxR, the essential Vibrio spp. gene that regulates toxins and affects biofilm formation. In contrast, phage KG6 could reduce the bacterial concentration but ultimately did not affect the toxR gene.
These results are consistent with previous bacteriophage findings that the virulent phages, phage KG6, inject their genome into the cell after being absorbed by the bacterial cell. Bacterial DNA is fragmented when phage nucleic acid replication and phage proteins are synthesized in the cell. The progeny virions are assembled and released from the cells at the end of a life cycle, leading to host death. Phage ST9 and phage ST10, in the lysogenic cycle, phage do not kill the host cell, but the viral genome attaches to the host cell genome, coexists for many generations, and could affect the host bacterial genome sequence [10]. It means that the phages could affect bacteria in several different pathways, such as directly killing bacteria, leading to a concentration decrease, and indirectly affecting the host bacteria by attachment to the gene sequence.
5. CONCLUSION
The prevalence and antibiotic resistance of Vibrio spp. are increasing, leading to the urgency of finding an alternative antibiotic therapy. One of them is the bacteriophage application method. In this study, the diversity between the isolated phages in terms of plaque characteristics, gene sequence-affected abilities, and the concentrations of host bacteria was found. The phage KG6 could reduce bacterial concentration; the others, phage ST9 and phage ST10 strains, did not significantly reduce bacterial concentration. However, they can attack and change Vibrio bacteria’s virulence regulation gene sequence, toxR. Study results show that the first step to define a new direction in bacteriophage research is to identify phage trains capable of affecting the coding sequences of the virulent genes of bacteria and the potential application of phages for several purposes in aquaculture, mainly controlling Vibrio spp.
6. ACKNOWLEDGMENT
The authors would like to thank the Department of Research Affairs, Institute of Food and Biotechnology, Can Tho University, Viet Nam for their valuable support.
7. AUTHOR CONTRIBUTION
All authors contributed to conceptualization and design, data collection or data analysis and interpretation; pertaining to writing or reviewing important intellectual content.
8. FUNDING
This study is funded by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number: 106.05-2019.335.
9. CONFLICTS OF INTEREST
The authors declared no financial or other conflicts of interest in this study.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
13. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Ina-Salwany MY, Al-saari N, Mohamad A, Mursidi FA, Mohd-Aris A, Amal MN, et al. Vibriosis in fish:A review on disease development and prevention. J Aquat Anim Health 2019;31:3-22. [CrossRef]
2. Midgett CR, Swindell RA, Pellegrini M, Jon Kull F. A disulfide constrains the ToxR periplasmic domain structure, altering its interactions with ToxS and bile-salts. Sci Rep 2020;10:9002. [CrossRef] [CrossRef]
3. Choudhury TG, Nagaraju VT, Gita S, Paria A, Parhi J. Advances in bacteriophage research for bacterial disease control in aquaculture. Rev Fish Sci Aquac 2017;25:113-25. [CrossRef]
4. Parasion S, Kwiatek M, Gryko R, Mizak L, Malm A. Bacteriophages as an alternative strategy for fighting biofilm development. Pol J Microbiol 2014;63:137-45. [CrossRef]
5. Schmelcher M, Loessner MJ. Application of bacteriophages for detection of foodborne pathogens. Bateriophage 2014;4:e28137.
6. Letchumana V, Chan KG, Pusparajah P, Saokaew S, Duangjai A, Goh BH, et al. Insights into bacteriophage application in controlling Vibrio species. Front Microbiol 2016;7:1114.
7. Kim SG, Jun JW, Giri SS, Yun S, Kim HJ, Kim SS, et al. Isolation and characterisation of pVa-21, a giant bacteriophage with anti-biofilm potential against Vibrio alginolyticus. Sci Rep 2019;9:6284. [CrossRef]
8. Matamp N, Bhat SG. Genome characterization of novel lytic Myoviridae bacteriophage ?VP-1 enhances its applicability against MDR-biofilm-forming Vibrio parahaemolyticus. Arch Virol 2020;165:387-96. [CrossRef]
9. Gummalla VS, Zhang Y, Liao YT, Wu VC. The role of temperate phages in bacterial pathogenicity. Microorganisms 2023;11:541. [CrossRef]
10. Cieslik M, Baginska N, Jonczyk-Matysiak E, Wegrzyn A, Wegrzyn G, Górski A. Temperate Bacteriophages-the powerful indirect modulators of eukaryotic cells and immune functions. Viruses 2021;13:1013. [CrossRef]
11. Zhou S, Liu Z, Song J, Chen Y. Disarm the bacteria:What temperate phages can do. Curr Issues Mol Biol2023;45:1149-67.
12. Jun JW, Han JE, Giri SS, Tang KF, Zhou X, Aranguren LF, et al. Phage application for the protection from acute hepatopancreatic necrosis disease (AHPND) in Penaeus vannamei. Indian J Microbiol 2017;43:114-7.
13. Wong HC, Wang TY, Yang CW, Tang CT, Ying CW, Wang CH, et al. Characterization of a lytic vibriophage VP06 of Vibrio parahaemolyticus. Res Microbiol 2019;170:13-23. [CrossRef]
14. Yang M, Liang Y, Huang S, Zhang J, Wang J, Chen H, et al. Isolation and Characterization of the novel phages vB_VpS_BA3 and vB_VpS_CA8 for lysing Vibrio parahaemolyticus. Front Microbiol 2020;11:259. [CrossRef]
15. Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol 1999;37:1173-7. [CrossRef]
16. Abdelaziz M, Ibrahem MD, Ibrahim MA, Abu-Elala NM, Abdel-Moneam DA. Monitoring of different Vibrio species affecting marine fishes in Lake Qarun and Gulf of Suez:Phenotypic and molecular characterization. Egypt J Aquat Res 2017;43:141-6. [CrossRef]
17. Montieri S, Suffredini E, Ciccozzi M, Croci L. Phylogenetic and evolutionary analysis of Vibrio parahaemolyticus and Vibrio alginolyticus isolates based on toxR gene sequence. New Microbiol 2010;33:359-72.
18. Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol 2009;501:69-76. [CrossRef]
19. Zhang Y, Hu L, Osei-Adjei G, Zhang Y, Yang W, Yin Z, et al. Autoregulation of toxr and its regulatory actions on major virulence gene loci in Vibrio parahaemolyticus. Front Cell Infect Microbiol 2018;8:291. [CrossRef]
20. Sander ER. Aseptic laboratory techniques:Plating methods. J Vis Exp 2012;63:e3064.
21. Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. Bacteriophages and phage-derived proteins--application approaches. Curr Med Chem 2015;22:1757-73. [CrossRef]
22. Jurczak-Kurek A, Gasior T, Nejman-Falenczyk B, Bloch S, Dydecka A, Topka G, et al. Biodiversity of bacteriophages:Morphological and biological properties of a large group of phages isolated from urban sewage. Sci Rep 2016;6:34338. [CrossRef]
23. Zaman SB, Hussain MA, Nye R, Mehta V, Mamun KT, Hossain N. A review on antibiotic resistance:Alarm bells are ringing. Cureus 2017;9:e1403.
24. Cai M, Pu B, Wang Y, Lv L, Jiang C, Fu X, et al. A plasmid with conserved phage genes helps Klebsiella pneumoniae defend against the invasion of transferable DNA elements at the cost of reduced virulence. Front Microbiol 2022;13:827545. [CrossRef]
25. Chen Y, Yang L, Yang D, Song J, Wang C, Sun E, et al. Specific integration of temperate phage decreases the pathogenicity of host bacteria. Front Cell Infect Microbiol 2020;10:14. [CrossRef]
26. Zhang M, Zhang T, Yu M, Chen YL, Jin M. The life cycle transitions of temperate phages:Regulating factors and potential ecological implications. Viruses 2022;14:1904. [CrossRef]
27. Monteiro R, Pires DP, Costa AR, Azeredo J. Phage therapy:Going temperate?Trends Microbiol 2019;27:368-78.
28. Dua P, Karmakar A, Ghosh C. Virulence gene profiles, biofilm formation, and antimicrobial resistance of Vibrio cholerae non-O1/non-O139 bacteria isolated from West Bengal, India. Heliyon 2018;4:e01040.
29. Chen L, Qiu Y, Tang H, Hu LF, Yang WH, Zhu XJ, et al. ToxR Is required for biofilm formation and motility of Vibrio parahaemolyticus. Biomed Environ Sci 2018;31:848-50.
30. Chen C, Wang QB, Liu ZH, Zhao JJ, Jiang X, Sun HY, et al. Characterization of role of the toxR gene in the physiology and pathogenicity of Vibrio alginolyticus. Antonie Van Leeuwenhoek 2012;101:281-8. [CrossRef]