1. INTRODUCTION
In humans, helminth infections are among the most prevalent diseases worldwide, widely distributed in tropical and subtropical areas, affecting approximately 24% of the world’s population, mainly impacting school-aged and preschool children (568 million and 267 million, respectively) [1]. On the other hand, intestinal parasitic infection by helminths in livestock and poultry is considered a severe problem, causing significant economic losses in these productive sectors, mainly in terms of mortality, growth retardation, reduction of animal fertility, and susceptibility to other infectious diseases [2].
The strategy against these diseases is based on the administration of antiparasitic drugs. In humans, the most used is albendazole (a benzimidazole) [3]. In livestock, benzimidazoles, along with other anthelmintic drugs such as macrocyclic lactones and imidazothiazoles, have been extensively used to control gastrointestinal nematodes in cattle [3]; for trematodes, the most effective drug is triclabendazole [4].
Due to the increase in helminth resistance to these drugs and the impact on the economy and human health, it is crucial to search for new molecular models to develop new drugs against these parasites. Finding candidate compounds from plants (medicinal or not) could be a valid alternative to the problem [5]. Only a small fraction of the world’s biodiversity has been explored for bioactivity. On the other hand, over the last century, several top-selling drugs have been developed from natural products [6]. The anthelmintic properties of plants are documented within the spectrum of biological activities associated with natural products, as outlined in the scientific literature [7,8].
The Campomanesia genus consists of trees or shrubs with opposite leaves, 3–15 flowers in dichasia or solitary flowers, and fruits that are berry-shaped, globose to ovoid-rhomboidal, and can be yellow, orange, or red. The sepals are persistent, and the fruits have 1–4 seeds [9].
Recent in vivo and in vitro studies involving various species of this genus have attributed several biological activities. A survey of Campomanesia phaea O. Berg demonstrated anti-inflammatory effects and was influential in improving glucose tolerance, ameliorating dyslipidemia, and reducing fasting insulin and glucose levels [10]. Several studies on Campomanesia xanthocarpa have shown anti-inflammatory activity [11-13], anti-ulcerogenic effects [11,14], hypotensive, antioxidant, and cardioprotective properties [11]. Another study evaluated the antidiabetic effects of C. xanthocarpa O. Berg, with results suggesting its potential usefulness in managing diabetes mellitus [15]. The species Campomanesia adamantium O. Berg exhibited antidiarrheal, antimicrobial, and anti-inflammatory activity [16,17] and the ability to reduce total cholesterol and triglyceride levels in the blood of Wistar rats, as well as cytotoxic potential in Jurkat leukemia cells [15]. Anti-inflammatory properties and vasodilatory effects have been reported for Campomanesia velutina (Cambess) O. Berg [18].
Despite the use of those plants for the cases described above, to the best of our knowledge, studies of the anthelmintic potential of the genus have not been described, so this study is innovative in this field.
In this context, three species of the Campomanesia genus have been selected to study their chemical composition and probable in silico and in vitro anthelmintic activity.
2. MATERIALS AND METHODS
2.1. Plant Material
Leaves and branches of Campomanesia guazumifolia (Cambess.) O. Berg (ñandú apysa), C. xanthocarpa O. Berg var. xanthocarpa (guavirá pyta), and Campomanesia guaviroba (DC.) Kiaersk (guavirá hovy) were collected on March 29, 2017, from the Garden of Acclimatization – Faculty of Chemical Sciences, located on the University Campus in the city of San Lorenzo, Central Department, Paraguay [19]. The collected materials were identified by the Department of Botany of the Faculty of Chemical Sciences; voucher specimens were deposited in the FCQ Herbarium (Herbarium specimen: G. Delmás, D. Bazán and G. González, No. 449; G. Delmás, D. Bazán and G. González, No. 450; and G. Delmás, D. Bazán and G. González, No. 451, respectively).
The samples were dried in an oven with surrounding air at 40°C for 5 days from the collection day. After drying, the materials were pulverized to a coarse powder (No. 10 sieve) in a blade mill. Methanolic extracts were obtained using methanol (analytical grade) as solvent. The solvent was removed in a rotary evaporator under reduced pressure.
The procedure involved macerating the pulverized plant material with the solvent in a proportion of 200 g/1,000 mL for 24 h, followed by filtration. This process was repeated twice, and the obtained filtrates were combined and concentrated in the rotary evaporator.
2.2. Increasing Polarity Fractions Obtention
The methanolic extracts obtained in the previous step, dried without solvent, were subjected to liquid-liquid partitioning. Each extract was individually suspended in distilled water and partitioned with solvents of increasing polarities (hexane, dichloromethane, ethyl acetate, and 1-butanol), allowing better division into fractions. The fractions obtained from the three Campomanesia species were used in the anthelmintic activity assay.
2.3. Preliminary Phytochemical Assay
The methanolic extracts were analyzed according to the method described by Sanabria-Galindo (1983) [20] to determine the main groups of secondary metabolites present in the samples based on precipitation/coloration reactions and thin-layer chromatography. According to the abundance of formed precipitates and the intensity of the reaction coloration, the results were tabulated, indicating the higher, medium, lower, or null presence of the secondary metabolites, expressing the results as (+++), (++), (+), or (−).
2.4. Chemical Fingerprint by Liquid Chromatography-Mass Spectrometry (LC-MS)
The chemical fingerprint of the methanolic extracts from the leaves of C. guazumifolia, C. xanthocarpa, and C. guaviroba were determined using ultra-high-performance liquid chromatography (UHPLC) coupled with a mass spectrometer detector.
The analysis was conducted on a Waters ACQUITY ultra-performance liquid chromatography, coupled with a triple quadrupole mass spectrometer (QqQ) Xevo TQD (Waters, Milford, MA, USA), utilizing a Phenomenex KINETEX core-shell EVO-C18 (2.1 × 100 mm, 1.7 μm, Torrance, CA, USA) column for separations. The run time was 13 min, with a sample injection volume of 10 μL, a flow rate of 0.3 mL/min, and an oven temperature of 40°C.
The mobile phase consisted of water (A) and methanol (B), both containing 0.1% formic acid and 10 mM ammonium formate as additives. The gradient was set as follows: 0–0.7 min, 80–80% A; 0.7–3.2 min, 80–60% A; 3.2–7.6 min, 60–20% A; 7.6–8.3 min, 20–0% A; 8.3–10.4 min, 0–80% A, 10.4–13 min, 80–80% A. The samples were dissolved in LC-MS MeOH, filtered through 0.22 μm nylon syringe filters, and injected at 5 mg/mL. Ultraviolet (UV) spectra were recorded using a diode array detector, scanning absorption between 200 and 500 nm, whereas chromatograms were recorded at 254 nm.
Electrospray ionization was used as the ionization source, operating in both negative and positive modes. The mass spectrometry conditions were as follows: Source temperature, 150°C; drying gas temperature, 500°C; drying gas (N2) flow rate, 900 L/h; cone flow, 50 L/h; cone voltage, 25 V; capillary voltage, 4,000 V; collision energy, 30 V; and Ar as collision gas. MS data were acquired in full scan mode (SCAN) within an m/z range of 80–800 (scan time 0.14 s). The MS/MS analyses were carried out in daughter ion mode (scan time 0.1 s), selecting the major compounds for fragmentation. The system was operated using Waters MassLynx V4.1 software for data and qualitative analysis.
2.5. In vitro anthelminthic activity assay
Three different concentrations of methanolic extracts from three species of the genus Campomanesia (10 mg/mL, 20 mg/mL, and 40 mg/mL), as well as the fractions derived from them at a concentration of 10 mg/mL, were used, according to the method described by Pawar et al. (2014) with some modifications [7].
The samples to be assayed were dissolved in a sterile saline solution containing 5% dimethyl sulfoxide (DMSO).
Groups of three Eisenia fetida worms with lengths ranging from 4 to 6 cm and diameters from 0.2 to 0.4 cm were prepared and placed in a Petri dish in contact with the concentration of extracts. Each treatment was performed in triplicate.
The reference substance used as a positive control was the drug albendazole 10 mg/mL, which was prepared under the same conditions as the samples and similarly brought into contact with the three worms per Petri dish.
Likewise, a negative control was used, following the same procedure but using only a sterile physiological solution with 5% DMSO.
Once confronted, any movement was monitored until each worm became paralyzed. The times of paralysis and death (in minutes) were recorded and timed. To ensure paralysis, the worms were vigorously shaken in the Petri dishes. Likewise, to record the time of death, the lack of reintegration of the worms was verified by vigorously shaking them in the Petri dish and then immersing them in warm water (50°C).
Data were analyzed using analysis of variance followed by Tukey’s multiple comparison test, employing GraphPad Prism software version 8.0.1. A p-value < 0.05 was considered statistically significant.
2.6. In silico Analysis of Anthelminthic Activity
2.6.1. Target selection and characterization of potential ligand binding sites
Acetylcholinesterase (AChE) (PDB: 4PQE), γ-aminobutyric acid receptor B (GABA B) (PDB: 4MS3), and β-tubulin (PDB: 1JFF) were selected as targets for in silico simulation; these proteins were previously employed and recommended in other studies of anthelmintic activities [21-23].
Water molecules, ion structure, and co-crystallized ligands were removed from target structures using Discovery Studio Visualizer v.20 (Dassault Systèmes BIOVIA, USA). The identification of amino acids with activity probability >50% was selected as a probable ligand binding site; this selection was performed with supervised machine learning methods using GRaSP software [24].
2.6.2. Molecular docking simulations
Target structures were obtained from Protein Data Bank databases and PubChem [21-23,25,26]. The energy minimization of the compound’s molecular structure was performed using MMFF94 force fields in four steps per update with a conjugate gradient algorithm employing a 50,000 cycle step and an energy convergence criteria of 0.001 kcal/mol.Å. Partial charges and polar hydrogen atoms were also added at a physiological pH of 7.4 using Avogadro software [27].
Molecular docking assays were performed with interaction boxes of 63 × 71 × 54 Å3 for AChE, 87 × 80 × 71 Å3 for GABA B receptor, and 61 × 59 × 61 Å3 for β-tubulin were carried out using AutoDock Vina v.1 software [28]. Rivastigmine was used as a control for AChE [21], Baclofen for the GABA B receptor [22], and Albendazole for β-tubulin [29]. Values of the dissociation constant (Kd) and ligand efficiency (LE) were estimated with equation (1) and equation (2) [30,31].
Kd = e (ΔGx1000/RT) (1)
LE = -ΔG/HA (2)
Where the temperature (T) value was 310 K (Kelvin), the ideal gas constant (R) value was 1.987207 cal/mol.K, and HA was the number of heavy atoms in the structure of the compounds. Complexes were analyzed on those with ΔG lower than controls using Discovery Studio Visualizer v.20 software (Dassault Systèmes BIOVIA, USA). Bioavailability predictions according to the modified Lipinski’s rule of five (molecular weight lower than 500 g/mol, Moriguchi’s water: Octanol participation ratio lower than 4.15, hydrogen bond acceptors lower than 10, hydrogen bond donors lower than 5) [32], pharmacokinetic and toxic properties (Absorption, Distribution, Metabolism, Excretion and Toxicity [ADMET]) of the compounds were performed using the SwissADME tool and ProTox 3.0 tool [33-36].
3. RESULTS AND DISCUSSION
3.1. Preliminary Phytochemical Analysis
Phytochemical analyses of methanolic leaf extracts indicated the presence of flavonoids, saponins, steroids, and triterpenes in all three species of the genus Campomanesia. Only in C. xanthocarpa was the presence of tannins additionally detected. Table 1 shows the phytochemical profile of the three species of Campomanesia.
Table 1: Phytochemical profile of methanolic extracts from leaves of three Campomanesia species.
| Secondary metabolites | Campomanesia guazumifolia (Cambess.) O. Berg | Campomanesia xanthorcarpa | Campomanesia guaviroba |
|---|---|---|---|
| Alkaloids | - | - | - |
| Flavonoids | +++ | +++ | +++ |
| Naphthoquinones and/or anthraquinones | - | - | - |
| Tannins | - | +++ | - |
| Saponins | +++ | +++ | +++ |
| Steroids and/or triterpenes | +++ | +++ | +++ |
| Coumarins | - | - | - |
| Cardiotonic glycosides | - | - | - |
| Triterpenic lactones | - | - | - |
The results are tabulated according to the highest, lowest, or absence of the secondary metabolite with (+++), (++), (+), or (−) in decreasing order of detection.
Other studies have revealed the presence of flavonoids, phenols, and tannins in the extract of C. xanthocarpa and different species of the same genus [12,37].
3.2. Chemical Fingerprint
The UHPLC-MS analyses identified seven compounds in the methanolic extracts of three species of the genus Campomanesia. These compounds include two glycosylated flavonols, two phenolic acids, one flavanone, and two flavanone derivatives. The compounds were tentatively identified based on their pseudomolecular ions, fragmentation patterns, and UV absorption spectra.
The fingerprint results of the methanolic extracts of the three Campomanesia species are presented in Table 2.
Table 2: Fingerprint of methanolic extracts from three Campomanesia species.
| N | tR (min) | UVmax (nm) | ESI mode | M-H (m/z) | Fragments (m/z) | Molecular formula | Identification | Gb | Gz | Xt |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0,69 (Gz) | 271 | - | 215 | 169 | C7H6O5 | Gallic acid (formate adduct) | X | X | |
| 2 | 4,79 (Gz) 5,12 (1Gb) | 351 | - | 463 | 301, 179 | C21H20O12 | Quercetin hexoside | X | X | |
| 3 | 5,08 | 253, 363 | - | 301 | 284, 253, 223 | C14H6O8 | Ellagic acid | X | ||
| 4 | 5,47 (Xt) 5,71 (Gz) 5,78 (Gb) | 353 | - | 447 | 301, 179 | C21H20O11 | Quercetin-3-O-rhamnoside | X | X | X |
| 5 | 8,52 | 286, 325sh | - | 271 | 189, 151, 119 | C15H12O5 | Naringenin | X | ||
| 6 | 9,14 | 290, 340sh | - | 285 | 239, 187, 147 | C16H15O6 | Methyl-naringenin | X | ||
| 7 | 9,63 | 274 | - | 299 | 270 | C17H18O7 | Dimethyl-naringenin | X |
*Chemical composition (fingerprint) of extracts from Campomanesia Guaviroba (Gb), Campomanesia guazumifolia (Cambess.) O. Berg (Gz) y Campomanesia. xanthocarpa (Xt). sh=shoulder. UV: Ultraviolet.
An early eluting compound (1) exhibited an [M+HCOOH]-ion at m/z 215, which, upon losing formic acid (46 amu), produced a base peak at m/z 169, suggesting the presence of gallic acid [38]. This compound was previously reported for C. xanthocarpa, supporting our identification [11].
Two quercetin derivatives (compounds 2 and 4) were identified based on their UVmax at 350 nm and characteristic MS² fragments at m/z 301 and 179 [39]. Compound 2 showed a neutral loss of one hexose (162 amu), resulting in the quercetin core at m/z 301. In contrast, compound 4 exhibited a neutral loss of one rhamnose (146 amu), yielding the quercetin core at m/z 301. Therefore, compounds 2 and 4 were assigned as quercetin hexoside and quercetin rhamnoside, respectively. Both compounds were previously described in C. adamantium, which is consistent with our results. In addition, quercetin rhamnoside was previously reported in C. lineatifolia leaves [39,40] and C. guazumifolia (Cambess.) O. Berg leaves [41]. Compound 3 exhibited a pseudomolecular ion at m/z 301 and MS/MS fragments at m/z 284, 253, and 223, characteristic of ellagic acid [39].
Finally, the most retained compounds showed UV spectra related to flavanones with UVmax around 280–285 nm. Peak 5 was identified as naringenin based on its characteristic ions at m/z 271, 151, and 119 [42]. Compound 6, with a pseudomolecular ion at m/z 285, was tentatively identified as methyl naringenin, a methylated derivative (15 amu) of compound 5. The last eluting compound (7), with [M-H]? at m/z 299, lost two methyl groups (15 amu each) to yield m/z 271, characteristic of naringenin [42] and was assigned as dimethyl naringenin. Methylated flavanones were previously reported in C. adamantium fruits, supporting our findings [39].
The distribution of the detected compounds in the studied Campomanesia species is presented in Table 2.
3.3. In vitro Anthelmintic Activity
The anthelmintic activity of the methanolic extracts of three species of the genus Campomanesia was evaluated at different concentrations, namely, 10 mg/mL, 20 mg/mL, and 40 mg/mL. All tested samples showed significant differences in the paralysis and death times of E. fetida compared to the positive control. The shortest paralysis and death times were obtained with C. xanthocarpa (40 mg/mL) at 7.05 and 10.81 min, respectively; the second shortest times were from C. guazumifolia (Cambess.) O. Berg (40 mg/mL) with paralysis at 7.59 min and death at 12.55 min [Figures 1-4].
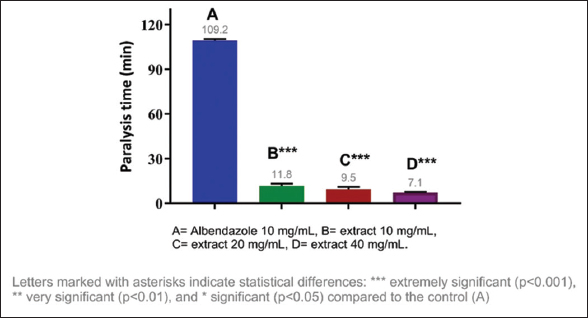 | Figure 1: Paralysis time of Eisenia fetida with the methanolic extract of Campomanesia xanthocarpa O. Berg var. xanthocarpa. [Click here to view] |
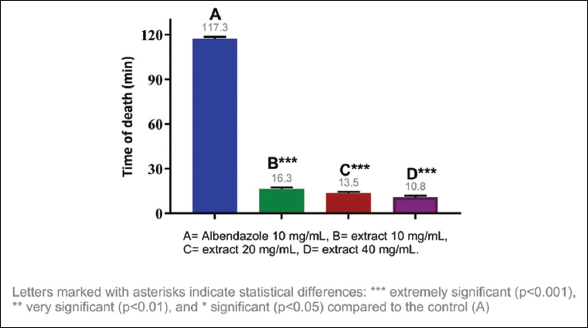 | Figure 2: Death time of Eisenia fetida with the methanolic extract of Campomanesia xanthocarpa O. Berg var. xanthocarpa. [Click here to view] |
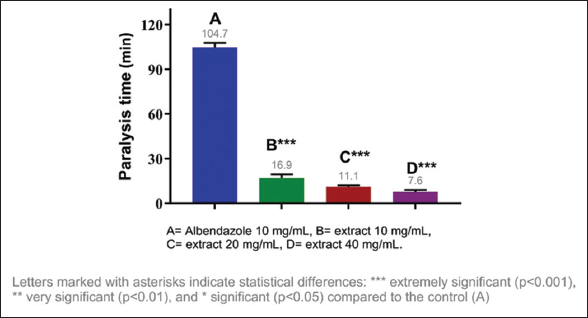 | Figure 3: Paralysis time of Eisenia fetida with the methanolic extract of Campomanesia guazumifolia. [Click here to view] |
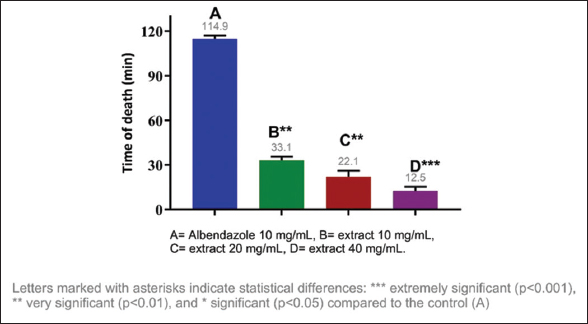 | Figure 4: Death time of Eisenia fetida with the methanolic extract of Campomanesia guazumifolia. [Click here to view] |
Considering the results obtained, it was decided to evaluate the anthelmintic activity of the fractions of each extract, identifying the most active fractions. The most active fraction was the ethyl acetate fraction (10 mg/mL) of C. guazumifolia (Cambess.) O. Berg, with paralysis times of 8.19 min and death at 9.30 min, followed closely by similar time values of the hexane and chloroform fractions of C. xanthocarpa, both at concentrations of 10 mg/mL. The second most active fraction that caused paralysis was the hexane fraction (9.36 min), and the second most active fraction that caused death was the chloroform fraction (12.41 min) [Figures 5-8]. The results of the anthelmintic activity are shown in Table 3.
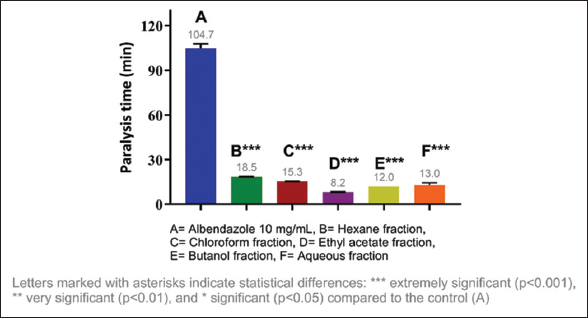 | Figure 5: Paralysis time of Eisenia fetida with the fractions of Campomanesia guazumifolia. [Click here to view] |
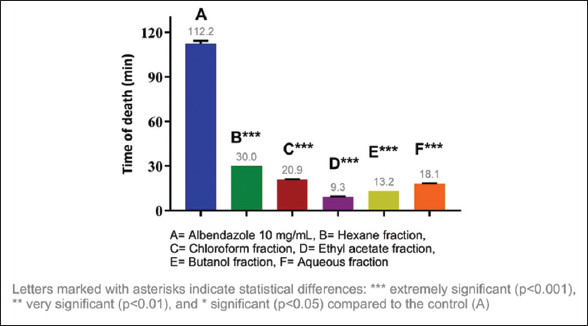 | Figure 6: Death time of Eisenia fetida with the fractions of Campomanesia guazumifolia. [Click here to view] |
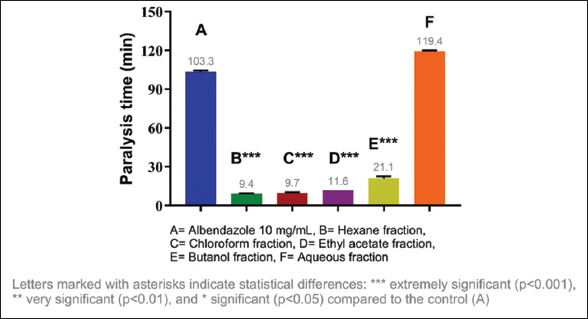 | Figure 7: Paralysis time of Eisenia fetida with the fractions of Campomanesia xanthocarpa O. Berg var. xanthocarpa. [Click here to view] |
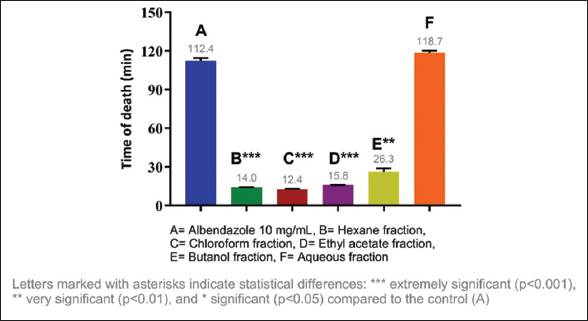 | Figure 8: Death time of Eisenia fetida with the fractions of Campomanesia xanthocarpa O. Berg var. xanthocarpa. [Click here to view] |
Table 3: Summary of the anthelmintic activity assay results of three Campomanesia species.
| Species | Extracts | Fractions | ||||
|---|---|---|---|---|---|---|
| Paralysis time | Death time | Concentration | Paralysis time | Death time | Fraction | |
| Campomanesia guaviroba | 12,5 | 21,9 | 40 mg/mL | 13,3 | 14,1 | Hexane fraction |
| Campomanesia guazumifolia (Cambess.) O. Berg | 7,6 | 12,5 | 40 mg/mL | 8,2 | 9,3 | Ethyl acetate fraction |
| Campomanesia xanthocarpa | 7,1 | 10,8 | 40 mg/mL | 9,4 | 12,4 | Hexane fraction/Chloroform fraction |
The literature review of works using the same model to test anthelmintic activity showed that a 2010 study conducted in India evaluated the anthelmintic potential of the ethanolic and aqueous extract of Holoptelea integrifolia bark against Eisenia fetida, determining the paralysis and death times of the worms [43].
The same methodology has been reported for determining the anthelmintic activity of Piper betel using adult Eisenia fetida, given that these earthworms have physiological and anatomical similarities to human intestinal parasites [44].
No anthelmintic tests were found for the species studied here in the literature, so the data reported here are novel.
The methanolic extract with the shortest anthelmintic action time was C. xanthocarpa. However, the most active fraction found was the ethyl acetate fraction of C. guazumifolia (Cambess.) O. Berg.
The three Campomanesia species showed excellent anthelmintic activity compared to the conventional drug, demonstrated by the drastic reduction in paralysis and death times relative to albendazole. This could open the possibility of developing new antiparasitic drugs.
3.4. In silico Analysis of Anthelminthic Activity
The structural analysis of AChE protein evidenced as a probable ligand binding site the residues Tyr72, Tyr119, Gly120, Gly121, Gly122, Tyr124, Glu202, Ser203, Trp236, Trp286, Leu289, Phe295, Arg296, Phe297, Tyr337, Phe338, Tyr341, His447, and Tyr449. Other investigations reported these residues in the catalytic site and its peripheral region in AChE [45,46] [Figure 9a].
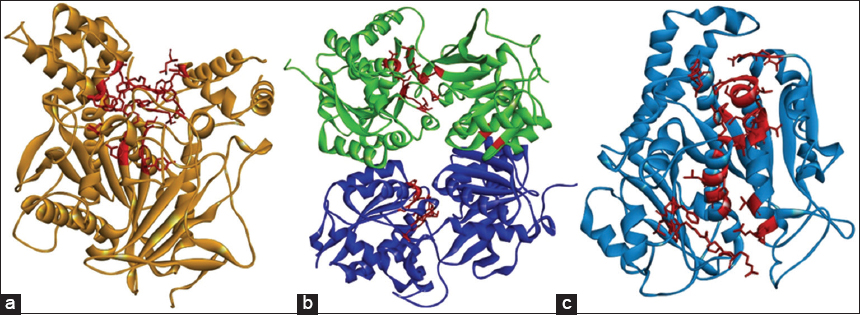 | Figure 9: Sites of highest ligand binding probability (red segment). (a) Acetylcholinesterase. (b) γ-aminobutyric acid receptor B receptor. (c) Tubulin β. [Click here to view] |
The residues recorded as active in GABA B receptor structure were Trp65, Cys103, Gly128, Cys129, Ser130, Ser131, Gly151, Ser152, Ser153, Ser154, Thr159, Arg168, Thr169, Arg174, Thr175, Ser178, Thr202, Ser231, Phe232, Phe319, Gly356, Tyr357, Asp360, Thr412, and Arg422. Furthermore, these residues were previously described in the active site of the receptor [47]. Regarding β-tubulin, the residues with a higher probability to interact with compounds were Ala9, Gly10, Gln11, Cys12, Gly13, Asn14, Gln15, Val23, Asp26, Glu27, Ala99, Gly100, Asn101, Thr138, Ser140, Gly142, Gly143, Gly144, Thr145, Gly146, Ser147, Gly148, Asp179, Glu183, Tyr224, Asn228, His229, Leu230, Ala233, Ser236, Phe272, Pro274, Leu275, Thr276, Arg278, Pro360, Arg369, and Gly370, these residues are located into an active site previously reported [48] [Figure 9b and c, Table 4].
Table 4: Results obtained from molecular docking simulations.
| Compounds | PubChem CID | ΔG (kcal.mol-1) | Kdcalc (μM) | LE (kcal.mol-1. heavy atom-1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AChE | GABA B | β-Tubulin | AChE | GABA B | β-Tubulin | AChE | GABA B | β-Tubulin | ||
| Rivastigmin | 77991 | −6.9 | - | - | 13.27 | - | - | - | - | - |
| Baclofen | 2284 | - | −6 | - | - | 57.41 | - | - | - | - |
| Albendazole | 2082 | - | - | −6.4 | - | - | 29.94 | - | - | - |
| Gallic acid | 46780424 | −9.4 | −9.7 | −8.9 | 0.23 | 0.14 | 0.51 | 0.29 | 0.30 | 0.28 |
| Quercetin-3-O-ramnoside | 5280459 | −8.3 | −9.3 | −8.3 | 1.36 | 0.27 | 1.36 | 0.26 | 0.29 | 0.26 |
| Ellagic acid | 5281855 | −9.8 | −7.8 | −8.1 | 0.12 | 3.07 | 1.88 | 0.45 | 0.35 | 0.37 |
| Naringenin | 439246 | −9.1 | −7.5 | −7.9 | 0.37 | 5.00 | 2.61 | 0.46 | 0.38 | 0.39 |
| Methyl-naringenin | 85037502 | −7.3 | −7.4 | −7.5 | 6.92 | 5.88 | 5.00 | 0.35 | 0.35 | 0.36 |
| 6,4’- Dimethyl-naringenin | 21721822 | −8.4 | −7.5 | −7.0 | 1.15 | 5.00 | 11.28 | 0.38 | 0.34 | 0.32 |
| Quercetin 3-D-glucoside | 5280804 | −8.1 | −9.7 | −7.3 | 1.88 | 0.14 | 6.92 | 0.25 | 0.29 | 0.22 |
Kdcalc: Calculated dissociation constant; LE: Ligand efficacy, AChE: Acetylcholinesterase.
Molecular docking simulations performed with AChE evidenced that all compounds present lower ΔG values than those detected with the Rivastigmin molecules, with ranges from −7.3 to −9.8 kcal/mol. Also, Kd values ranged from 0.12 to 6.92 μM, being lower than rivastigmine; it is essential to note that the dissociation constant gives quantitative information about the interaction affinity of the ligand to the protein target [30]. LE values were recorded between 0.25 and 0.46, with the optimal LE values being those lower than 0.3. However, it is important to denote that this parameter strictly depends on the molecular structure of compounds and ΔG values [31,49] [Table 4].
Complexes selected with favorable ΔG, Kd, and LE values were those formed with the ligands gallic acid, quercetin-3-O-rhamnoside, and ellagic acid [Table 4]. The AChE: Gallic acid complex showed the formation of π-π stacking interactions between the compound and Trp286 residue, as the formation of π-anion interactions with Asp74 residue, and Van der Waals type interactions with residues present in the binding pocket site [Figure 10a].
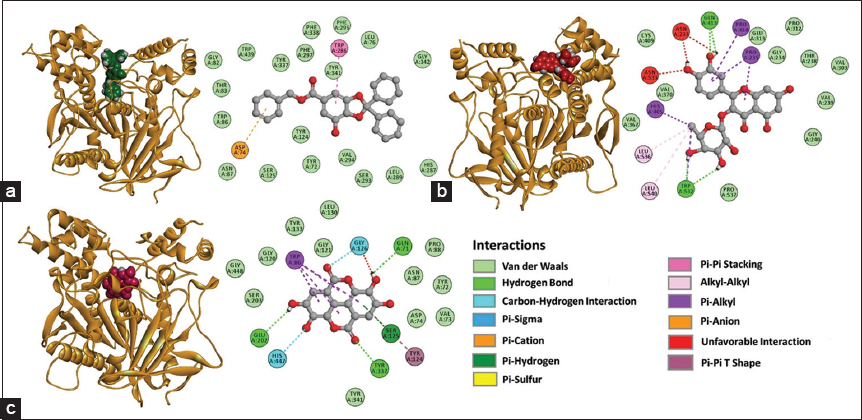 | Figure 10: 3D and 2D representations of the complexes with favorable ΔG values formed with the acetylcholinesterase (AChE), (a) AChE: Gallic acid, (b) AChE: Quercetin-3-O-rhamnoside, (c) AChE: Ellagic acid. [Click here to view] |
In AChE: quercetin-3-O-rhamnoside complex, hydrogen bonds between compound, Gln413, and Trp532 residues were recorded, and interactions between alkyl groups with Leu536 and Leu540 were evidenced. Furthermore, the formation of π-alkyl interactions with Pro410, Pro235, and His405 residues were registered. Van der Waals interactions with residues of the pocket site and unfavorable interactions between hydrogen bond donor groups of compound Asn233 and Asn533 were identified [Figure 10b].
The AChE: Ellagic acid complex evidenced a hydrogen bond between the compound and Glu202, Tyr337, and Gln71 residues, unconventional carbon-hydrogen interactions with Gly126 and His447 residues, and the formation of interactions between π orbitals of Trp86 and the aliphatic segment of the compound, and T-shaped interactions between π orbitals with Tyr124 were recorded. Unfavorable interactions with Gly126, interactions between π orbitals of the compound and the hydroxyl group (-OH) of the side chain of Ser125, and Van der Waals interactions with residues of the binding pocket site were also detected [Figure 10c].
Simulations performed with the GABA B receptor showed that all compounds evaluated had lower ΔG values than the baclofen (control), with ranges between −7.4 and −9.7 kcal/mol. The estimated Kd values registered ranged between 0.14 and 5.88 μM; these values were lower than the control. Likewise, the ligand efficacy values were maintained between 0.29 and 0.38. Complexes formed with gallic acid, quercetin-3-O-rhamnoside, and ellagic acid were selected in [Table 4].
In the GABA B receptor: Gallic acid complex, the presence of hydrogen bonds between compound and Ser140, Lys168, and Gln197 residues, π-stacking interactions between compound, Tyr250, and Phe227 residues, interactions between π orbitals and the aliphatic side chain of residues Pro105, Leu135, and Lys168, interactions between π orbitals and the sulfhydryl group of Cys103 were detected. Furthermore, Van der Waals interactions with residues of the pocket site were observed and registered in [Figure 11a]. In the GABA B receptor: Quercetin-3-O-rhamnoside complex were recorded hydrogen bonds with residues Arg162 and Glu200, unconventional carbon-hydrogen interactions with Thr199, interactions between the π orbitals and aliphatic side chain of Val162, interactions between π orbitals and hydrogens of the side chain of Ser140 and Gln206, Van der Waals interactions with residues of the pocket binding site were also detected [Figure 11b].
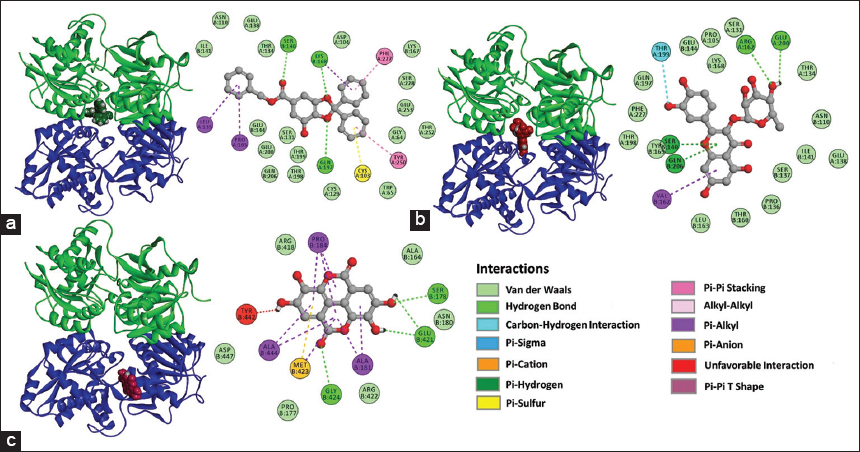 | Figure 11: 3D and 2D representations of the complexes with favorable ΔG values formed with the γ-aminobutyric acid receptor B (GABA B) receptor, (a) GABA B receptor: Gallic acid (b) GABA B receptor: Quercetin-3-O-rhamnoside, (c) GABA B receptor: Ellagic acid. [Click here to view] |
The GABA B receptor: ellagic acid complex evidenced hydrogen bonds between the compound Ser178, Glu421, and Gly424, interactions between alkyl chains with Pro184, Ala444, and Ala181, π-anion interactions with Met423, unfavorable interactions with Tyr442, and Van der Waals interactions with residues of the binding site were also recorded [Figure 11c].
Simulations with compounds and β-tubulin showed that all of them showed lower ΔG values than albendazole, ranging between −7.0 and −8.9 kcal/mol. Kd estimated values ranged from 0.51 to 11.28 μM, and LE parameter values ranged from 0.22 to 0.39 [Table 4].
The β-tubulin: Gallic acid complex evidenced hydrogen bonds with Thr145, Gly144, and Ser140, unconventional carbon-hydrogen interactions with residue Ala99, π-anion interactions with Asp179, interactions between π orbitals of the compound and aliphatic chain of Cys12. Furthermore, unfavorable interactions with Gly13 and Van der Waals interactions with residues of the binding site were recorded. However, in β-tubulin: Quercetin-3-O-rhamnoside complex, hydrogen bonds with Glu183, Ser140, Gly146, Thr145, Gln11, and Glu71 were registered; on the other hand, Van der Waals interactions with residues of the binding pocket site were also recorded [Figure 12a and b].
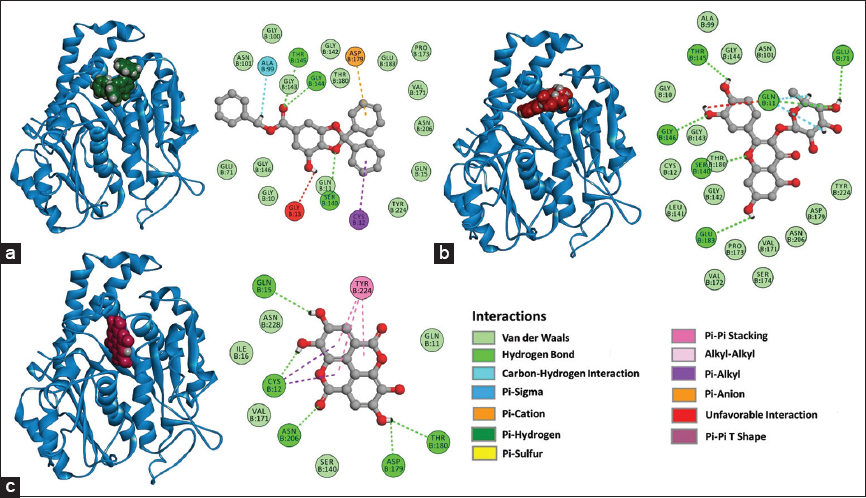 | Figure 12: 3D and 2D representations of the complexes with favorable ΔG values formed with the tubulin β, (a) Tubulin β: Gallic acid, (b) Tubulin β: Quercetin-3-O-rhamnoside, (c) Tubulin β: Ellagic acid. [Click here to view] |
In the β-tubulin: Ellagic acid complex, the hydrogen bond formation with Gln15, Cys12, Asn206, Asp179, and Thr180 was identified, and π-π stacking interactions were also detected with Tyr224. Interactions between π orbitals of the compound and the aliphatic chain of Cys12 were recorded, and Van der Waals interactions with residues of the binding pocket site were also observed in [Figure 12c].
AChE enzyme plays a crucial role in acetylcholine hydrolysis in neuromuscular junctions to maintain the physiology of the nervous system for normal motility of organisms; due to this, it is considered a biomarker related to neurotoxicity [50,51]. Furthermore, both the GABA B receptor and β-tubulin have been reported that inhibitions of their protein activities could lead to paralysis of the body wall muscle and thus to the death of the organism [21,52,53].
Results obtained in this study suggest that gallic acid, quercetin-3-O-rhamnoside, and ellagic acid molecules could bind to the AChE, GABA B receptor, and β-tubulin structures, destabilizing the protein’s functional conformation and generating a possible neurotoxic effect due to the loss of its physiological function [22].
Bioavailability predictions according to the modified Lipinski’s rule demonstrated that gallic acid, quercetin-3-O-rhamnoside, and quercetin-3-O-glucoside present violations to this rule, where the gallic acid MLOGP value was higher than 4.15. However, quercetin-3-O-rhamnoside and quercetin-3-O-glucoside showed a number of hydrogen bond acceptors >10 and a number of hydrogen bond donors higher than five, [Table 5].
Table 5: Compliance of the compounds with the modified Lipinski’s rule of five.
| Compounds | Molecular weight (g.mol-1) | H acceptors | H donors | MLOGP | Number of violations |
|---|---|---|---|---|---|
| Gallic acid | 424.44 | 5 | 1 | 4.16 | 1 |
| Quercetin-3-O-ramnoside | 448.38 | 11 | 7 | −1.84 | 2 |
| Ellagic acid | 302.19 | 8 | 4 | 0.14 | 0 |
| Naringenin | 272.25 | 5 | 3 | 0.71 | 0 |
| Methyl-naringenin | 286.28 | 5 | 3 | 0.96 | 0 |
| 6,4’-Dimethyl-naringenin | 300.31 | 5 | 1 | 1.2 | 0 |
| Quercetin 3-D-glucoside | 464.38 | 12 | 8 | −2.59 | 2 |
Moriguchi coefficient of water: Octanol partition.
According to the modified Lipinski’s rule of five, two violations could mean that the molecule may be less effective for oral use as a possible drug [54].
Regarding pharmacokinetic properties (ADMET), most compounds show high gastrointestinal absorption, except quercetin-3-O-rhamnoside and quercetin-3-O-glucoside. All the compounds show inhibitory capacities of any cytochrome P450 isoforms, except quercetin-3-O-rhamnoside and quercetin-3-O-glucoside, which did not show any predicted inhibition activities. As for the predicted toxicities, most of them showed possible carcinogenic activities, naringenin showed cytotoxic activities, and quercetin-3-O-glucoside did not show any predicted toxic activities [Table 6].
Table 6: Predictions of pharmacokinetic and toxic properties (ADMET) of the compounds.
| Compounds | GI Abs. | Cytochrome P450 Inhibitor | Toxicity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CYP1A2 | CYP2C19 | CYP2C9 | CYP2D6 | CYP3A4 | Mut | Carc | Cyt | Hep | ||
| Gallic acid | High | No | Yes | Yes | Yes | No | Inactive | Active | Inactive | Inactive |
| Quercetin-3-O-ramnoside | Low | No | No | No | No | No | Inactive | Active | Inactive | Inactive |
| Ellagic acid | High | Yes | No | No | No | No | Inactive | Active | Inactive | Inactive |
| Naringenin | High | Yes | No | No | No | Yes | Inactive | Inactive | Active | Inactive |
| Methyl-naringenin | High | Yes | No | No | No | Yes | Inactive | Inactive | Inactive | Inactive |
| 6,4’- Dimethyl-naringenin | High | Yes | Yes | No | Yes | Yes | Inactive | Active | Inactive | Inactive |
| Quercetin 3-D-glucoside | Low | No | No | No | No | No | Inactive | Inactive | Inactive | Inactive |
GI Abs.: Gastrointestinal absorption, Mut: Mutagenicity, Carc: Carcinogenicity, Cyt: Cytotoxicity, Hep: Hepatotoxicity.
4. CONCLUSION
Secondary metabolite groups were identified through phytochemical analysis employing precipitation and coloration reactions. Results indicated the presence of flavonoids, saponins, steroids, and triterpenes in methanolic extracts derived from the leaves of three species within the genus Campomanesia. Notably, tannins were exclusively detected in C. xanthocarpa, highlighting its potential pharmacological significance and warranting further investigation as a promising source for novel drug development.
LC/MS facilitated the determination of the chemical fingerprint of the extracts, revealing two glycosylated flavonols, two phenolic acids, one flavanone, and two derivatives based on their molecular characteristics. Among these, quercetin-3-O-rhamnoside was consistently identified across all three species.
The extracts demonstrated considerable anthelmintic activity against Eisenia fetida, evidenced by significant variances in both paralysis and mortality times compared to the positive control, with C. xanthocarpa exhibiting the most pronounced effect. Furthermore, the ethyl acetate fraction of C. guazumifolia (Cambess.) O. Berg exhibited the highest activity among the samples tested. In silico analysis suggested that gallic acid, quercetin-3-O-rhamnoside, and ellagic acid may interact with AChE, GABA B receptor, and β-tubulin, proposing potential mechanisms underpinning their anthelmintic action.
All three Campomanesia species revealed robust anthelmintic activity compared to albendazole, indicating their potential utility as sources for the development of new anti-parasitic agents.
5. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
6. FUNDING
There is no funding to report.
7. CONFLICTS OF INTEREST
The authors report no financial or other conflicts of interest in this work.
8. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
9. DATA AVAILABILITY
All the data are available with the authors and shall be provided upon request.
10. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors, and the reviewers. This journal remains neutral about jurisdictional claims in published institutional affiliation.
11. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declare that they have not used artificial intelligence (AI)-tools for writing and editing the manuscript, and no images were manipulated using AI.
REFERENCES
1. WHO. Available from:https://www.who.int/news-room/fact-sheets/detail/soil-transmitted-helminth-infections [Last accessed on 2022 Jan 10].
2. Fernandes MZ, Fernandes RM, Brito DR, Borba HR. Anthelmintic effect of aqueous and ethanolic extracts of Annona squamosa L. (sugar apple) on the nematode Ascaridia galli. Rev Brasil Plant Med 2009;11:124-9.
3. De Fátima Leal Pereira L, Duarte ER, Bastos GA, De Oliveira Vasconcelos V, Leite Costa EG, De Jesus Mendes L, et al. Helminthiasis control in calves raised in a hot Semi-arid area. Rev Mex Cien Pecu 2019;10:30-51.
4. Chávez VA, Sánchez RL, Arana DC, Suárez AF. Anthelmintic resistance and prevalence of bovine fasciolosis in dairy cattle in Jauja, Perú. Rev Investig Vet Perú2012;23:90-7.
5. Ferreira SB, Dantas IC, Catão RM. Evaluation of the antimicrobial activity of the essential oil of. Rev Brasil Plant Med 2015;16:225-30.
6. Sarker SD, Latif Z, Gray A. Natural Product Isolation. 2nd ed. New Jersey:Humana Press;2005. 6.
7. Pawar SD, Patil YB, Premchandani LA, Borse SL, Borse LB, Pawar SP. Study of anthelmintic activity of chloroform extract of Tinospora cordifolia. World J Pharm Pharm Sci 2014;3:2253-68.
8. Bazán D, Lopez E, Caceres A, Degen R, Alvarenga N. In vitro anthelmintic activity of methanol extracts and fractions of two amphilophium species against Eisenia fetida. J Appl Biol Biotechnol 2020;8:98-102.
9. De Oliveira MI, Funch LS, Landrum LR. Flora of bahia:Campomanesia (Myrtaceae). sitientibus series. Biol Sci 2012;12:91-107.
10. Lorençoni MF, Figueira MM, Toledo e Silva MV, Pimentel Schmitt EF, Endringer DC, Scherer R, et al. Chemical composition and anti-inflammatory activity of essential oil and ethanolic extract of Campomanesia phaea (O. Berg.) Landrum leaves. J Ethnopharmacol 2020;252:112562.
11. Sant'Anna LS, Merlugo L, Ehle CS, Limberger J, Fernandes MB, Santos MC, et al. Chemical composition and hypotensive effect of Campomanesia xanthocarpa. Evid Based Complement Altern Med 2017;2017:1591762.
12. Da Silva ÉR, Salmazzo GR, da Silva Arrigo J, Oliveira RJ, Kassuya CA, Cardoso CA. Anti-inflammatory evaluation and toxicological analysis of Campomanesia xanthocarpa. Inflammation 2016;39:1462-8.
13. Klafke JZ, Pereira RL, Hirsch GE, Parisi MM, Porto FG, de Almeida AS, et al. Study of oxidative and inflammatory parameters in LDLr-KO mice treated with a hypercholesterolemic diet:Comparison between the use of Campomanesia xanthocarpaand acetylsalicylic acid. Phytomedicine 2016;23:1227-34.
14. Markman BE, Bacchi EM, Kato ET. Antiulcerogenic effects of Campomanesia xanthocarpa. J Ethnopharmacol 2004;94:55-7.
15. Espindola PP, Rocha PS, Carollo CA, Schmitz WO, Pereira ZV, Vieira MC, et al. Antioxidant and antihyperlipidemic effects of Campomanesia adamantium. Oxidat Med Cell Longev 2016;2016:7910340.
16. Ferreira LC, Grabe-Guimarães A, de Paula CA, Michel MC, Guimarães RG, Rezende SA, et al. Anti-inflammatory and antinociceptive activities of Campomanesia adamantium. J Ethnopharmacol 2013;145:100-8.
17. Santos SM, Nascimento KF, Pereira ZV, Line JD, Junior PC, Marangoni JA, et al. The ethnopharmacological literature:An Analysis of the scientific landscape in the Cerrado in Central-Western Brazil. J Agric Sci 2020;12:307.
18. Michel MC, Guimarães AG, Paula CA, Rezende SA, Sobral ME, Saúde Guimarães DA. Extracts from the leaves of Campomanesia velutina inhibits production of LPS/INF-g induced inflammatory mediators in J774A.1 cells and exerts anti-inflammatory and antinociceptive effects in vivo. Rev Brasil Farmacogn 2013;23:927-36.
19. Martins-da-Silva RC. In:Oriental EA, editor. Coleta e Identificação de Espécimes Botânicos. Primera. Belén:Embrapa Eastern Amazon;2002. 40.
20. Sanabria-Galindo AS. Análisis Fitoquímico Preliminar:Metodología y su Aplicación en La Evaluación de 40 Plantas de la Familia Compositae. Colombia:Universidad Nacional de Colombia (Bogota), Departamento de Farmacia;1983. 111.
21. Choudhary N, Khatik GL, Choudhary S, Singh G, Suttee A. In vitro anthelmintic activity of Chenopodium album and in-silico prediction of mechanistic role on Eisenia foetida. Heliyon 2021;7:e05917.
22. He F, Liu R, Tian G, Qi Y, Wang T. Ecotoxicological evaluation of oxidative stress-mediated neurotoxic effects, genetic toxicity, behavioral disorders, and the corresponding mechanisms induced by fluorene-contaminated soil targeted to earthworm (Eisenia fetida) brain. Sci Total Environ 2023;871:162014.
23. Mowla TE, Zahan S, Sami SA, Naim Uddin SM, Rahman M. Potential effects and relevant lead compounds of Vigna mungo (L.) Hepper seeds against bacterial infection, helminthiasis, thrombosis and neuropharmacological disorders. Saudi J Biol Sci 2022;29:3791-805.
24. Santana CA, Silveira SA, Moraes JP, Izidoro SC, de Melo-Minardi RC, Ribeiro AJ, et al. GRaSP:A graph-based residue neighborhood strategy to predict binding sites. Bioinformatics 2020;36(Supplement_2):i726-34.
25. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein data bank. Nucl Acids Res 2000;28:235-42.
26. Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucl Acids Res 2016;44:D1202-13.
27. Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro:An advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 2012;4:17.
28. Trott O, Olson AJ. AutoDock Vina:Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455-61.
29. Horton J. Albendazole:A review of anthelmintic efficacy and safety in humans. Parasitology 2000;121(S1):S113-32.
30. Choudhury A, Das NC, Patra R, Bhattacharya M, Ghosh P, Patra BC, et al. Exploring the Binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis:An in silico approach. Future Virol 2021;16:277-91.
31. Onawole AT, Kolapo TU, Sulaiman KO, Adegoke RO. Structure based virtual screening of the Ebola virus trimeric glycoprotein using consensus scoring. Comput Biol Chem 2018;72:170-80.
32. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2012;64:4-17.
33. Daina A, Michielin O, Zoete V. ILOGP:A simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J Chem Inform Model 2014;54:3284-301.
34. Daina A, Michielin O, Zoete V. SwissADME:A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7:42717.
35. Daina A, Zoete V. A Boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. Chem Med Chem 2016;11:1117-21.
36. Banerjee P, Kemmler E, Dunkel M, Preissner R. ProTox 3.0:A webserver for the prediction of toxicity of chemicals. Nucl Acids Res 2024;2024:gkae303.
37. Alqethami A, Aldhebiani AY. Medicinal plants used in Jeddah, Saudi Arabia:Phytochemical screening. Saudi J Biol Sci 2021;28:805-12.
38. Leandro FD, Cabral LD, Machado TM, Koolen HH, da Silva FM, Guilhon-Simplicio F, et al. Dereplication and evaluation of the antinociceptive and anti-inflammatory activity of hydroethanolic extract of leaves from Campomanesia xanthocarpa. Nat Prod Res 2021;35:5549-53.
39. Araújo LC, Leite NR, Rocha PS, Baldivia DS, Agarrayua DA, Ávila DS, et al. Campomanesia adamantium O Berg. fruit, native to Brazil, can protect against oxidative stress and promote longevity. PLOS One 2023;18:e0294316.
40. Neves NC, de Mello MP, Zaidan I, Sousa LP, Braga AV, Machado RR, et al. Campomanesia lineatifolia Ruiz and Pavón (Myrtaceae):Isolation of major and minor compounds of phenolic-rich extract by high-speed countercurrent chromatography and anti-inflammatory evaluation. J Ethnopharmacol 2023;310:116417.
41. Catelan TB, Santos Radai JA, Leitão MM, Branquinho LS, Vasconcelos PC, Heredia-Vieira SC, et al. Evaluation of the toxicity and anti-inflammatory activities of the infusion of leaves of Campomanesia guazumifolia (Cambess.) O. Berg. J Ethnopharmacol 2018;226:132-42.
42. Gómez-Romero M, Zurek G, Schneider B, Baessmann C, Segura-Carretero A, Fernández-Gutiérrez A. Automated identification of phenolics in plant-derived foods by using library search approach. Food Chem 2011;124:379-86.
43. Kaur S, Kumar B, Puri S, Tiwari P, Divakar K. Comparative study of anthelmintic activity of aqueous and ethanolic extract of bark of Holoptelea integrifolia. Int J Drug Dev Res 2010;2:758-63.
44. Sudrik S, Fegade S, Shinde M. Anthelmintic activity of Piper betle Linn, (Paan/Nagavalli) aqueous extract. Res J Pharm Biol Chem Sci 2012;3:467-70.
45. Dvir H, Silman I, Harel M, Rosenberry TL, Sussman JL. Acetylcholinesterase:From 3D structure to function. Chem Biol Interact 2010;187:10-22.
46. Silman I, Sussman JL. Acetylcholinesterase:How is structure related to function?Chem Biol Interact 2008;175:3-10.
47. Geng Y, Bush M, Mosyak L, Wang F, Fan QR. Structural mechanism of ligand activation in human GABAB receptor. Nature 2013;504:254-9.
48. Löwe J, Li H, Downing KH, Nogales E. Refined structure of ab-tubulin at 3.5 Åresolution1. J Mol Biol 2001;313:1045-57.
49. Hopkins AL, KeserüGM, Leeson PD, Rees DC, Reynolds CH. The role of ligand efficiency metrics in drug discovery. Nat Rev Drug Discov 2014;13:105-21.
50. Gao B, Zhao S, Zhang Z, Li L, Hu K, Kaziem AE, et al. A potential biomarker of isofenphos-methyl in humans:A chiral view. Environ Int 2019;127:694-703.
51. Karthivashan G, Park SY, Kim JS, Cho DY, Ganesan P, Choi DK. Comparative studies on behavioral, cognitive and biomolecular profiling of ICR, C57BL/6 and its sub-strains suitable for scopolamine-induced amnesic models. Int J Mol Sci 2017;18:1735.
52. Chakraborty S, Mandal C, Bhattacharya S, Pal R, Ghosh B, Saha U, et al. An Overview on albendazole:Anthelmintic agent. Int J Res Appl Sci Eng Technol 2022;10:985-8.
53. Hondebrink L, Hermans EJ, Schmeink S, van Kleef RG, Meulenbelt J, Westerink RH. Structure-dependent inhibition of the human a1b2g2 GABAA receptor by piperazine derivatives:A novel mode of action. Neurotoxicology 2015;51:1-9.
54. Roskoski R. Rule of five violations among the FDA-approved small molecule protein kinase inhibitors. Pharmacol Res 2023;191:106774.