1. INTRODUCTION
Centella asiatica, which belongs to the family Apiaceae, is a herbaceous and perennial plant. The plant has immense commercial and economical significance due to its complex biochemical and phytochemical compositions. The phytochemical composition of the plant consists of pentacyclic triterpenoids, asiaticoside, brahmoside, asiatic acid, brahmic acid centellose, centelloside, and madecassoside [1-3]. These phytochemicals (phytochemicals are a group of bioactive chemical substances synthesized by plants) are of medicinal significance and play an important role to treat various diseases such as headache, drowsiness, wound healing lactation, and anti-aging [4,5] and are known to have anxiolytic (agents or drugs used to decrease anxiety level) properties as well [6]. The plant has significant dermatological applications [7]. The plant also contributes to the agricultural field as weed control and biomonitoring agent of soil for heavy metals.
The plant body has many aerial or above-ground parts. The most dominant aerial part of the plant is the leaf. The aerial body parts of plants such as stems, buds, flowers, and leaves provide a suitable habitat for the growth and development of a wide variety of microorganisms [8]. Such all the above-ground body parts of plants that are densely populated with microorganisms are altogether termed as phyllosphere whereas the surface area specifically associated with plant leaves is called phylloplane. Bacteria are the most commonly found inhabitants of the phyllosphere [9]. Phyllospheric bacteria can be found as epiphytes and entophytes. Epiphytic bacteria are those who colonize the external surface of the plant whereas endophytic bacteria grow inside the plant body.
These phyllosphere bacteria are closely associated with the host plant and affect the plant’s growth and development in varieties of ways [10,11]. These bacteria can affect the host plant positively or negatively. Some of the phyllosphere bacteria are known to play beneficial roles in various biological activities of host plants such as stress alleviation mechanism, carbon, nitrogen cycle, phosphate solubilization, and disease resistance. The study of phyllosphere bacteria helps to evaluate their role as plant growth-promoting bacteria which provides a platform for use of such bacteria for sustainable agriculture which helps to fulfill the food and commercial needs of society without leaving any harmful impact on the environment.
Like all living systems, bacterial growth and development are also affected by various abiotic parameters (it includes various non-living components such as physical and chemical factors of an ecosystem) of its surrounding. High salt concentration or salinity is one of the important abiotic parameters which induces abiotic stress and adversely affects the growth and survival of bacteria. High salt concentration induces osmotic shock (lysis of the cell induced by rapid alteration in salt concentrations in the medium surrounding the cell) which alters the osmotic potential gradient. This altered osmotic potential gradient decreases water and mineral uptake by living cells which leads death of the cell. Some bacteria have an inbuilt biological mechanism that helps them to resist osmotic shock and survive in a high-salt environment. Such bacteria are known as osmotolerant (salt-tolerant bacteria).
In today’s anthropogenic era (any change in the environment or natural resources caused by human interventions), most agricultural lands are degraded and destroyed by the continuous increase of various abiotic factors such as salinity, temperature, and soil erosion. Agricultural productivity is greatly threatened by salinity stress. This salinity stress can be alleviated by increasing the salt tolerance potential of the plants. Various microorganisms, which inhabited the plant body as a shelter, help to increase the stress tolerance mechanism of the host plant [12,13]. Certain bacteria have the potential to contribute to the stress tolerance mechanism by regulating the intracellular concentrations of various secondary metabolites in the host plants [14,15]. This ability of such plant commensal microorganisms makes them significant for sustainable agriculture and crop production.
2. MATERIALS AND METHODS
The methodology used for the present study included many steps. These steps were as follows:
2.1. Sample Collection and Preparation
The plant sample, C. asiatica, was collected and authenticated from the Botanical Garden of Indian Republic, NOIDA, UP. The fresh leaves of the plant were collected and washed with tap water to remove all dust particles from the leaf surface. Then these leaves were dried at room temperature. Dried leaves were crushed to make a homogenous powder. 1 g of this powder was homogenized into 1000 ml of distilled water. This leaf suspension was used as the sample for bacterial isolation [16].
2.2. Isolation of Bacteria
For the isolation of phyllosphere bacteria, 0.5 ml of the above leaf suspension was spread on the surface of plates containing sterile and solidified nutrient agar media. The inoculated plates were kept inside the incubator overnight. After incubation, bacterial colonies were grown and observed on the plates. Morphologically distinct bacterial colonies were further purified on similar media. The pure cultures of different bacterial isolates were used to perform various tests for their characterization [17].
2.3. Isolation of Osmotolerant Bacteria
The bacterial isolates were examined for their osmotic tolerance potential by inoculating the bacterial culture on the plates containing nutrient media with gradually increased concentrations of salts such as sodium chloride, D-mannitol, and sorbitol. The control plate was prepared by inoculating the same bacterial culture on the plate containing nutrient media without any salt concentrations. All the inoculated plates were kept in the incubator at 37°C for 24–48 h. After incubation, every plate was observed for bacterial growth. The bacteria that possessed osmotolerant capacity were able to grow on the media containing high salt concentrations and were considered osmotolerant bacteria [18].
2.4. Characterization of the Osmotolerant Bacterial Isolates
Isolated osmotolerant bacteria were identified and characterized using various methods of analysis described below.
2.4.1. Morphological analysis
The morphological characteristics of the isolated bacteria were tested by Gram’s staining. It was done using the standard protocol. A thin bacterial smear was fixed on a clean glass slide. The smear was stained with crystal violet for 60 s, then wash with water. After this, the smear was treated with Lugol’s iodine for 30 s, then washed with water, further with decolorization agent 70% of alcohol for 15 s, and washed with water. Finally, the smear was stained with safranin (counterstain), then washed with water and air-dried. The Gram’s-stained smear was observed under a microscope. The bacterial cells appeared purple, called Gram’s positive, and the bacterial cells appeared pink, referred to as Gram’s negative [19].
2.4.2. Motility test
The motility of the bacterial isolates was tested by inoculating 24 h old culture of the test bacteria into a stab of nutrient agar media in the test tube. The tubes were incubated for 24–48 h at 37°C. The non-motile bacteria had growth restricted to the inoculation line while motile bacteria had diffused growth spreading to the surrounding area from the inoculation line [20].
2.4.3. Biochemical analysis
According to the classification system published in Bergey’s Manual of Systematic Bacteriology, a series of biochemical tests were conducted to identify the biochemical profile of the isolated osmotolerant bacteria. These biochemical tests were –
2.4.3.1. Sugar fermentation test
This test was carried out to test the sugar fermentation ability of the test bacteria for various sugars such as glucose, lactose, and sucrose [21]. Certain bacteria can ferment a carbohydrate component such as sugar and produce acid and gas as products. For this test, a sugar indicator broth was prepared. The media was dispensed into test tubes, a durham tube was placed inside the tube in an inverted position and autoclaved all the tubes. The tubes were inoculated with a loopful culture of the test bacteria and incubated at 37°C for 2–7 days. The tubes were observed daily for any change in the color of the broth (that indicates acid production) and displacement of the broth in the durham tube (that indicates gas production). The test was performed for three sugars which were as follows –
2.4.3.1.1. Glucose utilization test
To test the glucose fermentation ability the test bacteria were inoculated in glucose indicator broth which contained peptone broth, 1% glucose, and 0.01% phenol red. After incubation, the yellow coloration of the broth indicates acid production, and displacement of broth in the durham tube indicates gas production.
2.4.3.1.2. Lactose utilization test
This test was performed using a similar protocol as the glucose fermentation test except that the broth used was the lactose indicator broth.
2.4.3.1.3. Sucrose utilization test
For this test, the media used was the sucrose indicator broth, rest protocol was similar to other sugar fermentation tests.
2.4.3.2. Catalase test
Catalase activity was tested by taking a small quantity of 24 h old bacterial culture on a clean glass slide, then a drop of hydrogen peroxide was added to the culture. The release of gas bubbles as white forth shows positive catalase activity [22].
2.4.3.3. Starch hydrolysis test
This test is used to test the ability of bacteria that can produce an enzyme called amylase that degrades starch. For this test, the plates were prepared by pouring the sterilized nutrient media containing 1% of soluble starch. The solidified plates were inoculated by streaking the bacterial culture across the media surface, the inoculated plates were kept in an incubator for 24 h. The starch-hydrolyzing bacteria were produced colonies that surrounded by the clear zone of starch hydrolysis due to the amylase enzyme released by the bacterial culture [21].
2.4.3.4. Urease test
The urease enzyme catalyzes the breakdown of urea into ammonia and carbon dioxide. The urease activity of the bacterial isolates was tested by inoculating the bacterial culture on the nutrient media containing urea and an indicator dye phenol red and then incubating at 37°C for 24 h. After incubation, the urease-producing bacteria changed the color of the media from yellow to reddish pink [23].
2.4.3.5. Casein digestion test
Caseinase is an enzyme that degrades milk protein into smaller peptides and free amino acids. The casein activity of the bacterial isolates was done by streaking the bacterial culture on the surface of the plates containing autoclaved casein media. After incubation, the plates were observed for the clear zone around the bacterial growth [24].
2.4.3.6. Indole test
The pure culture of the test organisms was mixed in the tube containing tryptone broth medium and then the tube was incubated at 37°C for 24–48 h. A few drops of Kovac’s indole reagent were added to the tube. The formation of a pink to cherry red color ring on the top of the tube indicated a positive test [25].
2.4.3.7. Methyl red and vogas proskauer (MR-VP)
5 ml of MR-VP broth was transferred into test tubes and autoclaved. The sterilized tubes were inoculated with test bacteria and incubated at 37°C for 48 h. After incubation, half of the media was transferred into a new sterile tube, and then a few drops of MR indicator were added to the tubes. The tubes were observed for any color change. A formation of red color shows a positive test [26]. The remaining quantity of the MR-VP broth was mixed with Berritt’s reagents 1 and 2, then the tube was shaken and allowed to rest for 30 min to 1 h. The development of red coloration on the top of the tube showed a positive test for VP [25].
2.4.3.8. Citrate utilization test
This test checks the ability of the bacteria to use citrate as a carbon source. It was performed by preparing slants in test tubes containing autoclaved citrate agar media. The media contains an indicator dye that indicates a change in PH by changing its color from green to blue. The tubes were inoculated with test bacteria and then incubated at 37°C. A change in media color from green to blue shows a positive test [21].
2.4.3.9. Nitrate reduction test
Some bacteria produce nitrate reductase enzyme that reduces nitrate into nitrite. For this test, a peptone broth containing potassium nitrate was prepared and its small quantity was dispensed in test tubes. The test tubes were autoclaved and inoculated with the culture of test bacteria. Incubate the test tubes at 37°C for 24 h. After incubation, a few drops of sulfanilic acid and alpha naphthylamine were added. Finally, zinc dust was added to the tubes, then tubes were observed for any change in the coloration of the broth [27].
2.4.4. Molecular Characterization of the Osmotolerant Bacteria
The osmotolerant bacterial isolates were characterized by 16s rRNA sequencing. The procedure includes standard protocols of genomic DNA isolation, gel electrophoresis, PCR amplification, DNA sequencing, and phylogenetic analysis. The genomic DNA was isolated by homogenizing the bacterial culture in extraction or lytic buffer. The resultant cell lysate was further treated by a series of solvent extraction and centrifugation. The genomic DNA was precipitated by adding a solution of 0.1 volume of 3 M Sodium acetate pH 7.0 and 0.7 volume of Isopropanol. The precipitated DNA pellet was washed with 70% ethanol and air-dried. Then, the genomic DNA was dissolved in TE buffer which was further subjected to RNAase treatment for the removal of RNA impurities. After a quality check, PCR amplification of the genomic DNA was carried out by using 16S rRNA forward primer (GGATGAGCCCGCGGCCTA) and reverse primer (CGGTGTGTACAAGGCCCGG). Thermos cycler was used for PCR amplification using 30 cycles at 94°C for 1 min (denaturation step), 50°C for 1 min (annealing step), and 72°C for 2 min (extension step) with a final extension of 7 min at 72°C. The amplified genomic DNA was tested by using agarose gel electrophoresis [Figures 1a and 2a]. The sequences of PCR products were analyzed using the ABI 3130xl platform. The resultant nucleotide sequences were further analyzed for homology and then the phylogenetic relationship was studied using the BLAST platform. Finally, the nucleotide sequences were submitted to NCBI (National Center for Biotechnological Information) for accession number [28,29].
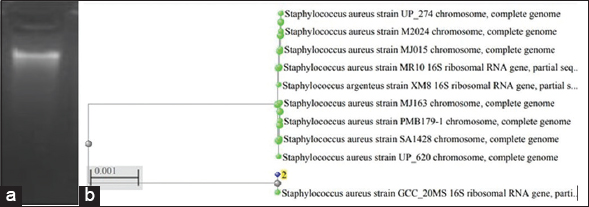 | Figure 1: (a) Identification of Isolated DNA of the bacterial isolate B-6 using gel electrophoresis and (b) Phylogenetic analysis of 16s rRNA sequence of the isolate B-6. [Click here to view] |
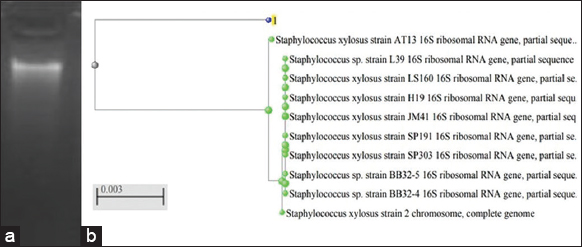 | Figure 2: (a) Identification of Isolated DNA of the bacterial isolate B-7 using gel electrophoresis and (b) Phylogenetic analysis of 16s rRNA sequence of the isolate B-7. [Click here to view] |
3. RESULTS
The results of the study were also recorded and studied in the same cascade-like methodology. The results were as follows –
3.1. Isolation of Bacteria from C. asiatica
In the present study, a total of 12 bacterial strains (B-1 to B-12) were isolated from the leaf extract of the C. asiatica plant. All the isolated bacterial strains were maintained as pure culture forms using pure culture techniques. These bacterial isolates were isolated using nutrient agar media plates [Figure 1].
3.2. The Osmotolerant Potential of the Bacterial Isolates
The data of the Osmotolerance study revealed that two out of the 12 bacterial isolates were found salt tolerant at varying concentrations (200mM, 400mM, 600mM,800mM, and 1000mM) of salts such as NaCl, mannitol, and sorbitol.
For NaCl salt, all the bacterial isolates (B-1 to B-12, except the B-9 isolate) showed good growth up to 200mM NaCl concentration but B-9 showed very slight growth at 200mM NaCl. The growth of all the bacterial isolates, except B-6, B-7, and B-8, were adversely affected by an increase in NaCl concentrations from 400 to 1000 mM. B-8 isolate showed good growth up to 600mM NaCl and the growth decreased up to slight and very slight at 800, and 1000 mM NaCl concentrations, respectively. The growth of B-9 was decreased with an increase in NaCl concentrations and showed no growth at 400mM to 1000mM NaCl concentration. The B-10 isolate was also adversely affected by increased salt concentration and showed no growth at higher concentrations of NaCl (800mM and 1000mM). the growth of B-11 and B-12 isolates was decreased with NaCl stress and showed no growth at 600 mM and above NaCl concentrations. The only two bacterial isolates, B-6 and B-7, showed good growth at the highest NaCl concentration (1000 mM), Table 1.
Table 1: The study of the osmotolerance potential of the bacterial isolates (B-1 to B-12) using various concentrations of NaCl (200 mM, 400 mM, 600 mM, 800 mM, and 1000 mM).
| S No. | Salt Conc  Bacterial isolates Bacterial isolates  | 0 mM | 200 mM | 400 mM | 600 mM | 800 mM | 1000 mM |
|---|---|---|---|---|---|---|---|
| 1. | B-1 | +++ | +++ | + | + | + | - |
| 2. | B-2 | +++ | +++ | ++ | + | + | - |
| 3. | B-3 | +++ | +++ | ++ | + | + | - |
| 4. | B-4 | +++ | +++ | ++ | - | - | - |
| 5. | B-5 | +++ | +++ | + | - | - | - |
| 6. | B-6 | +++ | +++ | +++ | +++ | +++ | +++ |
| 7. | B-7 | +++ | +++ | +++ | +++ | +++ | +++ |
| 8. | B-8 | +++ | +++ | +++ | +++ | ++ | + |
| 9. | B-9 | +++ | + | - | - | - | - |
| 10. | B-10 | +++ | +++ | + | ++ | - | - |
| 11. | B-11 | +++ | +++ | +++ | - | - | - |
| 12. | B-12 | +++ | +++ | + | - | - | - |
+++ = Good growth, ++ = Slight growth, + = Very slight growth, − = No growth.
The data of the osmotolerance study of the bacterial isolates using varied concentrations (200 mM to 100 mM) of mannitol are shown in Table 2. The data revealed that the bacterial isolates B-1 to B-8 showed good growth up to 600 mM concentration of mannitol, except the B-5 isolate showed slight growth at 600mM mannitol, then the growth of bacterial isolates B-1 to B-5 was decreased at higher concentrations of mannitol (800 mM, 1000 mM). the B-5 isolate showed no growth at a 1000mM concentration of mannitol. The growth of B-9 isolate was adversely affected by the gradual increase in mannitol concentration from 200 mM to 1000 mM. The bacteria isolate B-10 showed good growth at 400 mM mannitol concentration, then the growth continuously decreased at 600mM, and 800 mM and showed no growth at the highest mannitol concentration (1000 mM). The growth of the B-11 and B-12 isolates was also decreased with the gradual increase in mannitol concentrations and showed no growth at higher concentrations of mannitol (800 mM and 1000 mM).
Table 2: The study of the osmotolerance potential of the bacterial isolates (B-1 to B-12) using various concentrations of mannitol (200 mM, 400 mM, 600 mM, 800 mM, and 1000 mM).
| S. No. | Salt Conc  Bacterial isolates Bacterial isolates  | 0 mm | 200 mM | 400 mM | 600 mM | 800 mM | 100 mM |
|---|---|---|---|---|---|---|---|
| 1. | B-1 | +++ | +++ | +++ | +++ | ++ | + |
| 2. | B-2 | +++ | +++ | +++ | +++ | ++ | + |
| 3. | B-3 | +++ | +++ | +++ | +++ | ++ | + |
| 4. | B-4 | +++ | +++ | +++ | +++ | ++ | + |
| 5. | B-5 | +++ | +++ | +++ | ++ | + | - |
| 6. | B-6 | +++ | +++ | +++ | +++ | +++ | +++ |
| 7. | B-7 | +++ | +++ | +++ | +++ | +++ | ++ |
| 8. | B-8 | +++ | +++ | +++ | +++ | ++ | - |
| 9. | B-9 | +++ | + | + | - | - | - |
| 10. | B-10 | +++ | +++ | +++ | ++ | + | - |
| 11. | b-11 | +++ | ++ | ++ | + | - | - |
| 12 | B-12 | +++ | ++ | ++ | + | - | - |
+++ = Good growth, ++ = Slight growth, + = Very slight growth, − = No growth.
The Osmotolerance potential (using sorbitol) of all the bacterial isolates (B-1 to B-12) is presented in Table 3. The data revealed that all the bacterial isolates except B-5, B-8, B-9, and B-10 showed good growth at initial concentrations of sorbitol (200 mM and 400 mM). The B-5 isolate showed decreased growth after 200 mM sorbitol concentration and showed no growth at 600 mM, 800mM, and 1000mM sorbitol concentrations. The B-8 isolates showed decreased growth with an increase in sorbitol concentrations from 200 mM to 800 mM and had no growth at 1000 mM sorbitol concentrations. The B-9 isolates showed no growth at all concentrations of sorbitol stress. The growth of B-10 also decreased with sorbitol stress from 400mM to 800mM and showed no growth at the highest sorbitol concentration (100 mM). The B-1 and B-2 isolated showed very slight growth at 600–1000 mM sorbitol concentrations. The growth of the B-3 and B-4 bacterial isolates was decreased at higher ranges of sorbitol stress. The B-3 had no growth at 100 mM sorbitol concentration. The B-4 showed no growth at 800 mM and 1000 mM sorbitol concentrations. The B-11 and B-12 isolates were also negatively affected by sorbitol stress and showed no growth at the highest sorbitol concentrations (800 mM and 1000 mM). two of the total 12 bacterial isolates (B-6 and B-7) were able to grow at all the ranges of the sorbitol stress and showed good growth at the highest sorbitol concentrations (800 mM and 1000 mM).
Table 3: The study of the osmotolerance potential of the bacterial isolates (B-1 to B-12) by using various concentrations of sorbitol (200 mM, 400 mM, 600 mM, 800 mM, and 1000 mM).
| S. No. | Salt Conc  Bacterial isolates Bacterial isolates  | 0 mM | 200 mM | 400 mM | 600 mM | 800 mM | 1000 mM |
|---|---|---|---|---|---|---|---|
| 1. | B-1 | +++ | +++ | +++ | + | + | + |
| 2. | B-2 | +++ | +++ | +++ | + | + | + |
| 3. | B-3 | +++ | +++ | +++ | + | + | - |
| 4. | B-4 | +++ | +++ | +++ | + | - | - |
| 5. | B-5 | +++ | +++ | + | - | - | - |
| 6. | B-6 | +++ | +++ | +++ | +++ | +++ | +++ |
| 7. | B-7 | +++ | +++ | +++ | +++ | +++ | ++ |
| 8. | B-8 | +++ | ++ | ++ | + | + | - |
| 9. | B-9 | +++ | - | - | - | - | - |
| 10. | B-10 | +++ | +++ | ++ | + | + | - |
| 11. | B-11 | +++ | +++ | +++ | ++ | - | - |
| 12. | B-12 | +++ | +++ | +++ | ++ | - | - |
+++ = Good growth, ++ = Slight growth, + = Very slight growth, − = No growth.
3.3. Morphological and Biochemical Characterization of the Osmotolerant Bacterial Isolates
The results of morphological and biochemical characterization are shown in Table 4. The morphological (microscopic) test indicated that both the isolated osmotolerant bacterial isolates (B-6 and B-7) were Gram-positive, cocci (round in shape), and often arranged in grape-like clusters. The motility test showed that both the isolates B-6 and B-7 were non-motile. Both of these isolates showed positive tests for MR, VP, catalase, urease, citrate utilization, and sugar fermentation tests for glucose, lactose, and sucrose. The isolates (B-6 and B-7) were negative for amylase (starch hydrolysis test) and indole test.
Table 4: Morphological and biochemical characteristics of the isolated osmotolerant bacteria.
| S. No. | Bacterial isolates  Biochemical Tests Biochemical Tests | B-6 | B-7 |
|---|---|---|---|
| 1. | Gram’s staining | + (Cocci in the cluster) | + (Cocci in the cluster) |
| 2. | Motility test | - | - |
| 3. | Starch hydrolysis test | - | - |
| 4. | Catalase test | + | + |
| 5. | Urease test | + | + |
| 6. | Indole test | - | - |
| 7. | Methyl red test | + | + |
| 8. | Vogas Proskeur test | + | + |
| 9. | Citrate test | + | + |
| 10. | Glucose fermentation test | + | + |
| 11. | Lactose fermentation test | + | + |
| 12. | Sucrose fermentation test | + | + |
+ = Positive test, − = Negative test.
3.4. Molecular Characterization of the Osmotolerant Bacterial Isolates
Molecular identification of the isolated osmotolerant bacteria (B-6 and B-7) was performed by PCR amplification of 16SrRNA sequences. For this 16 S rRNA gene of both the isolates was amplified by PCR successfully, then the amplified products were checked with agarose gel electrophoresis [Figures 1a and 2a]. The results of the gel electrophoresis revealed that the size of the PCR-amplified products of both isolates was approximately 2500bp (2.5 kb). The amplified products were sequenced and the sequences were generated in FASTA format. These resultant sequences were compared with the NCBI nucleotide database using BLAST-N comparison. The BLAST-N comparison study revealed 100% similarity of the isolate B-6 with Staphylococcus aureus strain GCC_20MS (NCBI accession number MZ305087.1) and 97.85% similarity of the isolate B-7 with Staphylococcus xylosus strain AT13 (NCBI accession number MW175729.1). The determined 16S rRNA sequences of both the isolates (B-6 and B-7) were submitted to the database of NCBI, GeneBank under the accession numbers (OM993290) (OM860203), respectively. The phylogenetic tree study of the isolated osmotolerant bacteria (B-6 and B-7) was performed and the data are shown in [Figures 1b and 2b].
4. DISCUSSION
Microorganisms are found in all the biomes of the blue planet. A huge and diverse microbiome is also associated with plants. Scientists have a long-lasting interest to study the rhizosphere (below-ground region) of plants and its associated microbiome; hence, plenty of reported work is available on the rhizospheric microflora of the plants. As the Phyllosphere covers approximately 60% of the total biomass of the biosphere, it provides a huge shelter for the growth of a diverse microbial community [30]. In this study, the leaves of C. asiatica were studied for the presence of inhabitant bacterial communities. A total of 12 bacterial isolates were recovered and maintained as pure culture from the leaves of the C. asiatica plant. Numerous studies were performed earlier on microbial diversity of the Phyllosphere of several plants such as maize [31], rice [32], oat [33], and sugarcane [34]. Bhore et al., 2010 cultivated the bacterial endophytes from the leaves of Gaynura procumbent plants [35]. Rakotoniriana et al. also reported the presence of bacterial communities in the leaves of the C. asiatica plant, in their study [36]. Xie et al. revealed a phyllospheric bacterial community in macrophytes (aquatic plants) found in paddy soil in the study [37].
In the present study, these 12 bacterial isolates were assessed for salt tolerance using varied concentrations (200 mM, 400 mM, 6000 mM, 800 mM, and 1000 mM) of NaCl, mannitol, and sorbitol. The previous studies reported the occurrence of salt-tolerant microflora in plant-associated habitats [38]. Kamalraj et al., 2008 showed the impact of NaCl on the growth rate of endophytic fungus isolated from the leaves of mangrove C. roxburghiana [39]. Another work was also reported by Gayathri et al., 2010 which revealed that the bacteria isolated from mangrove and salt marsh leaves were found to tolerate salt up to 10% of NaCl [40]. The result of the present study showed that only two bacterial isolates (B-6 and B-7) were found to maintain their growth at higher concentrations of NaCl, mannitol, and sorbitol. Hence, both of these bacterial isolates were considered salt tolerant.
In the present study, the preliminary characterization of the two salt-tolerant bacterial isolates (B-6 and B-7) was done by morphological and various biochemical tests. Both isolates were chosen for Gram’s staining to evaluate their morphological or microscopic characteristics. In the results of this study, both the salt-tolerant bacteria (B-6 and B-7) were found Gram’s positive, round (cocci) in shape, and arranged in grapes-like clusters. Zinniel et al. (2002) also found the presence of Gram-positive endophytic bacteria in agronomic crop plants [41]. In the present study, these Gram-positive cocci were further characterized by various biochemical tests. The results of the biochemical test revealed that both the isolated salt-tolerant bacteria (B-6 and B-7) were shown negative tests for motility and indole tests and positive tests for MR, VP, citrate utilization, nitrate reduction, sugar fermentation tests (for glucose, lactose, and sucrose), catalase, and amylase [42]. In this study, both the salt-tolerant bacterial isolates were shown morphological and biochemical properties of Streptococcus Sps based on Bergey’s manual studies [43].
In the present work, both the isolated bacteria (B-6 and B-7) were finally identified on a molecular basis using 16 S rRNA sequencing. Based on the 16S rRNA sequencing results, phylogenetic trees were formed for both the isolates B-6 and B-76. The studies revealed that both the salt-tolerant bacterial isolates (B-6 and B-7) belong to the Staphylococcus group, containing S. aureus strain GCC_20MS and S. xylosus strain AT13, respectively [44].
The results of the present study were supported by various earlier studies, which showed the presence of salt-tolerant staphylococcus sps (namely, S. aureus and S. xylosus) in the Phyllosphere region of plants [45]. Liu et al., 2015 isolated the endophytic bacteria from the various parts of a medicinal plant, Noni (Morinda citrifolia L.), and confirmed the presence of S. aureus in the above-ground parts like leaves in the plant [46]. In another previous study, Swamy et al. 2020 reported the inhabitance of S. aureus sps in some plants such as the Ashok tree, Lablab, and Noni [47]. Adeniyi et al. 2020, also studied the phyllosphere microbial profile of the Ficus vogelii and confirmed the predominance of S. aureus in the plant leaf extract [48]. Similarly, Oluyemi et al., 2013 also isolated various multiple drug resistance staphylococcal species such as S. aureus and S. xylosus from the Phyllosphere of the Ficus sycomorus Linn [49].
In the present study, salt-tolerant Staphylococcal sps were explored in the leaf extract of the C. asiatica plants. This study will provide a reference to the phyllosphere bacterial diversity of the plant. In this study, salt-tolerant bacterial sps which were isolated from the plant leaves may have the potential to contribute to plant health by contributing to important plant metabolic activity like nitrogen fixation [50] or by alleviating various abiotic stressors like salinity [51]. The research works are widely documented on the role of plant-associated microorganisms in salinity tolerance mechanism [52,53]. Bian et al., 2011 demonstrated that salt-tolerant bacterial sps like Staphylococcal sps cultivated from plant sources were able to produce plant growth hormones like indole acetic acid and contribute to the salinity tolerance mechanism of the host plant under high salt conditions. More studies were also performed which supported the findings of the present studies [54-56]. Komaresofla et al. 2019 also reported the presence of S. aureus as a rhizospheric and endophytic bacteria of the Salicornia plant and also found their positive role in the host plant growth and salinity tolerance ability under salinity stress conditions. Shultana et al., 2022 also reviewed the role of S. xylosus as a plant growth-promoting rhizobacteria and its role in the promotion of the host plant’s growth by influencing salt tolerance adaptation mechanism [57]. Earlier, Afrasayab et al., 2010 performed a study that confirmed that the S. xylosus influenced the growth characteristics of the Triticum aestivum under salinity stress by increasing the seedling length of the plant by 25% at 100mM NaCl concentration [58]. Nagaraju et al., 2021 also revealed in their study that the S. xylosus inoculants increased the enzyme activity (like superoxide dismutase, and catalase) and proline content in the leaves of chickpea plants and increased the productivity under the salt stress conditions [59].
The main objective of the present study was to reveal the presence of osmotolerant bacterial species as natural inhabitants of the photospheric region of the plants. These osmotolerant bacterial species can be used as a biocontrol agent under salinity stress conditions and helped to induce tolerance mechanisms against high salinity for the host plants.
5. CONCLUSION
The present study explained the presence of a culturable diverse population of bacteria in the leaves of the C. asiatica plant. The results of this study reveal that the twelve bacterial isolates were cultured in pure forms, among which only two isolates, namely, S. aureus strain GCC_20MS and S. xylosus strain AT13 were found salt tolerant. The phyllosphere is an important habitat scientifically and economically as well. It is crowded with a great diversity of microorganisms that can contribute to various vital processes of the host plant and affect the growth of the host plant in a positive manner. Hence, the study of the phyllosphere microflora of plants always provides a better understanding of plant-microbial interactions and their practical applications in the sustainable growth of plants. The remarkable presence of such salt-tolerant bacteria in the phyllosphere region of the plant (C. asiatica) suggests that such bacteria can be used as biocontrol agents in the future application and helped in more effective stress management in the host plants under saline field conditions. The present study provides a strong base for further research to find out the positive role of such salt-tolerant bacteria in salinity stress management for plants and their commercial utility in sustainable agricultural production as a biocontrol agent.
6. ACKNOWLEDGMENT
The authors would like to thank and acknowledge the Department of Biotechnology, AKS University, Satna (M.P.), Rupture Biotech Laboratory, NOIDA (U.P.), and BGIR, NOIDA (U.P.) who provided all the infrastructural and laboratory assistance for this study.
7. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. Funding
There is no funding to report.
9. Ethical approvals
This study does not involve experiments on animals or human subjects.
10. Data availability
All the data is available with the authors and shall be provided upon request.
11. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Brinkhaus B, Lindner M, Schuppan D, Hahn EG. Chemical, pharmacological and clinical profile of the East Asian medical plant Centella asiatica. Phytomedicine2000;7:427-48. [CrossRef]
2. James JT, Dubery IA. Pentacyclic triterpenoids from the medicinal herb, Centella asiatica (L.) Urban. Molecules 2009;14:3922-41. [CrossRef]
3. Hashim P, Sidek H, Helan M, Sabery A, Palanisamy UD, Ilham M. Triterpene composition and bioactivities of Centella asiatica.Molecules2011;16:1310-22. [CrossRef]
4. Gohil KJ, Patel JA, Gajjar AK. Pharmacological review on Centella asiatica:A potential herbal cure-all. Indian J Pharm Sci 2010;72:546-56. [CrossRef]
5. Singh S, Gautam A, Sharma A, Batra A. Centella asiatica (L.):A plant with immense medicinal potential but threatened. Int J Pharm Sci Rev Res2010;4:9-19.
6. Wijeweera P, Arnasona JT, Koszycki, Merali Z. Evaluation of anxiolytic properties of Gotukola-(Centella asiatica) extracts and asiaticoside in rat behavioral models. Phytomedicines2006;13:668-76. [CrossRef]
7. Bylka W, Awizen ZP, Studzi?ska SE, Pazdrowska D, Brzezi?ska A. Centella asiatica in dermatology. An overview of phytotherapy.2014;28:1117-24. [CrossRef]
8. Andrews JH, Harris RF. The ecology and biogeography of microorganisms on plant surfaces. Ann Rev Phytopathol2000;38:145-80. [CrossRef]
9. Muller T, Muller M, Behrendt U, Stadler B. Diversity of culturable phyllosphere bacteria on beech and oak:The effects of lepidopterous larvae. Microbiol Res2003;158:291-7. [CrossRef]
10. Kembel SW, Connor T, Hubbell SP, Wright SJ, Green JL. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc Natl Acad Sci U S A2014;111:13715-20. [CrossRef]
11. Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol 2012;10:828-40. [CrossRef]
12. Lata R, Chowdhury S, Gond SK. Induction of abiotic stress tolerance in plants by endophytic microbes. Lett Appl Microbiol 2018;66:268-76. [CrossRef]
13. Numan M, Bashir S, Khan Y, Mumtaz R, Shinwari ZK, Khan AL, et al. Plant growth-promoting bacteria as an alternative strategy for salt tolerance in plants:A review. Microbiol Res2018;209:21-32. [CrossRef]
14. Bharti N, Yadav D, Barnawal D, Maji D, Kalra A. Exiguobacterium oxidotolerans, a halotolerant plant growth-promoting rhizobacteria, improves the yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World J Microbiol Biotechnol2013;29:379-87. [CrossRef]
15. Jalili F, Khavazi K, Pazira E, Nejati A, Rahmani HA, Sadaghiani HR, et al. Isolation and characterization of ACC deaminase-producing fluorescent pseudomonads, to alleviate salinity stress on canola (Brassica napus L.) growth. J Plant Physiol 2009;166:667-74. [CrossRef]
16. Ruszymah BH, Chowdhury SR, Manan NA, Fong OS, Adenan MI, Saim AB. Aqueous extract of Centella asiatica promotes corneal epithelium wound healing in vitro. Ethnopharmacology2012;140:333-8. [CrossRef]
17. Mazinani Z, Zamani M, Sardari S. Isolation and identification of phyllospheric bacteria possessing antimicrobial activity from Astragalus obtusifolius, Prosopis juliflora, Xanthium strumarium, and Hippocrepisunisiliqousa. AvicennaJ Med Biotechnol2017;9:31-7.
18. Arora S, Patel PN, Vanza MJ, Rao GG. Isolation and characterization of endophytic bacteria colonizing halophyte and other salt tolerant plant species from coastal Gujarat. Afr J Microbiol Res 2014;8:1779-88. [CrossRef]
19. O'Toole GA. Classic spotlight:How the gram stain works. J Bacteriol 2016;198:3128. [CrossRef]
20. Murinda ES, Nguyen LT, Ivey SJ, Almeida RA, Oliver SP. Novel single-tube agar-based test system for motility enhancement and immunocapture of Escherichia coli O157:H7 by H7 flagellar antigen-specific antibodies. J Clin Microbiol 2002;40:4685-90. [CrossRef]
21. Hemraj V, Diksha S, Avneet G. A review on commonly used biochemical test for bacteria. Innov J Life Sci2013;1:1-7.
22. Naik S, Naik GB, Patil B, Revathi RM. Biochemical characterization of Xanthomonas campestris pv. arecae causing leaf stripe in areca nut. J Pharmacol Phytochem2017;7:1168-70.
23. Christensen WB. Urea decomposition as a means of differentiating Proteus and paracolon cultures from each other and from Salmonella and Shigella types. J Bacteriol1946;52:461-6. [CrossRef]
24. William AS. Joseph JL. Hydrolysis of casein:A differential aid for the identification of Serratia marcescens. JClin Pathol1972;25:1083-5. [CrossRef]
25. Bhutia MO, Thapa N, Tamang JP. Molecular characterization of bacteria, detection of enterotoxin genes and screening of antibiotic susceptibility pattern in traditionally processed meat products of Sikkim, India. Front Microbiol2021;11:599606. [CrossRef]
26. Cowan ST. Micromethod for the methyl red test. Gen J Microbiol 1953;9:101-9. [CrossRef]
27. MacFaddin JF. Biochemical Tests for Identification of Medical Bacteria. 3rd ed. Philadelphia, PA:Lippincott Williams and Wilkins;2000.
28. Rastogi G, Coaker GL, Leveau JH. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol Lett2013;348:1-10. [CrossRef]
29. Andleeb S, Shafique I, Naseer A, Abbasi WA, Ejaz S, Liaqat I, et al. Molecular characterization of plant growth-promoting vermibacteria associated with Eisenia fetida gastrointestinal tract. PLoS One 2022;17:6. [CrossRef]
30. Koskella B. The phyllosphere. Curr Biol 2020;30:R1143-6. [CrossRef]
31. Manching HC, Carlson K, Kosowsky S, Smitherman CT, Stapleton AE. Maize phyllosphere microbial community niche development across stages of host leaf growth. F1000 Res2018;6:1698. [CrossRef]
32. Mano H, Morisaki H. Endophytic bacteria in the rice plant. Microbes Environ 2008;23:109-17. [CrossRef]
33. Burlaga HR, Grabolinska M. Characterization of the selected group of microorganisms occurring in soil rhizosphere and phyllosphere of Oat. Polish J Microbiol2006;55:227-3.
34. Magnani GS, Didonet CM, Cruz LM, Picheth FO, Pedrosa FO, Souza EM. Diversity of endophytic bacteria in Brazilian sugarcane. Genet Mol Res2010;9:250-8. [CrossRef]
35. Bhore SJ, Nithya R, Chye YL. Screening of endophytic bacteria isolated from leaves of Sambung Nyawa [Gynura procumbens (Lour.) Merr.] for cytokinin-like compounds. Bioinformation2010;5:191-7. [CrossRef]
36. Rakotoniriana EF, Rafamantanana M, Randriamampionona D, Rabemanantsoa C, Urveg-Ratsimamanga S, El Jaziri M, et al. Study in vitro of the impact of endophytic bacteria isolated from Centella asiatica on the disease incidence caused by the hemibiotrophic fungus Colletotrichum higginsianum. Antonie van Leeuwenhoek2013;103:121-33. [CrossRef]
37. Xie WI, Su JQ, Zhu YG. Phyllosphere bacterial community of floating macrophytes in paddy soil environments as revealed by illumina high-throughput sequencing. Appl Environ Microbiol 2015;81:522-32. [CrossRef]
38. Upadhyay SK, Singh DP, Saikia R. Genetic diversity of plant growth promoting rhizobacteria isolated from rhizospheric soil of wheat under saline condition. Curr Microbiol 2009;59:489-96. [CrossRef]
39. Kamalraj S, Sridevi S, Gangodevi V, Venkatesan A, Muthumary J. Effect of NaCl on biochemical changes and endophytic assemblages in the leaves of a mangrove, Ceriops roxburghiana Arn. Indian J Sci Technol2008;1:1-7. [CrossRef]
40. Gayathri S, Saravanan D, Radhakrishnan M, Balagurunathan R, Kathiresan K. Bioprospecting potential of fast-growing endophytic bacteria from leaves of mangrove and salt-marsh plant species. Indian J Biotechnol2010;9:397-402.
41. Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P, et al. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol2002;68:2198-208. [CrossRef]
42. Silpa RS, Justin DK, Kumar VK, Mathew M, Kumar VK. Isolation and molecular characterization of Staphylococcus aureus from pigs in Thrissur district, Kerala. Pharm Innov J2018;7:139-41.
43. Martin SE, Landolo JJ. Staphylococcus Introduction. Netherlands:Elsevier;1999. 2062-5. [CrossRef]
44. El-Razik AK, Abdelrahman KA, Ahmed YF, Gomaa AM, Eldebaky HA. Direct identification of major pathogens of the bubaline subclinical mastitis in Egypt using PCR. J Anim Sci2010;10:652-60.
45. Rajpoot D S, Jha R, Goswami MP, Jha A, Godbole M, Goswami M. An isolation study of microorganisms in different stages of in vitro propagation. J Plant Dev Sci2019;11:257-64.
46. Liu Y, Li Y, Yao S, Wang H, Cao Y, Li J, et al. Diversity and distribution of endophytic bacterial community in the Noni (Morinda citrifolia L.) plan. Afr J Microbiol Res 2015;9:1649-57. [CrossRef]
47. Swamy AL, Ananth R, Yamuna SP. Phyllosphere microflora of few medicinal, garden, terrestrial and aquatic plants in and around Mysuru Districts, Karnataka. Int J Vet Sci Agric Res 2020;2:2.
48. Adeniyi O, Damilola A. Oluwasegun D, Ifeoluwa O, Olugbenga P, Olubunmi A. Microbial profile of the phyllosphere and the antimicrobial potency of Ficus vogelii extracts. J Pharm Sci Res 2020;12:191-5.
49. Oluyemi BM. Multidrug resistant Staphylococcus isolates from the phyllosphere and rhizosphere of Ficus sycomorus Linn. World Rural Observ2013;5:3.
50. Furnkranz M, Wanek W, Richter A, Abell G, Rasche F, Sessitsch A. Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. ISMEJ 2008;2:561-70. [CrossRef]
51. Ruppel S, Franken P, Witzel K. Properties of the halophyte microbiome and their implications for plant salt tolerance. Funct Plant Biol 2012;40:940-51. [CrossRef]
52. Nagaraju Y, Swamy M, Gundappagol RC, Naik NM. Response of black gram to seed biopriming with facultative halophilic bacteria under salinity. Int J Environ Climate Change 2020;10:561-71. [CrossRef]
53. Bian G, Zhang Y, Qin S, Xing K, Xie H, Jiang J. Isolation and biodiversity of heavy metal tolerant endophytic bacteria from halotolerant plant species located in coastal shoal of Nantong. Acta Microbiol Sin 2011;51:1538-47.
54. Aslam F, Ali B. Halotolerant bacterial diversity associated with Suaeda fruticosa (L.) forssk. Improved growth of maize under salinity stress. Agronomy 2018;8:131. [CrossRef]
55. Hernandez-Canseco J, Bautista-Cruz A, Mendoza SS, Aquino-Bolanos T, Sanchez-Medina PS. Plant growth-promoting Halobacteria and their ability to protect crops from abiotic stress:An eco-friendly alternative for saline soils. Agronomy 2022;12:804. [CrossRef]
56. Komaresofla BR, Alikhani HA, Etesami H, Khoshkholgh-Sima NA. Improved growth and salinity tolerance of the Halophyte salicornia Sp. by co-inoculation with endophytic and rhizosphere bacteria. Appl Soil Ecol 2019;138:160-70. [CrossRef]
57. Shultana,R, Zuan AT, Naher UA,Islam AK, Rana MM, Rashid MH, et al. The PGPR mechanisms of salt stress adaptation and plant growth promotion. Agronomy 2022;12:2266. [CrossRef]
58. Afrasayab S, Faisal M, Hasnain M. Comparative study of wild and transformed salt tolerant bacterial strains on Triticum aestivum growth under salt stress. Brazil J Microbiol 2010;41:4. [CrossRef]
59. Nagaraju Y, Mahadevswamy, Naik NM, Gowdar SB, Narayanarao K, Satyanarayanarao K. ACC deaminase-positive halophilic bacterial isolates with multiple plant growth-promoting traits improve the growth and yield of Chickpea (Cicer arietinum L.) under salinity stress. Front Agron 2021;3:681007. [CrossRef]