1. INTRODUCTION
Increased population growth rates in poor and developing markets negatively affect food access and availability for poor households increasing the need to provide overall food and diet security. It is an extremely high priority in the developing regions of the world, where population growth is coupled with the increased intensity of climate change [1]. However, deliberate initiatives include orphan crops, which are plant varieties whose output and use are restricted to a few locations or niche markets. Agricultural importance in industrialized economies receives minimal attention in terms of research and development [2,3]. Neglected crops provide food and income for farmers in the poorest nations worldwide because they are tailored to local conditions and serve as key mainstays in local feeds [4].
Orphan crops were particularly crucial for promoting economical, sustainable, and diversified agricultural systems they consume little water than other agricultural crops and may benefit soils via nitrogen fixation and organic matter insertion. Orphan crops are defined as crops that have either originated in a geographic location or those that have become “indigenized” over many years (> 10 decades) of cultivation as well as natural and farmer selection. Furthermore, most orphan crops are robust and can withstand adverse environmental circumstances including drought, cold, salinity, insects, and disease [5]. Orphan crop species can benefit from speed breeding techniques by increasing genetic diversity in breeding populations and shortening the time it takes to reach breeding goals by synchronizing the flowering of farmed and wild populations of the species [6–8]. Orphan crops are only now beginning to profit from new breeding techniques, and also the advantages are projected to outweigh those of most traditional crops [9,10].
Despite the development, a fundamental bottleneck remains the inability to generate the necessary crossings for hybridization, and period required for successive selfing gives rise to homozygous lineages. The current germplasm pool has the issue of nonsynchronous flowering, which is a bottleneck despite the fact that most orphan crops are still in the early phases of cultivation and have prolonged immature phases (interfering with hybridization and the transmission of desirable features). Some individuals are also vegetatively propagated. Speed breeding techniques will aid in overcoming this barrier and allowing contemporary breeding methods to be used. For instance, in a speed breeding program, the quick generation of recombinant inbred lines might permit rapid genomics-aided breeding [6,11]. Several reports have recorded and examined orphan crops, including their origins, production locations, specific characteristics, traditional and modern applications, and breeding development [12,13].
2. TYPES OF ORPHAN CROPS
Orphan crops are also known as underutilized crops [14], lost crops (NRC 1996, 2006, 2008), and neglected crops or crops for the future. According to Crops for the Future (CFF 2019), the diverse names given to these crops reflect the following characteristics: “neglected” (by science and development), “orphan” (without champions or crop experts), “minor” (relative to global crops), “promising” (for emerging markets, or because of previously unrecognized value traits), “niche” (of marginal importance in production systems and economies), and “traditional” (used for centuries or even millennia). Orphan crops belong to the major groups of crops, which include cereals, legumes, and fruit as well as root crops (Table 1).
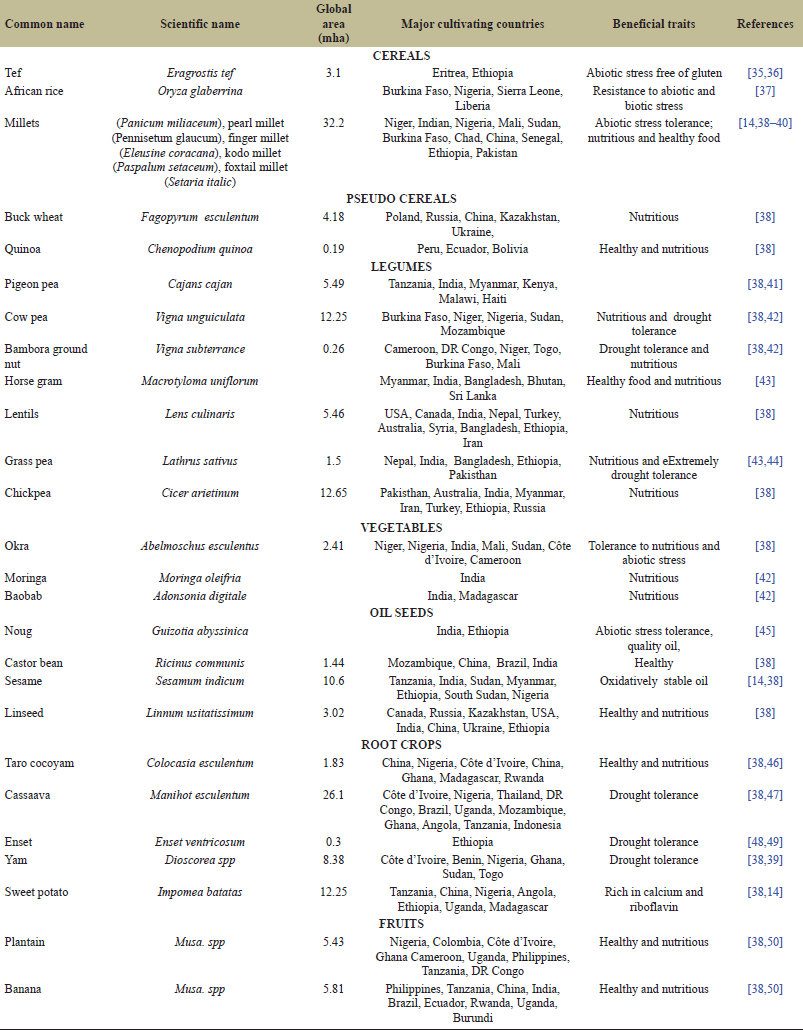 | Table 1. Orphan food crops and their geographical significance. [Click here to view] |
Cereals are excellent suppliers of nutrition for animals and humans and are a great source of iron, potassium, magnesium, zinc, calcium, and other nutrients [15]. Millets represent different types of millets which include Barnyard millet (Echinochloa crusgalli), finger millet (Eleusine coracana), foxtail millet (Setaria italica), kodo millet (Paspalum scrobiculatum), little millet (Panicum sumatrense), pearl millet (Pennisetum glaucum), and prosomillet (Panicum miliaceum) as well as tef (Eragrostis tef) and fonio (Digitaria sp.). Pearl millet, one of the main varieties of millets, is primarily grown as a food crop because of its extraordinary resistance to moisture shortage. Due to its low glycemic index and gradual release of glucose into the bloodstream resulting from its slow digestion and high fiber content, finger millet is also a favorite food among diabetics [16]. Tef is regarded as a lifestyle crop and has gained popularity recently because it does not contain gluten, which is the cause of celiac disease.
Pseudocereals are a class of crops that, in contrast to grasses, have two cotyledons instead of one, making them not members of the grass family. [17]. However, the nutritional makeup of pseudocereals, particularly in terms of the composition of carbohydrates, demonstrates their strong resemblance to “true cereals”. Amaranths (Amaranthus spp.), buckwheat (Fagopyrum esculentum), and quinoa (Chenopodium quinoa) are the major representatives of this category [18]. Pseudocereals, besides being gluten-free, have several health advantages such as lowering oxidative stress, preventing cardiovascular illnesses, preventing cancer, reducing diabetes, reducing inflammation, and reducing hypertension [19].
Legume crops benefit soil because they fix atmospheric nitrogen and transform it into ammonium [20]. While Bambara groundnut (Vigna subterranean) and cowpea (Vigna unguiculata) are extensively cultivated in Africa, horse gram (Macrotyloma uniflorum) is mainly cultivated in Asia. Bambara groundnut seeds are regarded as a complete food since they have sufficient amounts of fat (6.5%), carbohydrates (63%), and protein (19%). In addition to being heat- and drought-tolerant, the crop outperforms many other crops in subpar soil [21]. This characteristic makes the crop regarded as an insurance crop since it yields consistently even in the event that all other crops fail owing to severe moisture scarcity.
In the developing globe, there are a lot of native or significant local veggies. Many of them have advantageous agronomic and/or nutritional characteristics such as baobab (Adansonia digitate), a multipurpose tree with iron-rich leaves and vitamin C-rich fruits, and okra (Abelmoschus esculentus), which grows quickly and is nutritious [15]. Sesame (Sesamum indicum L), one of the oilseeds, is grown on more than 10 million hectares of land each year, mostly in Tanzania, India, Sudan, and Myanmar [22]. Other oilseeds that are somewhat significant in developing nations include noug (Guizotia abyssinica), castor bean (Ricinus communis), and linseed (Linum usitatissimum).
A significant portion of the population in developing countries is fed by root crops such as yam (Dioscorea sp.), sweet potato (Ipomoea batatas), and cassava (Manihot esculenta) [23,24]. Cassava is tolerant to drought and performs better than other crops on soils with poor nutrients [25]. Whereas sweet potatoes are grown all throughout the world, yams are only grown in Africa. It is worthwhile to mention the plant called enset (Ensete ventricosum), which is also known as “false banana” because it resembles the domesticated banana plant [26]. Ensete is a staple food for over 20 million people in the densely populated regions of Ethiopia. In contrast to bananas, where the fruit is eaten, in enset the underground corm and pseudo-stem are edible [27]. Ensete is an extremely drought-tolerant crop that adapts to different soil types.
In underdeveloped countries, banana and plantain (Musa spp.) are significant fruit crops, despite the scarcity of genetic resources and research [28]. It is often considered to be a vegetable rather than a fruit since the plantain is cooked like a vegetable. Bananas, particularly the orange-pulped kind with high carotenoid and iron content, can help lower vitamin A deficiency and iron deficiency anemia [29]. Plantains and bananas are regarded as healthful foods because they are high in vital elements for human consumption.
3. SIGNIFICANCE OF ORPHAN CROPS
3.1. Tolerance to Biotic Stress
It has been possible to successfully introduce resistance genes into important crops by using other orphan crops as donors. Coming to the Solanaceae family, resistance genes to wilt of eggplant (Fusarium oxysporium f. sp. melongenae) [30,31] and has been found in the Solanum aethiopicum (African eggplant). Even some of the orphan crops rootstocks were also used for improvising the Solanaceae family crops such as S. aethiopicum and Solanum torvum are selected rootstocks to enhance disease resistance in (brinjal) Solanum melongena [32,33] and (tomato) Solanum lycopersicum [34]. Fusarium spp. resistance genes are found when grafting of Cucumis melo (watermelon) onto (bottle guard) Lagenaria siceraria [51,52].
3.2. Tolerance to Abiotic Stress
At present the major problem is drought and heat waves. Some of the examples are briefed below. Tef is a waterlogging-tolerant crop [35]. Noug is an example of an abiotic stress-tolerant crop [45]. Enset and Yam were examples of drought resistance. Pearl millet is widely grown and it is resistant to moisture deficit [53]. Lathyrus sativus (grasspea) is highly resistance to drought [6]. Vigna unguiculata is a drought and heat-tolerance crop [54].
3.3. Alternate Names of Orphan Crops
Orphan crops are also called as “neglected crops” [55] or “underutilized crops” [14] or “crops for the future” (Fig. 1). The following traits are reflected in the given name to these crops: by science and development (“neglected”), without leaders or agricultural specialists (“orphan”). Compared to world crops (“minor”), due to developing markets or previously unnoticed value characteristics (“promising”), of little consequence to economies and manufacturing systems (“niche”), used for hundreds, perhaps even generations (“traditional”) [15].
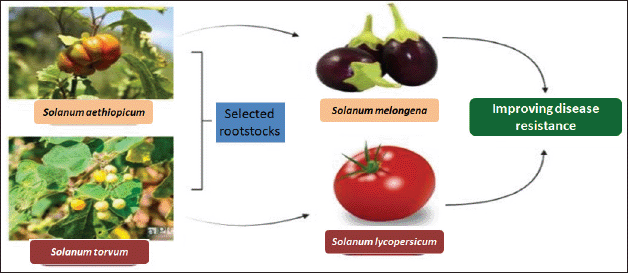 | Figure 1. Improving the existing crops using neglected crops. [Click here to view] |
4. METHODS FOR IMPROVING ORPHAN CROP RESISTANCE TO ABIOTIC STRESS
At present, there are so many technologies for analyzing genomes that are accessible to the general public. These tools make it possible to transfer and use genes (resources) to orphan crops (minor crops) for commercial production from major crops [52]. Recent research has shown that for many African crops, the actual yield that farmers achieve falls significantly short of the potential output [56], implying that increased crop yields are possible with the help of improved varieties and smarter agricultural practices. Recent studies have highlighted the tremendous potential of modern breeding and gene sequencing approaches in the creation of crops that are resistant or tolerant to abiotic stresses [57,58].
5. MODERN BREEDING METHODS AND GENOMIC APPROACHES
5.1. Molecular Breeding Steps
In the initial stage of molecular breeding, molecular markers are used to understand the genetic variation present in crop types and the possible benefits supplied by their wild forms (Fig. 2). The 2nd way is through the exchange of genetic material between different genotypes and “Genetic engineering” is commonly used to describe this method. This allows for the introduction of really unique features into a crop. When genetic engineering is used to improve the effectiveness with which native genes are throughout a gene pool [12].
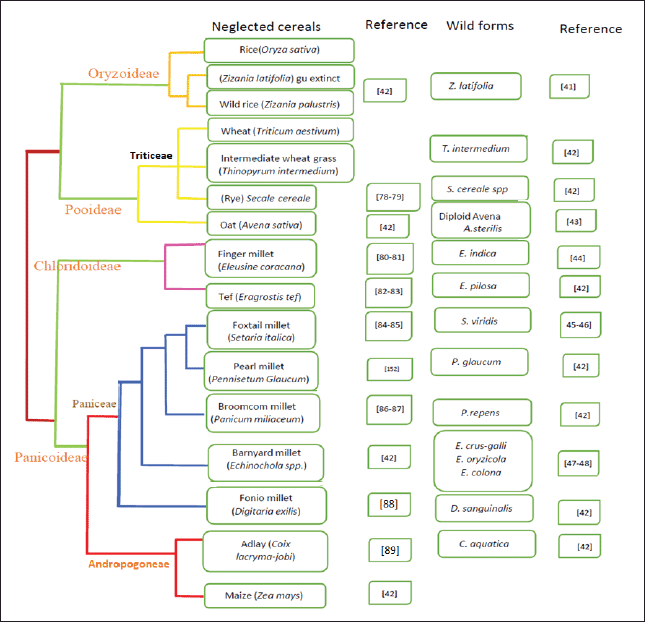 | Figure 2. Neglected crops and respective wild forms. [Click here to view] |
5.2. Marker-Assisted Selection
Marker assisted selection (MAS) refers to the process of identifying DNA sequences that are situated in close proximity to genes, molecular markers can be utilized to selectively breed for characteristics that are challenging to observe [13]. Single nucleotide polymorphisms (SNPs) and microsatellites (or) simple sequence repeats are two types of markers frequently used in plant breeding. Genome-wide association studies (GWASs) [59] and genotyping-by-sequencing (GBS) [60] are other SNP-based techniques that have developed recently. The feasibility of GBS has recently been explored in a variety of crops with varying sizes of genomes and breeding techniques [61]. Wheat varieties with high resistance to aluminum were successfully identified using GWASs for the identification of abiotic stress tolerance [62]. DArT diversity arrays technology [63], hybridization-based molecular marker creation in orphan crops has proven to be an effective strategy because it does not necessitate the availability of sequence information. DArT-sequencing (DArT-seq) is a relatively new technique that combines DArT with next generation sequencing (NGS) [64]. It allows for high throughput genotyping and speeds up the process of discovering SNPs in many neglected crops. Many orphan crops that are able to withstand extreme weather conditions are now being characterized using DArT-seq. This includes the Eleusine coracana (Finger millet) [65], Kerstingiella geocarpa (Kersting’s groundnut) [66], L. sativus (grass pea) [67], and Vigna subterranean (Bambara groundnut) [68].
5.3. Marker-Assisted Genetic Mapping
The concept of “quantitative traits” has proven challenging to comprehend and control in traditional crop breeding programmes. Quantitative trait locus (QTL) defines the chromosomal areas of genes that regulate quantitative characteristics [12]. Goff et al. [69] predicted that roughly 2,000 cereal QTLs had been mapped. For MAS in crop development, undisclosed markers associated with QTL have been utilized in some cases, the significant degree of uncertainty in mapping preferred sites of minor effects might restrict the value of such markers. “Candidate genes” allow for the considerably more exact localization of ideal genetic mutations [12]. Multiple studies have reported the identification of plausible candidate genes that co-locate with QTLs for various characteristics, encompassing quantitative disease resistance in Triticum aestivum (Wheat) [70], Solanum tuberosum (potato) [71], Phaseolus (bean) [72], and Oriza sativa (rice) [73].
5.3.1. Hybridization
It is a process of mating or crossing two plants or lines of closely related plant species together to increase genetic diversity for improve desirable characteristics. NERICA is a popular new variety of Rice in Africa, which developed through interspecific crossing between Oryza glaberrima Steudel (African rice) and O. sativa L. (Asian rice). NERICA inherits the best qualities of its parents such as low soil fertility, early maturing and drought-tolerant from O. glaberrima, and high protein content and seed yield from O. sativa [37]. Because of the early maturing character of O. glaberrima, it is able to survive the final drought that often strikes after the flowering stage. Several nations in Africa are now cultivating NERICA [73].
5.3.2. Genome editing
Genome editing is used to modify an organism genotypically and phenotypically [74] and uses mutations (both induced and natural) to enhance crops. Site-specific nucleases (SSNs) direct gene editing, are enables accurate mutagenesis of a target genome without permanently introducing DNA to the target organism. Various types of SSNs, such as transcription activator-like effector nucleases, Clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins (CRISPR/Cas), zinc finger nucleases and meganucleases, have the ability to cause targeted double-strand breaks in DNA [75–77]. There are several genome editing tools are there, and out of them, the most common tool is CRISPR/Cas 9 [78]. Genome editing techniques, such as CRISPR-Cas9, hold promise for speeding up the domestication process of orphan plants by selectively modifying their undesirable characteristics [79,80] in both polyploids [80,81], and diploids [82]. Genome editing with CRISPR/Cas9 was effectively performed by a researcher in Physalis pruinose (groundcherry), a wild relative of Lycopersicon esculentum (tomato) [52]. Musa acuminata (Banana) is an understudied polyploid that serves as a model for other neglected/orphan crops because of its complicated genome and successful application of CRISPR-Cas9-based genome editing [83].
5.3.2.1. Omics Tools (Proteomics, Transcriptomics, and Genomics)
Thirty orphan crops, spanning thirteen families, have been subjected to genome sequencing in the last 5 years [52]. A minority of the genomes (8 out of 30) that were sequenced were found to be polyploids, suggesting a potential bias towards simpler genomes. This observation may be influenced by the predominant use of 2nd-generation sequencing platforms. In spite of the fact that a few of these draft-genomes will be suitable for application for molecular breeding, to enhance complicated genomes in the same way as tef was a, 3rd -generation sequencing technology will be required [84]. In order to answer specific biological issues, other orphan crop transcriptomes have been created, and RNA sequencing has emerged as the method of interest [85]. As an illustration, [86] identified 2,416 genes that were altered when profiling the response to salt stress in C. quinoa (quinoa). A transcriptional study was performed on Corchorus olitorius (Jute-mallow) to locate genes involved with drought stress response [87]. Prior to NGS, microarrays were the preferred method for analyzing transcriptomes, even being used in some orphan crops like Solanum nigrum [88], Fagopyrum esculentum [89], Sinapis arvensis [90], Eragrostis tef (Tef) [91], and Lupinus albus [92] to identify expression profiles associated with resistance to abiotic stress. Proteomic methods have also been used to investigate the mechanisms behind rice’s drought tolerance [93] and in cereals (like drought and salinity tolerance) [94].
5.3.3. Transgenic
The second method of “molecular breeding” for plant characteristics, direct gene transfer, makes use of recombinant DNA technology to introduce (one (or) several) genes into the plant genome. The potential for extending the uses of this technique to neglected (orphan) crop improvement may be substantial. Despite the fact, that most transgenic research and implementations to date have centered on some major crops [95]. Public research centers in atleast 10 poor nations are now conducting field tests of propitious transgenic lines for about 20 various crops including Piper nigrum (Pepper), Ipomoea batatas (Sweet potato), and (Cucurbita) squash [96]. These lines are genetically modified and exhibit desirable characteristics, such as resistance to viruses and pests. Instead of using promoters from bacteria or other organisms, as is done in transgenesis, plant-specific promoters are employed to drive the gene of interest in cisgenesis [97].
5.3.4. High-throughput methods
Eco targeting induced local lesions in genomes (TILLING) from wild species and TILLING from induced mutagenized populations are two high-throughput approaches that have been used to find alleles in orphan crops [15]. Eco TILLING and TILLING have both been used for the betterment of native crops like Eragrostis tef (Tef) [98,99].
5.3.5. Speed breeding
When applied to orphan or neglected crop species, speed breeding methods may be used to increase the extent of variation in breeding individuals and hasten the accomplishment of breeding objectives by synchronizing the blooming of wild relatives and cultivated species. Optimization of the plant growth environment including temperature, plant density, and photoperiod, genetic engineering to the target blooming pathway, using plant growth regulators, grafting young plants onto mature rootstocks, and harvesting premature seed are all viable methods for rapid cycling [6,100,101]. Speed breeding protocols have been improved for legumes (Cicer arietinum) (Chickpea) [102], Arachis hypogaea (Groundnut) [6] and cereals O. sativa (Rice) [103], Triticum aestivum (Wheat) [104].
5.3.6. Speed breeding centres among various countries
Potential collaborators for speed breeding centers that are well-positioned may include: Taiwan (The World Vegetable Center), Ghana (The West Africa Center for Crop Improvement), Malaysia (Crops for the Future), UAE (The Global Pulse Confederation), worldwide (CGIAR Center and Research Programs), Kenya (The African Orphan Crops Consortium). Researchers at IITA (International Institute of Tropical Agriculture), Ibadan, Nigeria: ICRISAT (International Crops Research Institute for Semi-Arid Tropics), Patancheru, Telangana; CGIAR centers and International Center for Agricultural Research in the Dry Areas, Beirut, Lebanon, have voiced an interest in building speed breeding centers to quicken the breeding process for the crops they are required to grow [101].
6. CHALLENGES
One of the most challenging aspects of adopting speed breeding is ensuring optimal growing conditions, including protection from pests and diseases. In addition to plant population assessments, drones can also be used for various agricultural applications, such as crop monitoring, irrigation management, and soil analysis. By collecting data from the above, drones provide a comprehensive view of crop health and growth patterns, allowing farmers to make informed decisions about their crops [87,136]. Trait analysis using drones is now affordable and practical [87,137]. Combining speed breeding with automated phenotypic screening enables fast evaluation of plants in orphan crops, allowing researchers to develop new cultivars with multiple desirable traits. The primary focus would be to provide training to breeders on the utilization of sophisticated breeding techniques, similar to the ongoing efforts of the African Plant Breeding Academy [52].
Traditional procedures, such as line selection from landraces, are used to better orphan crops. Introgressions using interspecific or intraspecific crossings are used in some orphan crop breeding initiatives. Through collaborations between institutions in rich and developing nations, innovative breeding technologies have recently been introduced for a number of orphan crops Table 2. Marker-based research, such as GWAS and GBS, are examples of sophisticated approaches (GWAS). EcoTILLING from wild species and TILLING from induced mutagenized populations are two high-throughput approaches that have been used to find alleles in orphan crops. The major goal of the AOCC is genome sequencing of one hundred and one African edible plants and improve the nutritional status of Africans by means of molecular breeding technologies and education [138]. Moreover, the partnership is committed to ensuring that its work is aligned with the United Nations Sustainable Development Goals, particularly those related to zero hunger, responsible consumption and production, and climate action. Transcriptomics and proteomics, among other omics methods, have been employed to explain gene transcription patterns of several underutilized crops. On Manihot esculenta (cassava) and, more recently, on groundcherry, genome alteration techniques such as CRISPR/Cas9 have been used [113,114]. Orphan crops such as cassava, millet, and teff can have their unwanted traits altered rapidly with the help of clustered regularly interspaced short palindromic technology [139].
 | Table 2. Advancing in orphan crops by modern breeding and genomics methods. [Click here to view] |
7. CONCLUSION
Food security, especially in underdeveloped areas, and genetic diversity depend critically on minor or neglected crops. Despite their relevance, few researchers have paid much attention to these plants. As a result, major agricultural changes are needed to raise agricultural production for understudied crops to meet the developing world’s increasing population density. These collaborations among important stakeholders are required to address the issues, particularly in light of changing climate. Orphan crop varieties can be developed, tested, and released to the market much sooner if speed breeding is used in conjunction with different breeding methods and low-cost automated phenotypic and genotypic. Field testing, as well as farmer participation in the examination and assessment of elite breeding lines, will be critical in hastening the creation and spread of better cultivars.
8. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. FUNDING
There is no funding to report.
11. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
12. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
13. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
14. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, et al. Food security: the challenge of feeding 9 billion people. Science 2010;327:812–8.
2. Varshney RK, Ribaut JM, Buckler ES, Tuberosa R, Rafalski JA, Langridge P. Can genomics boost productivity of orphan crops? Nat Biotechnol 2012;30:1172–6.
3. Sogbohossou E, Achigan-Dako EG, Maundu P, Solberg S, Deguenon E, Mumm RH, et al. A roadmap for breeding orphan leafy vegetable species a case study of Gynandropsis gynandra (Cleomaceae). Hortic Res 2018;5:2; doi: 10.1038/s41438-017-0001-2
4. Ebert AW. Potential of underutilized traditional vegetables and legume crops to contribute to food and nutritional security, income and more sustainable production systems. Sustainability 2014;6:319–35.
5. Massawe F, Mayes S, Cheng A. Crop diversity: an unexploited treasure trove for food security. Trends Plant Sci 2016;21:365–8.
6. O’Connor D, Wright G, Dieters M, George D, Hunter M, Tatnell J, et al. Development and application of speed breeding technologies in a commercial peanut breeding program. Peanut Sci 2013;40:107–14.
7. Van Nocker S, Gardiner SE. Breeding better cultivars, faster: applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic Res 2014;1:14022; doi: 10.1038/hortres.2014.22
8. Ceballos H, Jaramillo J, Salazar S, Pineda LM, Calle F, Setter T. Induction of flowering in cassava through grafting. J Plant Breed Crop Sci 2017;9:19–29.
9. Pazhamala L, Saxena RK, Singh VK, Sameerkumar C, Kumar V, Sinha P, et al. Genomics-assisted breeding for boosting crop improvement in pigeonpea (Cajanus cajan). Front Plant Sci 2015;6:50.
10. Kumar V, Khan AW, Saxena RK, Garg V, Varshney RK. First-generation HapMap in Cajanus spp. reveals untapped variations in parental lines of mapping populations. Plant Biotechnol J 2016;14:1673–81.
11. Li H, Rasheed A, Hickey LT, He Z. Fast-forwarding genetic gain. Trends Plant Sci 2018;23:184–6.
12. Naylor RL, Falcon WP, Goodman RM, Jahn MM, Sengooba T, Tefera H, et al. Biotechnology in the developing world: a case for increased investments in orphan crops. Food Policy 2004;29:15–44.
13. Nelson RJ, Naylor RL, Jahn MM. The role of genomics research in improvement of” orphan” crops. Crop Sci 2004;44:1901.
14. Dawson I, Jaenicke H. Underutilised plant species: the role of biotechnology. Position Paper 2006;1:27.
15. Tadele Z. Orphan crops: their importance and the urgency of improvement. Planta 2019;250:677–94.
16. Tadele Z. African orphan crops under abiotic stresses: challenges and opportunities. Scientifica 2018;2018:1451894.
17. Serna Saldívar SO, Hernández DS. Dietary fiber in cereals, legumes, pseudocereals and other seeds. In: Welti-Chanes J, Serna-Saldívar SO, Campanella O, Tejada-Ortigoza V (eds.). Science and technology of fibers in food systems, Springer International Publishing, Cham, Switzerland, pp 87–122, 2020.
18. Janssen F, Pauly A, Rombouts I, Jansens KJ, Deleu LJ, Delcour JA. Proteins of amaranth (Amaranthus spp.), buckwheat (Fagopyrum spp.), and quinoa (Chenopodium spp.): a food science and technology perspective. Compr Rev Food Sci Food Saf 2017;16:39–58.
19. Mir NA, Riar CS, Singh S. Nutritional constituents of pseudo cereals and their potential use in food systems: a review. Trends Food Sci Technol 2018;75:170–80.
20. Meena RS, Lal R. Legumes and sustainable use of soils. In: Meena R, Das A, Yadav G, Lal R (eds). Legumes for soil health and sustainable management, Springer, Singapore, pp 1–31, 2018; doi: 10.1007/978-981-13-0253-4_1
21. Khatun M, Sarkar S, Era FM, Islam AM, Anwar MP, Fahad S, et al. Drought stress in grain legumes: effects, tolerance mechanisms and management. Agronomy 2021;11:2374.
22. Myint D, Gilani SA, Kawase M, Watanabe KN. Sustainable sesame (Sesamum indicum L.) production through improved technology: an overview of production, challenges, and opportunities in Myanmar. Sustainability 2020;12:3515.
23. Escobar-Puentes AA, Palomo I, Rodríguez L, Fuentes E, Villegas-Ochoa MA, González-Aguilar GA, et al. Sweet potato (Ipomoea batatas L.) phenotypes: from agroindustry to health effects. Foods 2022;11:1058.
24. Ferraro V, Piccirillo C, Tomlins K, Pintado ME. Cassava (Manihot esculenta Crantz) and yam (Dioscorea spp.) crops and their derived foodstuffs: safety, security and nutritional value. Crit Rev Food Sci Nutr 2016;56:2714–27.
25. de Oliveira EJ, Morgante CV, de Tarso Aidar S, de Melo Chaves AR, Antonio RP, Cruz JL, et al. Evaluation of cassava germplasm for drought tolerance under field conditions. Euphytica 2017;213:188.
26. Acero M, Mukasa S, Baguma Y. Morphotypes, distribution and uses of false banana in Uganda. Afr Crop Sci J 2018;26:575–85.
27. Berhanu H, Kiflie Z, Miranda I, Lourenço A, Ferreira J, Feleke S, et al. Characterization of crop residues from false banana/Ensete ventricosum/in Ethiopia in view of a full-resource valorization. PLoS One 2018;13:e0199422.
28. Campos H, Caligari PD, Brown A, Tumuhimbise R, Amah D, Uwimana B, et al. Bananas and plantains (Musa spp.). In: Campos H, Caligari P (eds.). Genetic improvement of tropical crops, Springer, Cham, Switzerland, pp 219–40, 2017
29. Amah D, van Biljon A, Brown A, Perkins-Veazie P, Swennen R, Labuschagne M. Recent advances in banana (Musa spp.) biofortification to alleviate vitamin A deficiency. Crit Rev Food Sci Nutr 2019;59:3498–510.
30. Rizza F, Mennella G, Collonnier C, Sihachakr D, Kashyap V, Rajam M, et al. Androgenic dihaploids from somatic hybrids between Solanum melongena and S. aethiopicum group gilo as a source of resistance to Fusarium oxysporum f. sp. melongenae. Plant Cell Rep 2002;20:1022–32.
31. Toppino L, Valè G, Rotino GL. Inheritance of Fusarium wilt resistance introgressed from Solanum aethiopicum Gilo and Aculeatum groups into cultivated eggplant (S. melongena) and development of associated PCR-based markers. Mol Breed 2008;22:237–0.
32. Sakata Y, Nishio T, Mon’ma S. Resistance of Solanum species to Verticillium wilt and bacterial wilt. In EUCARPIA VIIth meeting on genetics and breeding on capsicum and eggplant, Jun 27–30, Kragujevac, Serbia, 1989
33. Ramesh R, Achari G, Asolkar T, Dsouza M, Singh N. Management of bacterial wilt of brinjal using wild brinjal (Solanum torvum) as root stock. Indian Phytopathol 2016;69:260–5.
34. Nkansah G, Ahwireng A, Amoatey C, Ayarna A. Grafting onto African eggplant enhances growth, yield and fruit quality of tomatoes in tropical forest ecozones. J Appl Hortic 2013;15:16–20.
35. Ketema S. Tef (Eragrostis tef (Zucc.) Trotter): promoting the conservation and use of underutilized and neglected crops. Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute, Rome, Italy, p 12, 1997.
36. Spaenij-Dekking L, Kooy-Winkelaar Y, Koning F. The Ethiopian cereal tef in celiac disease. N Engl J Med 2005;353:1748–49.
37. Linares OF. African rice (Oryza glaberrima): history and future potential. Proc Nat Acad Sci 2002;99:16360–5.
38. Fao F. Food and agriculture organization of the United Nations. Rome, Italy, p 403, 2018.
39. Williams JT. Global research on underutilized crops: an assessment of current activities and proposals for enhanced cooperation. ICUC, Southampton, UK, p 50, 2000.
40. Gupta SM, Arora S, Mirza N, Pande A, Lata C, Puranik S, et al. Finger millet: a “certain” crop for an “uncertain” future and a solution to food insecurity and hidden hunger under stressful environments. Front Plant Sci 2017;8:643.
41. Pal D, Mishra P, Sachan N, Ghosh AK. Biological activities and medicinal properties of Cajanus cajan (L) Mill sp. J Adv Pharm Technol Res 2011;2:207–14.
42. Council NR. Lost crops of Africa: volume I: grains. National Academies Press, Washington, DC, 1996.
43. Singh M, Upadhyaya HD, Bisht IS. Genetic and genomic resources of grain legume improvement. Newnes 2013;1:305.
44. Campbell CG. Grass pea. Lathyrus sativus L. Promoting the conservation and use of underutilized and neglected crops, 18. Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute, Rome, Italy, 1997.
45. Getinet A, Sharma SM. Niger-Guizotia abyssinica (L. f.) Cass. Promoting the conservation and use of under-utilized and neglected crops, 5. International Plant Genetic Resources Institute, Rome, Italy, p 59, 1996.
46. Otekunrin OA, Sawicka B, Adeyonu AG, Otekunrin OA, Racho? L. Cocoyam [Colocasia esculenta (L.) Schott]: exploring the production, health and trade potentials in Sub-Saharan Africa. Sustainability 2021;13:4483.
47. Ceballos H, Iglesias CA, Pérez JC, Dixon AG. Cassava breeding: opportunities and challenges. Plant Mol Biol 2004;56:503–16.
48. Spring A, Diro M, Brandt SA, Tabogie E, Wolde-Michael G, McCabe JT, et al. Tree against hunger: enset-based agricultural systems in Ethiopia. American Association for the Advancement of Science, Washington, DC, 1997.
49. Olango TM, Tesfaye B, Catellani M, Pè ME. Indigenous knowledge, use and on-farm management of enset (Ensete ventricosum (Welw.) Cheesman) diversity in Wolaita, Southern Ethiopia. J Ethnobiol Ethnomed 2014;10:1–18.
50. Heslop-Harrison JS, Schwarzacher T. Domestication, genomics and the future for banana. Ann Bot 2007;100:1073–84.
51. Davis AR, Perkins-Veazie P, Sakata Y, Lopez-Galarza S, Maroto JV, Lee SG, et al. Cucurbit grafting. Crit Rev Plant Sci 2008;27:50–74.
52. Kamenya SN, Mikwa EO, Song B, Odeny DA. Genetics and breeding for climate change in orphan crops. Theor Appl Genet 2021;134:1787–815.
53. Kholova J, Hash C, Kumar PL, Yadav RS, Ko?ová M, Vadez V. Terminal drought-tolerant pearl millet [Pennisetum glaucum (L.) R. Br.] have high leaf ABA and limit transpiration at high vapour pressure deficit. J Exp Bot 2010;61:1431–40.
54. Sanginga N, Lyasse O, Singh B. Phosphorus use efficiency and nitrogen balance of cowpea breeding lines in a low P soil of the derived savanna zone in West Africa. Plant Soil 2000;220:119–28.
55. Bermejo JEH, León J. Neglected crops: 1492 from a different perspective. Plant production and protection series. Food & Agriculture Organization, Rome, Italy, vol. 26, 1994.42.
56. Tadele Z. Role of crop research and development in food security of Africa. Int J Plant Biol Res 2014;2(3):1019.
57. Barabaschi D, Tondelli A, Desiderio F, Volante A, Vaccino P, Valè G, et al. Next generation breeding. Plant Sci 2016;242:3–13.
58. Schaart JG, van de Wiel CC, Lotz LA, Smulders MJ. Opportunities for products of new plant breeding techniques. Trends Plant Sci 2016;21:438–49.
59. Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun 2011;2:467.
60. Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 2011;6:e19379.
61. Kim C, Guo H, Kong W, Chandnani R, Shuang LS, Paterson AH. Application of genotyping by sequencing technology to a variety of crop breeding programs. Plant Sci 2016;242:14–22.
62. Raman H, Stodart B, Ryan PR, Delhaize E, Emebiri L, Raman R, et al. Genome-wide association analyses of common wheat (Triticum aestivum L.) germplasm identifies multiple loci for aluminium resistance. Genome 2010;53:957–66.
63. Jaccoud D, Peng K, Feinstein D, Kilian A. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 2001;29:e25.
64. Sansaloni C, Petroli C, Jaccoud D, Carling J, Detering F, Grattapaglia D, et al. Diversity arrays technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc 2011;5:P54; doi: 10.1186/1753-6561-5-S7-P54
65. Dida MM, Oduori CA, Manthi SJ, Avosa MO, Mikwa EO, Ojulong HF, et al. Novel sources of resistance to blast disease in finger millet. Crop Sci 2021;61:250–62.
66. Kafoutchoni KM, Agoyi EE, Agbahoungba S, Assogbadjo AE, Agbangla C. Genetic diversity and population structure in a regional collection of Kersting’s groundnut (Macrotyloma geocarpum (Harms) Maréchal & Baudet). Genet Resour Crop Evol 2021;68:3285–300.
67. Almeida NF, Gonçalves L, Lourenço M, Julião N, Aznar-Fernández T, Rubiales D, et al. The pursuit of resistance sources to biotic stresses in Lathyrus sativus. In 2nd International Legume Society Conference, Tróia, Portugal, 2016, pp 11–4.
68. Redjeki ES, Ho W, Shah N, Molosiwa O, Ardiarini N, Kuswanto, et al. Understanding the genetic relationships between Indonesian bambara groundnut landraces and investigating their origins. Genome 2020;63:319–27.
69. Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002;296:92–100.
70. Faris J, Li W, Liu D, Chen P, Gill B. Candidate gene analysis of quantitative disease resistance in wheat. Theor Appl Genet 1999;98:219–25.
71. Trognitz F, Manosalva P, Gysin R, Niño-Liu D, Simon R, del Rosario Herrera M, et al. Plant defense genes associated with quantitative resistance to potato late blight in Solanum phureja× dihaploid S. tuberosum hybrids. Mol Plant Microbe Interact 2002;15:587–97.
72. Geffroy V, Sévignac M, De Oliveira JC, Fouilloux G, Skroch P, Thoquet P, et al. Inheritance of partial resistance against Colletotrichum lindemuthianum in Phaseolus vulgaris and co-localization of quantitative trait loci with genes involved in specific resistance. Mol Plant Microbe Interact 2000;13:287–96.
73. Wang Z, Taramino G, Yang D, Liu G, Tingey S, Miao GH, et al. Rice ESTs with disease-resistance gene-or defense-response gene-like sequences mapped to regions containing major resistance genes or QTLs. Mol Genet Genomic 2001;265:302–10.
74. Zhang L, Li X, Ma B, Gao Q, Du H, Han Y, et al. The tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol Plant 2017;10:1224–37.
75. Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Nat Acad Sci 1996;93:1156–60.
76. Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010;186:757–61.
77. Venezia M, Creasey Krainer KM. Current advancements and limitations of gene editing in orphan crops. Front Plant Sci 2021;12:742932.
78. Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346:1258096.
79. Jørgensen K, Bak S, Busk PK, Sørensen C, Olsen CE, Puonti-Kaerlas J, et al. Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol 2005;139:363–74.
80. Østerberg JT, Xiang W, Olsen LI, Edenbrandt AK, Vedel SE, Christiansen A, et al. Accelerating the domestication of new crops: feasibility and approaches. Trends Plant Sci 2017;22:373–84.
81. Zaman QU, Li C, Cheng H, Hu Q. Genome editing opens a new era of genetic improvement in polyploid crops. Crop J 2019;7:141–50.
82. Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, et al. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat Plants 2018;4:766–70.
83. Kaur N, Alok A, Shivani N, Kaur N, Pandey P, Awasthi P, et al. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct Integr Genomics 2018;18:89–99.
84. VanBuren R, Man Wai C, Wang X, Pardo J, Yocca AE, Wang H, et al. Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat Commun 2020;11:884.
85. Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 2011;12:87–98.
86. Ranasinghe R, Maduwanthi S, Marapana R. Nutritional and health benefits of jackfruit (Artocarpus heterophyllus Lam.): a review. Int J Food Sci 2019;2019:4327183.
87. Yang Z, Dai Z, Lu R, Wu B, Tang Q, Xu Y, et al. Transcriptome analysis of two species of jute in response to polyethylene glycol (PEG)-induced drought stress. Sci Rep 2017;7:16565.
88. Schmidt S, Baldwin IT. Down-regulation of systemin after herbivory is associated with increased root allocation and competitive ability in Solanum nigrum. Oecologia 2009;159:473–82.
89. Golisz A, Sugano M, Fujii Y. Microarray expression profiling of Arabidopsis thaliana L. in response to allelochemicals identified in buckwheat. J Exp Bot 2008;59:3099–109.
90. Srivastava S, Srivastava AK, Sablok G, Deshpande TU, Suprasanna P. Transcriptomics profiling of Indian mustard (Brassica juncea) under arsenate stress identifies key candidate genes and regulatory pathways. Front Plant Sci 2015;6:157215.
91. Degu HD. Analysis of differentially expressed genes induced by drought stress in tef (Eragrostistef) root. Nigerian J Biotechnol 2019;36:167–87.
92. Zhu YY, Zeng HQ, Dong CX, Yin XM, Shen QR, Yang ZM. microRNA expression profiles associated with phosphorus deficiency in white lupin (Lupinus albus L.). Plant Sci 2010;178:23–9.
93. Ali GM, Komatsu S. Proteomic analysis of rice leaf sheath during drought stress. J Proteome Res 2006;5:396–403.
94. Ngara R, Ndimba BK. Understanding the complex nature of salinity and drought-stress response in cereals using proteomics technologies. Proteomics 2014;14:611–21.
95. James C. Preview: global status of commercialized transgenic crops: 2003. ISAAA Briefs 2003.
96. Toenniessen GH, O’Toole JC, DeVries J. Advances in plant biotechnology and its adoption in developing countries. Curr Opin Plant Biol 2003;6:191–8.
97. Jacobsen E, Schouten HJ. Cisgenesis strongly improves introgression breeding and induced translocation breeding of plants. Trends Biotechnol 2007;25:219–23.
98. Esfeld K, Uauy C, Tadele Z. Application of TILLING for orphan crop improvement. In: Jain S, Dutta Gupta S (eds.). Biotechnology of neglected and underutilized crops, Springer, Dordrecht, The Netherlands, pp 83–113, 2013; doi: 10.1007/978-94-007-5500-0_6
99. Tadele Z, Mba C, Till BJ. TILLING for mutations in model plants and crops. In: Jain S, Brar D (eds.). Molecular techniques in crop improvement, Springer, Dordrecht, The Netherlands, pp 307–32, 2009; doi: 10.1007/978-90-481-2967-6_13
100. Lulsdorf MM, Banniza S. Rapid generation cycling of an F2 population derived from a cross between Lens culinaris Medik. and Lens ervoides (Brign.) Grande after aphanomyces root rot selection. Plant Breed 2018;137:486–91.
101. Chiurugwi T, Kemp S, Powell W, Hickey LT. Speed breeding orphan crops. Theor Appl Genet 2019;132:607–16.
102. Samineni S, Sen M, Sajja SB, Gaur PM. Rapid generation advance (RGA) in chickpea to produce up to seven generations per year and enable speed breeding. Crop J 2020;8:164–9.
103. Ohnishi T, Yoshino M, Yamakawa H, Kinoshita T. The biotron breeding system: a rapid and reliable procedure for genetic studies and breeding in rice. Plant cell Physiol 2011;52:1249–57.
104. Alahmad S, Dinglasan E, Leung KM, Riaz A, Derbal N, Voss-Fels KP, et al. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods 2018;14:1–15.
105. Kumar A, Sharma D, Tiwari A, Jaiswal J, Singh N, Sood S. Genotyping-by-sequencing analysis for determining population structure of finger millet germplasm of diverse origins. Plant Genome 2016;9;1–15, doi: 10.3835/plantgenome2015.07.0058
106. Girma D, Cannarozzi G, Weichert A, Tadele Z. Genotyping by sequencing reasserts the close relationship between tef and its putative wild Eragrostis progenitors. Diversity 2018;10:17.
107. Rabbi I, Hamblin M, Gedil M, Kulakow P, Ferguson M, Ikpan AS, et al. Genetic mapping using genotyping-by-sequencing in the clonally propagated cassava. Crop Sci 2014;54:1384–96.
108. Rabbi IY, Kulakow PA, Manu-Aduening JA, Dankyi AA, Asibuo JY, Parkes EY, et al. Tracking crop varieties using genotyping-by-sequencing markers: a case study using cassava (Manihot esculenta Crantz). BMC Genetics 2015;16:1–11.
109. Moumouni K, Kountche B, Jean M, Hash C, Vigouroux Y, Haussmann B, et al. Construction of a genetic map for pearl millet, Pennisetum glaucum (L.) R. Br., using a genotyping-by-sequencing (GBS) approach. Mol Breed 2015;35:1–10.
110. Deokar AA, Ramsay L, Sharpe AG, Diapari M, Sindhu A, Bett K, et al. Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly. BMC Genomics 2014;15:1–19.
111. Xiong H, Shi A, Mou B, Qin J, Motes D, Lu W, et al. Genetic diversity and population structure of cowpea (Vigna unguiculata L. Walp). PLoS One 2016;11:e0160941.
112. Fatokun C, Girma G, Abberton M, Gedil M, Unachukwu N, Oyatomi O, et al. Genetic diversity and population structure of a mini-core subset from the world cowpea (Vigna unguiculata (L.) Walp.) germplasm collection. Sci Rep 2018;8:16035.
113. Bull SE, Seung D, Chanez C, Mehta D, Kuon JE, Truernit E, et al. Accelerated ex situ breeding of GBSS-and PTST1-edited cassava for modified starch. Sci Adv 2018;4:eaat6086.
114. Gomez MA, Lin ZD, Moll T, Chauhan RD, Hayden L, Renninger K, et al. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF 4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol J 2019;17:421–34.
115. Odipio J, Alicai T, Ingelbrecht I, Nusinow DA, Bart R, Taylor NJ. Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front Plant Sci 2017;8:1780.
116. Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, et al. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol J 2018;16:1275–82.
117. Cannarozzi G, Plaza-Wüthrich S, Esfeld K, Larti S, Wilson YS, Girma D, et al. Genome and transcriptome sequencing identifies breeding targets in the orphan crop tef (Eragrostis tef). BMC Genomics 2014;15:1–21.
118. Verma M, Kumar V, Patel RK, Garg R, Jain M. CTDB: an integrated chickpea transcriptome database for functional and applied genomics. PLoS One 2015;10:e0136880.
119. Hittalmani S, Mahesh H, Shirke MD, Biradar H, Uday G, Aruna Y, et al. Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genomics 2017;18:1–16.
120. Hatakeyama M, Aluri S, Balachadran MT, Sivarajan SR, Patrignani A, Grüter S, et al. Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res 2018;25:39–47.
121. Varshney RK, Shi C, Thudi M, Mariac C, Wallace J, Qi P, et al. Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol 2017;35:969–76.
122. Mahato AK, Sharma AK, Sharma TR, Singh NK. An improved draft of the pigeonpea (Cajanus cajan (L.) Millsp.) genome. Data Brief 2018;16:376–80.
123. Varshney RK, Saxena RK, Upadhyaya HD, Khan AW, Yu Y, Kim C, et al. Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat Genetics 2017;49:1082–8.
124. Yemataw Z, Muzemil S, Ambachew D, Tripathi L, Tesfaye K, Chala A, et al. Genome sequence data from 17 accessions of Ensete ventricosum, a staple food crop for millions in Ethiopia. Data Brief 2018;18:285–93.
125. Parween S, Nawaz K, Roy R, Pole AK, Venkata Suresh B, Misra G, et al. An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.). Sci Rep 2015;5:12806.
126. Muñoz-Amatriaín M, Mirebrahim H, Xu P, Wanamaker SI, Luo M, Alhakami H, et al. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J 2017;89:1042–54.
127. Ghatak A, Chaturvedi P, Nagler M, Roustan V, Lyon D, Bachmann G, et al. Comprehensive tissue-specific proteome analysis of drought stress responses in Pennisetum glaucum (L.) R. Br.(Pearl millet). J Proteomics 2016;143:122–35.
128. Kamies R, Farrant JM, Tadele Z, Cannarozzi G, Rafudeen MS. A proteomic approach to investigate the drought response in the orphan crop Eragrostis tef. Proteomes 2017;5:32.
129. Bhushan D, Pandey A, Choudhary MK, Datta A, Chakraborty S, Chakraborty N. Comparative proteomics analysis of differentially expressed proteins in chickpea extracellular matrix during dehydration stress. Mol Cell Proteomics 2007;6:1868–84.
130. Bajaj D, Srivastava R, Nath M, Tripathi S, Bharadwaj C, Upadhyaya HD, et al. EcoTILLING-based association mapping efficiently delineates functionally relevant natural allelic variants of candidate genes governing agronomic traits in chickpea. Front Plant Sci 2016;7:450.
131. Cannarozzi G, Weichert A, Schnell M, Ruiz C, Bossard S, Blösch R, et al. Waterlogging affects plant morphology and the expression of key genes in tef (Eragrostis tef). Plant Direct 2018;2:e00056.
132. Kudapa H, Garg V, Chitikineni A, Varshney RK. The RNA-Seq-based high resolution gene expression atlas of chickpea (Cicer arietinum L.) reveals dynamic spatio-temporal changes associated with growth and development. Plant Cell Environ 2018;41:2209–25.
133. Zhang S, Chen X, Lu C, Ye J, Zou M, Lu K, et al. Genome-wide association studies of 11 agronomic traits in cassava (Manihot esculenta Crantz). Front Plant Sci 2018;9:503.
134. Jia G, Huang X, Zhi H, Zhao Y, Zhao Q, Li W, et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat Genetics 2013;45:957–61.
135. Sharma D, Tiwari A, Sood S, Jamra G, Singh N, Meher PK, et al. Genome wide association mapping of agro-morphological traits among a diverse collection of finger millet (Eleusine coracana L.) genotypes using SNP markers. PLoS One 2018;13:e0199444.
136. Shi Y, Thomasson JA, Murray SC, Pugh NA, Rooney WL, Shafian S, et al. Unmanned aerial vehicles for high-throughput phenotyping and agronomic research. PLoS One 2016;11:e0159781.
137. Reynolds D, Baret F, Welcker C, Bostrom A, Ball J, Cellini F, et al. What is cost-efficient phenotyping? Optimizing costs for different scenarios. Plant Sci 2019;282:14–22.
138. Hendre P, Muchugi A, Chang Y, Fu Y, Song Y, Liu M, et al. Generation of open-source genomics resources for African orphan tree crops by African orphan crops consortium (AOCC), a public-private partnership for promoting food and nutritional security in Africa. Acta Hortic 2020;615–22.
139. Mollins J. Smallholder farmers to gain from targeted CRISPR-Cas9 crop breeding. México-Veracruz: International Maize and Wheat Improvement Centre, 2019.