1. INTRODUCTION
Breastmilk is a complex biological nutrient that supports babies’ growth and development. The success of lactation requires the expansion and differentiation of extensive breast tissues during pregnancy, followed by a sufficient amount of breast milk production post-labor. These two processes require mechanism coordination in nutrition transport, breastmilk production, and mammary gland secretion and are supported by several molecular events regulated by reproduction hormones [1].
The prolactin hormone plays a key role in the mammary glands’ growth and development, particularly in mammogenesis, lactogenesis, and galactopoesis [2]. In lactogenesis, prolactin stimulates the absorption of some amino acids, milk and protein synthesis (such as α-lactalbumin and casein), glucose absorption, and lactose and milk fat synthesis [2-4]. Prolactin, together with progesterone and other metabolic hormones like insulin, is needed in the differentiation process of mammary epithelial cells that can synthesize and secrete the specific components of breastmilk [5]. High prolactin level during breast feeding can influence metabolism, to supply glucose and fat for milk production [6].
One of the prolactin’s functions during breastfeeding is to provide nutrition in the breast milk’s composition [7-10]. Prolactin can enhance arginase’s activity and the mammary tissue’s polyamine transport rate and stimulate decarboxylase activities. That mechanism can improve polyamine synthesis, which is necessary for milk production. The polyamine stabilizes the membrane structure, improves transcriptional and translational activities, and regulates enzymes [1].
Prolactin also activates the STAT5 pathway, which induces β-casein gene expression and the morphogenesis and differentiation of alveolar cells in the mammary gland [7-9]. Thus, prolactin is the primary hormone that controls the function and growth process of the mammary gland [9]. The prolactin gene in the mammary gland is expressed in the mid to late gestation period as the mammary gland develops, then continuously expressed throughout the lactation period, and finally decreases in the early weaning phase [11].
Various factors influenced the success of the lactation process. Internal factors, such as genetic variations, affect breast milk quantity and quality, while external factors consist of nutritional status, partner support, stress levels, and the baby’s attachment to the mother [5]. The frequently emerging issues causing the failure in immediate breastfeeding after labor and the lack of breastmilk production are caused by nutritional and non-nutritional factors, including hormonal issues, parity, pregnancy, age, and psychological factors [12-14].
Forinash et al. [15] recommend non-pharmacological therapy using herbal galactagogues (plants that increase breast milk production) to boost breast milk production. The most commonly used herbs are fenugreek (Trigonella foenum-graecum), milk thistle (Silybum marianum), Shatavari (Asparagus racemosus), alfalfa (Medicago sativa), blessed thistle (Cnicus benedictus), goat’s rue (Galega officinalis), fennel (Foeniculum vulgare), and brewer’s yeast (Saccharomyces cereviceae).
Using galactagogue in diet during lactation can be an alternative [16] to increase breast milk production [17]. Moreover, it can also affect breast milk’s nutritional composition [13-16]. In various cultures, the knowledge of herbal galactagogues is passed down from generation to generation. Papaya (Carica papaya) leaves have been used by Indonesian society to increase breast milk production. However, the scientific empirical evidence and the mechanism of action have yet to be elucidated. Papaya leaves contain alkaloids (carpain and pseudocarpain), enzymes (papain, chymopapain, cystatin), tocopherols, ascorbic acid, tannins, nicotinic acid, saponins, peonidin, chlorogenic acid, coumarin compounds, phenolic compounds (caffeic acid, p-coumaric acid), and protocatechuic acid as the major phytochemicals and flavonoids [18-24]. The extract of juvenile papaya leaves contains alkaloids, phenols, flavonoids, and several amino acids [25]. According to Miean and Mohamed [26], the flavonoid level in juvenile papaya leaves was 126 mg/100 g, with quercetin and kaempferol as the main compounds.
Flavonoids are widely found in vegetables, fruits, tea, and chocolate; these components act as nutraceuticals [27]. The previous studies reported that galactagogue herbs, such as fenugreek and malunggay (Moringa oelifera), are known to contain flavonoids as the main phytochemical [28,29]. Based on the liquid chromatography mass spectroscopy (LCMS)/MS analysis, it is known that papaya leaves also contain flavonoids (unpublished data). An in vivo study has shown that papaya leaves can increase plasma prolactin levels in lactating rats [30]. This study aims to explain the molecular mechanism of active compounds from papaya leaves in regulating or influencing the production of the prolactin hormone using a docking technique as a tool for virtual screening and optimizing the combination of active compounds (ligands) with estrogen, leptin, and aromatase as the target proteins.
Estrogen is a hormone produced by the ovaries, soluble in the membrane, and interacts with the intracellular receptors that stimulate the development of the epithelium in the mammary glands. This hormone is needed to produce lactation-competent glands [31]. Leptin is a peptide that regulates food intake, body mass, and reproductive function through a negative feedback mechanism between the adipose tissue and the hypothalamus [32]. Meanwhile, aromatase is an estrogen synthetase [33] that converts androgen into estrogen [34].
2. MATERIALS AND METHODS
2.1. Bioactive Compounds Selection
C. papaya leaf extract was analyzed using the LCMS method. Then, data were compared with the list of bioactive compounds from the KnapSack Kanaya database [http://www.knapsackfamily.com/KNApSAcK/] and Dr. Duke’s Phytochemical and Ethnobotanical (https://phytochem.nal.usda.gov/phytochem/search) to explore the various aspects of metabolite relationship as well as other detailed information on the metabolites [35]. Furthermore, the list of the bioactive compounds found in the database and LCMS results were compared for further bioactive selection [Supplementary Data].
2.2. Sample Preparation
C. papaya leaves were dried in the air, extracted with hexane and ethanol, and stored in the freezer at −20°C (this process produces 15.6% ethanol extract). To obtain C. papaya leaf juice extract, fresh C. papaya leaves were mixed with a juice extractor (Panasonic, Kobe), then filtered and frozen to get lyophilized leaf juice extract and stored in the freezer at −20°C (this process produces 14% ethanol extract).
2.3. LCMS Analysis
0.5 g of sample was put into a 10 mL measuring flask, then added methanol for 30 min and homogenized. Then, filter with a filter membrane and inject into the ultra-performance liquid chromatography (LC) system. LC settings were carried out on column C18 with column temperature 40°C and autosampler temperature 15°C, injection volume was 10 mL, with the flow rate at 0.6 mL/min, and mobile Phase A 0.1% formic acid in acetonitrile, mobile Phase B 0.1% formic acid in aquabides [36].
2.4. Target Protein Prediction
The target proteins were obtained from SEA Search Server (https://sea.bkslab.org/) and STITCH [http://stitch.embl.de/] database. An analysis using Swiss Target Prediction and SEA Search Server requires each selected compound to predict its target protein separately with a SMILE structure as the keyword. The TC value or Tanimoto Similarity (SEA) of >50% (0.5) was determined as the basis for selecting the target protein for each bioactive compound. Meanwhile, the list of bioactive compounds can be used as the input in STITCH. The additional settings, such as Rattus norvegicus in the type of organism and the minimum required interaction score, were set to 0.700.
2.5. Pharmacology Network Analysis
The compiled target proteins from two databases were used as the input of the STRING database [https://string-db.org/] for analysis by the multiple protein menu. Homo sapiens was selected as the type of organism. The network edges were used as confidence, and the minimum required interaction score was set to 0.700. After updating the network based on the additional settings, the visualization results were downloaded as TSV (tab-separated values). The downloaded file (.tsv) was then imported into Cytoscape v.3.8.2 software for network analysis and biological process prediction involving the target proteins using BINGO on the GOlororize app menu.
2.6. Docking Analysis
The 3D structures of the selected target proteins were obtained from the RSCB PDB database [https://www.rcsb.org/], namely, estrogen receptor 1 (ESR1) (PDB ID: 1ERE), LEP (GDP ID: 3V6O), and CYP19A1 (GDP ID: 5JKW). Meanwhile, the 3D structure of each papaya leaf’s active compound (quercetin, kaempferol, linoleic acid, caffeic acid, and beta carotene) and control (estradiol and testosterone) was obtained from the PubChem database [https://www.pubchem.ncbi.nlm.nih.gov]. Furthermore, the protein was prepared by removing the water molecules using Discovery Studio 2019 Software, while the energy of the ligand was minimized using the PyRx v.0.9.8 software. The docking was conducted using Autodock Vina integrated into PyRx v.0.9.8 [37]. The docking resulted in binding affinity from interacting with the compounds and proteins [38]. Furthermore, the interaction was visualized using the BioVia Discovery Studio 2019.
Besides BINGO, a PPI network analysis was also conducted. The variable consists of Stress, Degree, Betweenness Centrality, and Closeness Centrality. The degree centrality variable is the number of interactions of one node with other nodes, where a high value indicates that the protein has many interactions with other nodes and could be the main protein or regulatory influencer. The betweenness centrality variable compared the shortest distances used by nodes in the entire pathway and could be interpreted as having a dominant function. Meanwhile, closeness centrality was the shortest relative distance to access nodes in the pathway, where a high value could be interpreted as easy to reach and easy to become the center of other protein regulators.
3. RESULTS
3.1. Bioactive Compounds Selection
154 C. papaya leaf bioactive compounds were obtained from the KnapSack Kanaya, and 31 C. papaya leaf bioactive compounds were obtained from Dr. Dukes’s Phytochemical and Ethnobotanical databases. Furthermore, these data were compared with the results of LCMS analysis. Five compounds were selected for further analysis [Table 1]. Table 1 shows the results of the analysis based on a comparison of the database and LCMS results for the bioactive components of C. papaya leaves. These compounds are classified based on the SMILES code in Pubchem to facilitate the pharmacology network analysis.
Table 1: Bioactive component.
| Bioactive component | Pubchem ID | Smiles structure |
|---|---|---|
| Caffeic acid | 689043 | C1=CC(=C(C=C1C=CC(=O)O)O)O |
| Beta carotene | 5280489 | CC1=C(C(CCC1)(C)C)C=CC(=CC=CC(=CC=CC=C(C)C=CC=C(C)C=CC2=C(CCCC2(C)C)C)C)C |
| Linoleic acid | 5280450 | CCCCCC=CCC=CCCCCCCCC(=O)O |
| Kaempferol | 5280863 | C1=CC(=CC=C1C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O)O |
| Quercetin | 5280343 | C1=CC(=C(C=C1C2=C(C(=O)C3=C(C=C(C=C3O2)O)O)O)O)O |
3.2. Pharmacology Network Analysis
The obtained target proteins from two databases [SEA Server and STITCH] were 177 proteins. STITCH results showed the potential of binding interaction, which is the basis of molecular dynamics [Figure 1]. The results of BINGO analysis from GOlorize showed the value of betweenness centrality [Figure 2] [39]. Each target protein that plays a role in biological processes related to breast milk is shown in Table 2. The targets with the most potential were marked in bold.
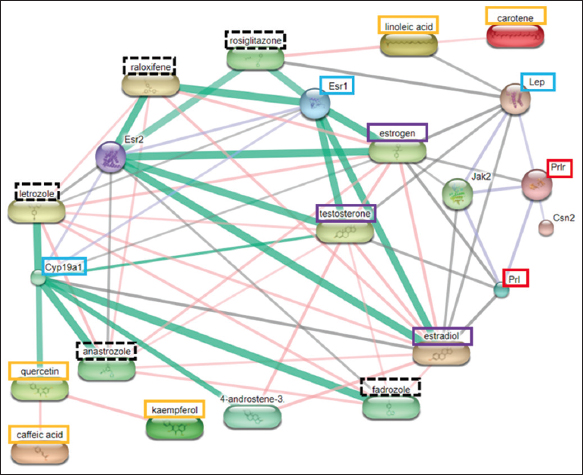 | Figure 1: Protein-compound interaction analysis result using STITCH (Rattus norvegicus). The orange box shows the most potent papaya compound; the purple square indicates the essential endogenous compound; the red box shows the protein acting directly on the prolactin biological process; the blue box shows the most potential target protein; the black dotted box indicates the control compound in the form of a drug. [Click here to view] |
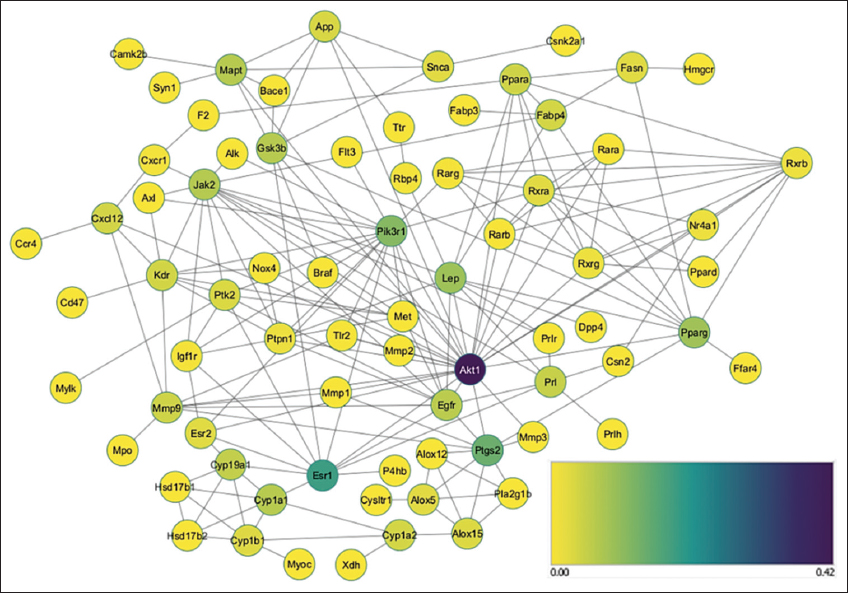 | Figure 2: Protein-protein interaction result of BINGO analysis from GOlorize. [Click here to view] |
Table 2: The biological process from the network analysis
| GO-ID | Description | P-value | Corr P-value | Cluster freq | Total freq | Genes |
|---|---|---|---|---|---|---|
| 7259 | JAK-STAT cascade | 2.02E-04 | 1.11E-03 | 3/72 4.1% | 23/14291 0.1% | LEP PRL JAK2 |
| 7631 | Feeding behavior | 5.49E-03 | 1.54E-02 | 3/72 4.1% | 71/14291 0.4% | APP LEP PRLH |
| 22612 | Gland Morphogenesis | 2.95E-03 | 9.80E-03 | 3/72 4.1% | 57/14291 0.3% | MMP2 ESR1 IGF1R |
| 30879 | Mammary gland development | 5.69E-05 | 3.98E-04 | 5/72 6.9% | 82/14291 0.5% | PRL MET XDH ESR1 IGF1R |
| 32355 | Response to estradiol stimulus | 3.21E-10 | 9.44E-09 | 10/72 13.8% | 117/14291 0.8% | MMP2 MMP3 CYP1A2 RARA PRL PIK3R1 PTGS2 MMP9 ESR1 PTK2 |
| 42698 | Ovulation Cycle | 6.03E-05 | 4.17E-04 | 5/72 6.9% | 83/14291 0.5% | MMP2 LEP PRL ESR1 EGFR |
| 42981 | Regulation of apoptosis | 4.11E-13 | 2.96E-11 | 23/72 31.9% | 723/14291 5.0% | APP GSK3B MMP2 ALOX15 ALOX12 BRAF PIK3R1 PTGS2 ESR1 EGFR PTK2 ESR2 NR4A1 RXRA AXL KDR AKT1 RARB NOX4 PPARG JAK2 PPARD SNCA |
| 43627 | Response to estrogen stimulus | 2.71E-09 | 6.74E-08 | 11/72 15.2% | 191/14291 1.3% | MMP2 MMP3 CYP1A2 RARA PPARG PRL PIK3R1 PTGS2 MMP9 ESR1 PTK2 |
| 46425 | Regulation of JAK-STAT cascade | 1.34E-02 | 3.01E-02 | 2/72 2.7% | 35/14291 0.2% | PRL JAK2 |
| 46545 | Development of primary female sexual characteristics | 3.78E-04 | 1.85E-03 | 4/72 5.5% | 68/14291 0.4% | RBP4 MMP2 LEP ESR1 |
| 48732 | Gland Development | 1.34E-07 | 2.22E-06 | 9/72 12.5% | 165/14291 1.1% | RXRA MMP2 RARA PRL MET CYP19A1 XDH ESR1 IGF1R |
| 60745 | Mammary gland branching in pregnancy | 2.00E-02 | 3.87E-02 | 1/72 1.3% | 4/14291 0.0% | ESR1 |
Figure 1 showed that the active compound of C. papaya leaves could modulate prolactin production through the Jak2 signaling pathway which is mediated by ESR1 and Cyp19a1. The compounds that play the roles are quercetin, kaempferol, and caffeic acid. The pathways that mediated by Lep are linoleic acid and carotene.
Figure 2 shows a cellular process illustrating protein interaction that can modulate prolactin production, a gene that has the strongest interaction is ESR1 and the most potential target proteins in the biological process related to breast milk are shown in Table 3.
Table 3: Selected target protein after network analysis using Cytoscape.
| Gene | Protein | Betweenness Centrality | Closeness Centrality | Degree | Stress |
|---|---|---|---|---|---|
| Esr1 | Estrogen Receptor 1 | 0.185737484 | 0.450617284 | 11 | 3582 |
| Lep | Leptin | 0.091525895 | 0.424418605 | 11 | 1638 |
| Jak2 | Tyrosine-protein kinase JAK2 | 0.069734321 | 0.39673913 | 14 | 1214 |
| Egfr | Epidermal growth factor receptor | 0.064367392 | 0.45625 | 11 | 1304 |
| Cyp19a1 | Aromatase | 0.05599585 | 0.330316742 | 6 | 1518 |
| Prl | Prolactin | 0.051988266 | 0.382198953 | 7 | 832 |
| Esr2 | Estrogen receptor beta | 0.027615266 | 0.412429379 | 5 | 730 |
3.3. Docking Analysis
The protein-compound interaction (PCI) was performed with the arrangement of the Homo sapiens organism to adjust as the organism of origin from the protein used as the target protein. The selected compounds as ligands were five bioactive compounds from C. papaya leaves that have the potential to interact with estrogen, Leptin, and Aromatase based on PCI prediction [Figure 3] and the results of network interaction analysis using Cytoscape. The control ligands, estradiol for ESR1 and testosterone for Aromatase, are also predicted for the PCI [Figure 1] and were obtained from the previous studies [40,41]. Figure 3 indicates that in the human body, quercetin has a strong interaction with Cyp19a1 to modulate prolactin production through the STAT5A signaling pathway and is mediated by ESR1. Meanwhile, linoleic acid modulate prolactin production through the STAT5B signaling pathway and mediated by Lep.
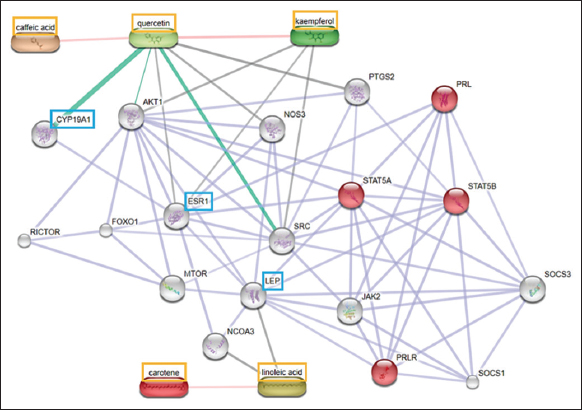 | Figure 3: Protein compound interaction analysis using STITCH (Homo sapiens). The orange box shows the compound with the most potential; the red circle indicates the protein acting on the biological lactation process; the blue box shows the most potential target protein. [Click here to view] |
3.4. ESR1 (ESR α)
The molecular docking results of the ESR1 protein with bioactive compounds from C. papaya leaves showed that two flavonoid compounds had the lowest binding affinity compared to other bioactive compounds. However, the binding affinity value for the quercetin-ESR1 complex and the kaempferol-ESR1 complex was higher than the control, which was −7.6 kcal/mol compared to estradiol [Table 4]. In addition to these two compounds, fatty acid compounds have a binding affinity lower than −7 kcal/mol. These results indicate that the bond formed between the quercetin, kaempferol, and linoleic acid complex to ESR1 is relatively stable, although they do not have a stronger bond than control.
Table 4: The binding affinity between ESR1 protein and bioactive compounds of Carica papaya leaves.
| Molecule | ID Ligand | Compound | Binding affinity (kcal/mol) |
|---|---|---|---|
| Control | 5757 | Estradiol | −10.7 |
| 6013 | Testosterone | −9.6 | |
| Sample | 5280343 | Quercetin | −7.6 |
| 5280863 | Kaempferol | −7.6 | |
| 5280450 | Linoleic acid | −7.1 | |
| 689043 | Caffeic acid | −6.6 | |
| 6419725 | Carotene | 17.4 |
Figure 4 showed the 3D visualization of the ESR1 protein complex with control ligands and the three most potent compounds. The figure also showed a surface hydrophobicity map. Surface hydrophobicity is displayed based on the color of the protein surface, with brown indicating hydrophobic, blue indicating hydrophilic, and white indicating the intermediate between those properties. The more hydrophobic the interaction, the more stable the complex conformation will be.
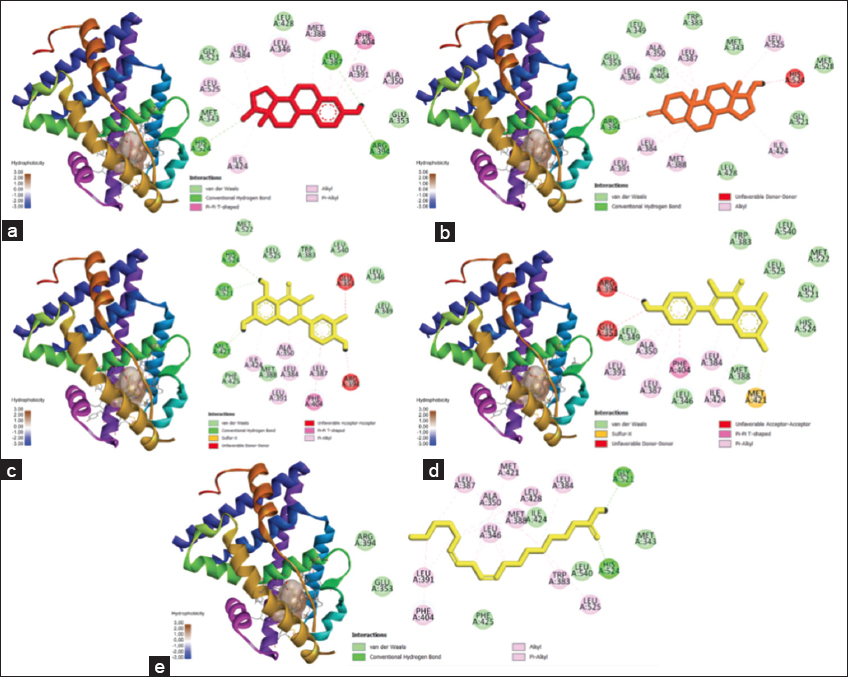 | Figure 4: Docking results visualization of (a) ESR1-estradiol protein, (b) ESR1-testosterone protein, (c) ESR1-quercetin protein, and (d) ESR1-linoleic acid protein. The left image shows a 3D visualization, and the right shows the type of bonding produced between the ligand-proteins, (e) Protein ESR1-linoleic acid. [Click here to view] |
3.5. Leptin (Lep)
The molecular docking of leptin with the selected compounds showed that three compounds from the flavonoid group had the lowest binding affinity values compared to others. In addition, the binding affinity values for the carotene-Lep (−6.9 kcal/mol) and quercetin-Lep (−6.7 kcal/mol) were lower compared to the control (−6.6 kcal/mol). At the same time, kaempferol-Lep complexes had equal binding affinity values with the control [Table 5]. However, the compound has a binding affinity value >−7 kcal/mol. This shows that the bond formed between carotene, quercetin, and kaempferol with Leptin is less stable even though it has a similar or stronger affinity than control. These results indicate that the network pathway is less precise in predicting the interaction of papaya compounds with leptin, especially for linoleic acid.
Table 5: The binding affinity between Leptin protein and the bioactive compounds of Carica papaya leaves.
| Molecule | Ligand ID | Compound | Binding affinity (kcal/mol) |
|---|---|---|---|
| Control | 6013 | Testosterone | −6.7 |
| 5757 | Estradiol | −6.6 | |
| Sample | 6419725 | Carotene | −6.9 |
| 5280343 | Quercetin | −6.7 | |
| 5280863 | Kaempferol | −6.6 | |
| 5280450 | Linoleic acid | −5.5 | |
| 689043 | Caffeic acid | −4.3 |
Figure 5 shows the 3D visualization of the leptin protein complex with control ligands and the three most potent compounds. The figure also shows a surface hydrophobicity map. The more hydrophobic interaction, the more stable the conformation complex will be.
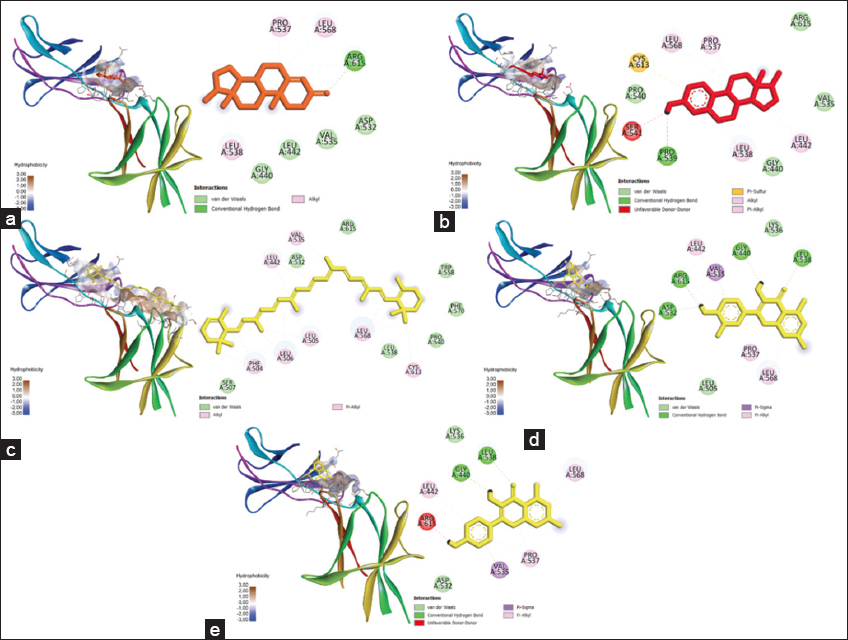 | Figure 5: Docking results visualization of (a) Lep-testosterone protein, (b) Lep-estradiol protein, (c) Lep-carotene, (d) Lep-quercetin, and (e) Lep-kaempferol protein. The left image shows a 3D visualization, while the right one shows the type of bonding produced between the ligand-proteins. [Click here to view] |
3.6. Aromatase (Cyp19a1)
The molecular docking of aromatase protein with bioactive compounds from C. papaya leaves showed that two compounds from the flavonoid group had the lowest binding affinity compared to other bioactive compounds. However, the binding affinity of the quercetin-CYP19A1 complex and the kaempferol-CYP19A1 complex was higher than the control [Table 6]. In addition, the two compounds also have a lower than −7 kcal/mol binding affinity value. This result indicates that the bond formed between the complex quercetin and kaempferol to aromatase is relatively stable. However, it has no potential to have a stronger bond than the control. This result also proves that the network pathway prediction is very accurate.
Table 6: The binding affinity between CYP19A1 protein and bioactive compounds of Carica papaya leaves.
| Molecule | Ligand ID | Compound | Binding affinity (kcal/mol) |
|---|---|---|---|
| Control | 6013 | Testosterone | −10.1 |
| 5757 | Estradiol | −8.3 | |
| Sample | 5280343 | Quercetin | −7.7 |
| 5280863 | Kaempferol | −7.7 | |
| 689043 | Caffeic acid | −6.1 | |
| 5280450 | Linoleic acid | −5.6 | |
| 6419725 | Carotene | 122.4 |
Figure 6 shows the results of 3D visualization of the CYP19A1 protein complex with control ligands and the two most potent compounds.
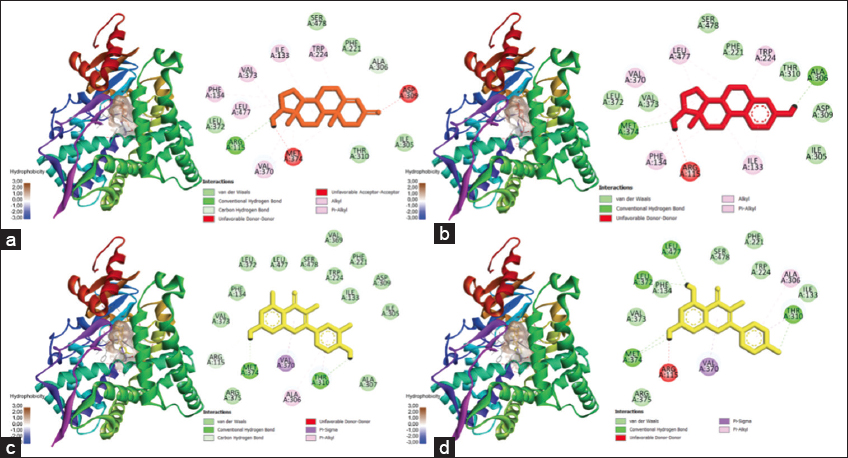 | Figure 6: Docking results visualization of (a) CYP19A1-testosterone protein, (b) CYP19A1-estradiol protein, (c) CYP19A1-quercetin protein, and (d) CYP19A1-kaempferol protein. The left image shows a 3D visualization, and the right one shows the type of bonding produced between the ligand-proteins. [Click here to view] |
4. DISCUSSION
PCI analysis using STITCH [Figure 1] showed that the most potent target proteins associated with prolactin production are Estrogen, Leptin, and Aromatase (CYP19A1), which is in agreement with previous studies related to prolactin metabolism and lactation [42-44]. The compound that has the most potential effect in this process is quercetin. Estrogen protein (ESR1) is involved in the prolactin metabolism (PRL) pathway, where both estrogen and JAK2 are required for the activation of STAT5A and STAT5B, which will induce ESR activation, especially ESR1 [45]. The scheme of the prolactin pathway is shown in Figure 7.
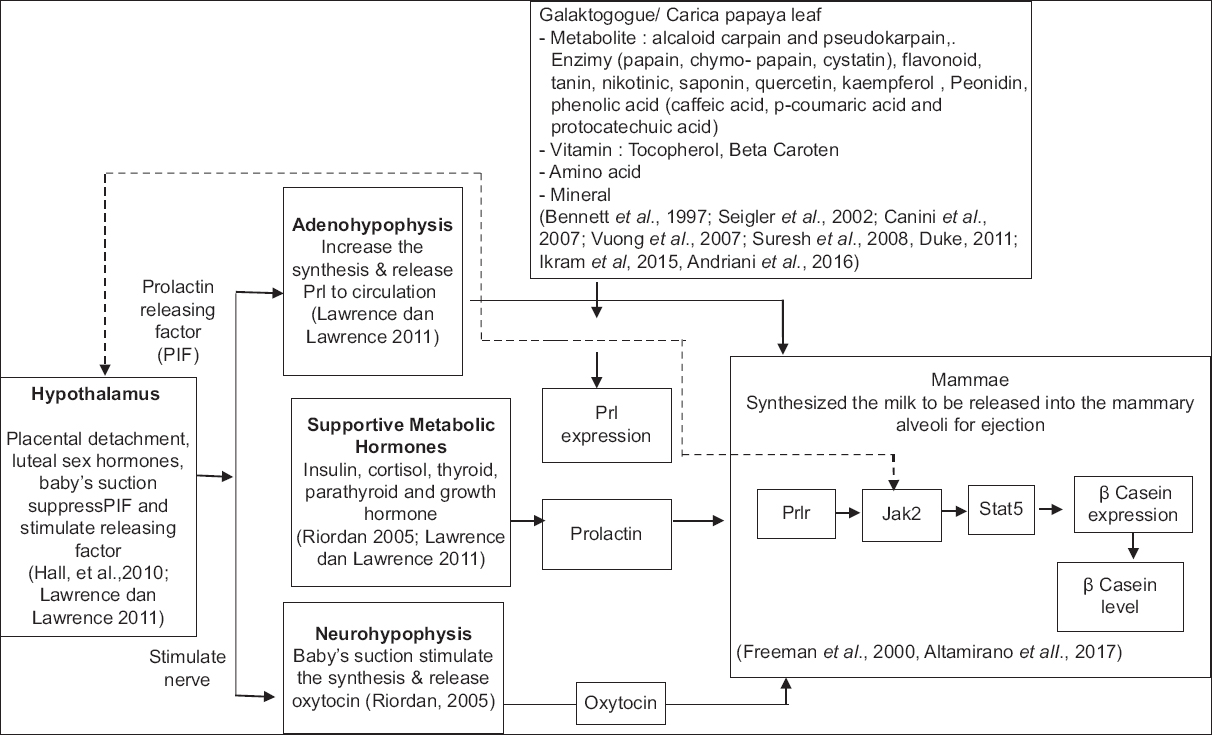 | Figure 7: Scheme of Prolactin pathway stimulated by phytochemical of Carica papaya leaf extract. [Click here to view] |
The leptin protein is known to have a high expression in the mammary gland during the lactation period, and it is found to be in insufficient quantity in the virgin mammary gland [42]. Meanwhile, aromatase protein was found to have the highest expression at the 148–149th days of gestation due to the aromatization process of androgens as the initial process of the estrogen formation in the mammary gland as a preparation for further pathways involving PRL, STAT5A, and STAT5B [46].
The result of the network analysis showed that the interaction between the active compounds of C. papaya leaves and ESR1 is involved in biological processes, including the branching of the mammary glands during pregnancy, differentiation, and morphogenesis of the mammary glands, estradiol response, and producing phenotypic trait [31,47]. There are two intracellular receptors for estrogen, alpha (α) and beta (β), encoded by the ESR1 and ESR2 genes, respectively. Furthermore, the interaction between the active compounds of C. papaya leaves and Leptin is involved in the JAK-STAT activation pathway. In contrast, the interaction with aromatase is involved in the mammary glands’ development [Table 2]. Hence, the target protein in this study is estrogen, leptin, and aromatase [Table 3].
One of the important things in determining the strength of interaction between the active compound and the target protein is a binding affinity value that showed the association of the compound and target receptor, so it can be calculated how the effectiveness of the interaction between these molecules [38]. Docking analysis showed that the bioactive compounds of C. papaya with the potential to have the most stable bond with ESR1 are quercetin, kaempferol, and linoleic acid which the binding affinity values were −7,6; −7,6, and −7,1, respectively [Table 4]. Bai et al. [48] showed that quercetin strongly interacts with ESR1 and kaempferol [49]. A study by Morselli et al. [50] proved that fatty acids act as potential mediators in the ESR signaling activation pathway and are commonly used as anti-cancer compounds [50,51]. Another study related to prolactin metabolism has shown that PRL can stimulate the expression of the ESR gene in the corpus luteum and decidua [45].
The docking analysis between the active compound of C. papaya leaf and leptin showed that beta carotene, quercetin, and kaempferol have the potential for interaction, which is noted by the binding affinity values were −6,9; −6,7; and −6,6, respectively [Table 5]. Leptin is encoded by the LEP gene, consisting of 167 amino acids, and plays an important role in maintaining energy homeostasis [52]. A study conducted by Getrude et al. [53] showed that a diet with a high beta-carotene content could affect leptin levels. Canas et al. [3] reported that carotenoid combination supplementation could increase beta carotene levels in the plasma, associated with BMI decrease, thereby preventing obesity.
Likewise, quercetin, which can suppress microglia-mediated inflammatory responses through HO-1 induction, can prevent obesity-induced hypothalamic inflammation [49]. Another study stated that quercetin administration could inhibit the leptin gene’s secretion and expression in cancer cells [54].
Meanwhile, kaempferol is an alternative therapy to prevent obesity and insulin resistance in rats with a high-fat diet [48]. Muni Swamy et al. [55] showed that kaempferol compounds isolated from M. oleifera could affect leptin gene expression by inducing lipolysis and suppressing adipogenesis.
Based on the docking analysis, the components of C. papaya leaf that can form stable bonds with aromatase are quercetin and kaempferol, with the binding affinity value of −7,7 of both compounds. Aromatase 450, encoded by the CYP19 gene, catalyzes the formation of multiple estrogens in several tissues under the control of specific promoters regulated by different signaling pathways [56]. Sanderson et al. [57] showed that a natural flavonoid, quercetin, can enhance aromatase activity up to 4 times, associated with the intracellular cAMP increase. Quercetin can inhibit the production of VEGF and change the redox status to affect the angiogenic process [58]. Due to its ability to influence the expression of the CYP19A1 gene, quercetin can be used as a safe chemotherapy agent for breast cancer [59].
Based on the docking analysis, the bioactive components of C. papaya leaves that have the most potential to increase prolactin production are flavonoid compounds, specifically quercetin, and kaempferol. These compounds play a role in the mechanism of estrogen inhibition by influencing the expression of the ESR1 and CYP19A1 genes. Flavonoids are compounds that play a role in various biological reactions [27] and have antioxidant, anti-inflammatory, anti-mutagenicity, and anti-carcinogenic properties, coupled with their capacity to modulate the function of intracellular enzymes [27,60]. Thus, many studies showed the role of flavonoids as an alternative therapy for breast cancer [61,62]. However, the studies that specifically relate to the role of flavonoids in breast milk, especially how they play a role in prolactin production, are still limited. Further testing is needed both in vivo and in vitro.
This study found that a potential pathway for papaya leaf extract to prolactin production is through ESR1. Estrogen and prolactin stimulate alveoli differentiation into specialized structures for synthesizing and secreting milk during lactation. The lack of ESR1 in the alveoli will inhibit ductal elongation, disrupting lobulo alveolar development and resulting in insufficient milk production [31]. The milk production pathway is shown in Figure 8.
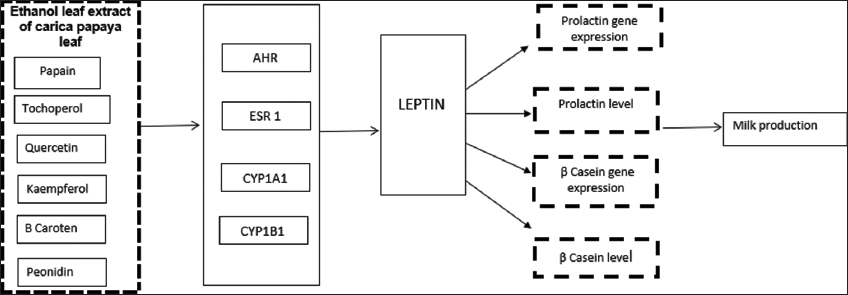 | Figure 8: Milk production pathway stimulated by phytochemical of Carica papaya leaf extract. [Click here to view] |
The potential pathway of papaya leaf extract through signals captured by ESR1 through Leptin is expressed into the PRL gene, then expressed into the prolactin and captured by PRLR. Leptin is a potent stimulator of PRL release, and its actions are mediated through ERK1/2 stimulation. As a highly potent PRL stimulant, leptin induces an initial increase in PRL release during the first 6 h, followed by an increase in two lower levels over 1–4 and 4–16 h [63].
Cytoscape visualization results show that the five active compounds from papaya leaves can interact with the ESR1, ESR2, ERBB, and EGFR genes to activate various cellular and molecular signaling pathways, activating leptin transcription. This presence can further modulate pituitary hormone secretion, including prolactin [64]. Leptin is also a critical compound that can modulate various molecular signals, such as the JAK/STAT, PI3K, and MAPK pathways, so it can trigger the transcription of several genes or exert different hormonal effects in the other parts of the cell [65]. Münzberg and Morrison [52] stated that leptin levels and circulation constantly fluctuate depending on the nutrients and the quality of those nutrients.
5. CONCLUSION
The abundant phytochemical components in C. papaya leaves are alkaloids and flavonoids. Alkaloids are widely used as nutraceuticals to manage oxidative stress and prevent inflammation, while flavonoids are known to affect metabolism through hydroxylation, methylation, and acylation reactions. We found that the flavonoid compounds from C. papaya leaves, namely, quercetin, and kaempferol, have the potential as multi-activators, especially for the Estrogen protein, Leptin, and Aromatase, which are involved in the prolactin pathway and the lactation process. However, the compounds from the fatty acid group, namely, linoleic acid, are predicted to have a reasonably stable interaction in binding to estrogen. Thus, it shows that C. papaya leaf has the potential to be used as a galactagogue herb to increase breast milk production through increasing prolactin levels.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
This research was funded by the Ministry of Education Through Domestic Postgraduate Education Scholarships (BPPDN) for doctoral scholarship and research funding assistance from STIKes Dharma Husada.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
The study protocol was approved by the Research Ethics Commission of Universitas Padjadjaran Bandung (No. 1340/UN6.KEP/EC/2019).
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Lee S, Kelleher SL. Biological underpinnings of breastfeeding challenges:The role of genetics, diet, and environment on lactation physiology. Am J Physiol Endocrinol Metab 2016;311:E405-22. [CrossRef]
2. Grattan DR. 60 years of neuroendocrinology:The hypothalamo-prolactin axis. J Endocrinol 2015;226:T101-22. [CrossRef]
3. Canas JA, Lochrie A, McGowan AG, Hossain J, Schettino C, Balagopal PB. Effects of mixed carotenoids on adipokines and abdominal adiposity in children:A pilot study. J Clin Endocrinol Metab 2017;102:1983-90. [CrossRef]
4. Lawrence RA, Lawrence RM. Breastfeeding:A Guide for the Medical Professional. 9th ed. New York:Elsevier Publication;2021.
5. Golan Y, Assaraf YG. Genetic and physiological factors affecting human milk production and composition. Nutrients 2020;12:1500. [CrossRef]
6. Nilsson L. Effects of Prolactin on Metabolism-changes Induced by Hyperprolactinemia. Doctoral Thesis. Gothenburg:University of Gothenburg;2009.
7. Trott JF, Schennink A, Petrie WK, Manjarin R, VanKlompenberg MK, Hovey RC. Triennial lactation symposium:Prolactin:The multifaceted potentiator of mammary growth and function. J Anim Sci 2012;90:1674-86. [CrossRef]
8. Gallego MI, Binart N, Robinson GW, Okagaki R, Coschigano KT, Perry J, et al. Prolactin, growth hormone, and epidermal growth factor activate Stat5 in different compartments of mammary tissue and exert different and overlapping developmental effects. Dev Biol 2001;229:163-75. [CrossRef]
9. Qian X, Zhao FQ. Interactions of the ubiquitous octamer-binding transcription factor-1 with both the signal transducer and activator of transcription 5 and the glucocorticoid receptor mediate prolactin and glucocorticoid-induced b-casein gene expression in mammary epithel. Int J Biochem Cell Biol 2013;45:724-35. [CrossRef]
10. Kobayashi K, Oyama S, Kuki C, Tsugami Y, Matsunaga K, Suzuki T, et al. Distinct roles of prolactin, epidermal growth factor, and glucocorticoids in b-casein secretion pathway in lactating mammary epithelial cells. Mol Cell Endocrinol 2017;440:16-24. [CrossRef]
11. Iwasaka T, Umemura S, Kakimoto K, Koizumi H, Osamura YR. Expression of prolactin mRNA in rat mammary gland during pregnancy and lactation. J Histochem Cytochem 2000;48:389-96. [CrossRef]
12. Zuppa AA, Sindico P, Orchi C, Carducci C, Cardiello V, Romagnoli C. Safety and efficacy of galactogogues:Substances that induce, maintain and increase breast milk production. J Pharm Pharm Sci 2010;13:162-74. [CrossRef]
13. Tabares FP, Jaramillo JV, Ruiz-Cortés ZT. Pharmacological overview of galactogogues. Vet Med Int 2014;2014:602894. [CrossRef]
14. Kent JC, Gardner H, Geddes DT. Breastmilk production in the first 4 weeks after birth of term infants. Nutrients 2016;8:756. [CrossRef]
15. Forinash AB, Yancey AM, Barnes KN, Myles TD. Uso de estimulantes de producción de leche materna en madres que amamantan [The use of galactogogues in the breastfeeding mother]. Ann Pharmacother 2012;46:1392-404. [CrossRef]
16. Asztalos EV. Supporting mothers of very preterm infants and breast milk production:A review of the role of galactogogues. Nutrients 2018;10:600. [CrossRef]
17. Liu H, Hua Y, Luo H, Shen Z, Tao X, Zhu X. An herbal galactagogue mixture increases milk production and aquaporin protein expression in the mammary glands of lactating rats. Evid Based Complement Alternat Med 2015;2015:760585. [CrossRef]
18. Bennett RN, Kiddle G, Wallsgrove RM. Biosynthesis of benzylglucosinolate, cyanogenic glucosides and phenylpropanoids in Carica papaya. Phytochemistry 1997;45:59-66. [CrossRef]
19. Seigler DS, Pauli GF, Nahrstedt A, Leen R. Cyanogenic allosides and glucosides from Passiflora edulis and Carica papaya. Phytochemistry 2002;60:873-82. [CrossRef]
20. Canini A, Alesiani D, D'Arcangelo G, Tagliatesta P. Gas chromatography-mass spectrometry analysis of phenolic compounds from Carica papaya L. leaf. J Food Compos Anal 2007;20:584-90. [CrossRef]
21. Liu Y, Perera CO, Suresh V. Comparison of three chosen vegetables with others from South East Asia for their lutein and zeaxanthin content. Food Chem 2007;101:1533-9. [CrossRef]
22. Ikram EH, Stanley R, Netzel M, Fanning K. Phytochemicals of papaya and its traditional health and culinary uses-a review. J Food Compos Anal 2015;41:201-11. [CrossRef]
23. Memudu AE, Oluwole TJ. The contraceptive potential of Carica papaya seed on oestrus cycle, progesterone, and histomorphology of the Utero-ovarian tissue of adult wistar rats. JBRA Assist Reprod 2021;25:34-43 [CrossRef]
24. Dr Duke's Phytochemical and Ethnobotanical Databases. Washington, DC:U.S. Department of Agriculture;c1995. Available from:https://phytochem.nal.usda.gov/phytochem/search [Last accessed on 2020 Oct 13].
25. Akhila S, Vijayalakshmi NG. Phytochemical studies on Carica papaya leaf juice. Int J Pharm Sci Res 2015;6:880-3.
26. Miean KH, Mohamed S. Flavonoid (myricetin, quercetin, kaempferol, luteolin, and apigenin) content of edible tropical plants. J Agric Food Chem 2001;49:3106-12. [CrossRef]
27. Alseekh S, Perez de Souza L, Benina M, Fernie AR. The style and substance of plant flavonoid decoration;towards defining both structure and function. Phytochemistry 2020;174:112347. [CrossRef]
28. Badgujar SB, Patel VV, Bandivdekar AH. Foeniculum vulgare Mill:A review of its botany, phytochemistry, pharmacology, contemporary application, and toxicology. Biomed Res Int 2014;2014:842674. [CrossRef]
29. Paikra BK, Dhongade HK, Gidwani B. Phytochemistry and pharmacology of Moringa oleifera Lam. J Pharmacopuncture 2017;20:194-200. [CrossRef]
30. Herawati Y, Kalsum U, Wiyasa IW, Yuniarti L, Sardjono TW. Ethanol extract of Carica papaya leaf can increase breast milk in lactating rat. Open Access Maced J Med Sci 2021;9:520-6. [CrossRef]
31. Macias H, Hinck L. Mammary gland development. Wiley Interdiscip Rev Dev Biol 2012;1:533-57. [CrossRef]
32. Obradovic M, Sudar-Milovanovic E, Soskic S, Essack M, Arya S, Stewart AJ, et al. Leptin and obesity:Role and clinical implication. Front Endocrinol (Lausanne) 2021;12:585887. [CrossRef]
33. Hong Y, Li H, Yuan YC, Chen S. Molecular characterization of aromatase. Ann N Y Acad Sci 2009;1155:112-20. [CrossRef]
34. Di Nardo G, Zhang C, Marcelli AG, Gilardi G. Molecular and structural evolution of cytochrome P450 aromatase. Int J Mol Sci 2021;22:631. [CrossRef]
35. Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, et al. KNApSAcK:A comprehensive species-metabolite relationship database. Biotechnol Agric For 2006;57:165-81. [CrossRef]
36. Yuriev E, Ramsland PA. Latest developments in molecular docking:2010-2011 in review. J Mol Recognit 2013;26:215-39. [CrossRef]
37. Qiao L, Lewis R, Hooper A, Morphet J, Tan X, Yu K. Waters Applicatin Note;using Natural Product Application Solution with UNIFI for the Identification of Chemical Ingredients of Green tea Extract. Massachusetts:Waters Corporation;2013. Available from:https://www.waters.com/content/dam/waters/en/app-notes/2013/720004837/720004837-en.pdf [Last accessed on 2020 Oct 13].
38. Yunta MJ. Docking and ligand binding affinity:Uses and pitfalls. Am J Model Optim 2016;4:74-114.
39. Maere S, Heymans K, Kuiper M. BiNGO:A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005;21:3448-9. [CrossRef]
40. Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engström O, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 1997;389:753-8. [CrossRef]
41. Ghosh D, Egbuta C, Lo J. Testosterone complex and non-steroidal ligands of human aromatase. J Steroid Biochem Mol Biol 2018;181:11-9. [CrossRef]
42. Feuermann Y, Mabjeesh SJ, Shamay A. Leptin affects prolactin action on milk protein and fat synthesis in the bovine mammary gland. J Dairy Sci 2004;87:2941-6. [CrossRef]
43. Caglar AS, Kapucu A, Dar KA, Ozkaya HM, Caglar E, Ince H, et al. Localization of the aromatase enzyme expression in the human pituitary gland and its effect on growth hormone, prolactin, and thyroid stimulating hormone axis. Endocrine 2015;49:761-8. [CrossRef]
44. Sato Y, Kaneko Y, Cho T, Goto K, Otsuka T, Yamamoto S, et al. Prolactin upregulates female-predominant P450 gene expressions and downregulates male-predominant gene expressions in mouse liver. Drug Metab Dispos 2017;45:586-92. [CrossRef]
45. Frasor J, Gibori G. Prolactin regulation of estrogen receptor expression. Trends Endocrinol Metab 2003;14:118-23. [CrossRef]
46. Peaker M. Recent advances in the study of monovalent ions movements across the mammary epithelium:Relation to onset of lactation. J Dairy Sci 1975;58:1042-7. [CrossRef]
47. Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest 2006;116:561-70. [CrossRef]
48. Bai X, Tang Y, Li Q, Chen Y, Liu D, Liu G, et al. Network pharmacology integrated molecular docking reveals the bioactive components and potential targets of Morinda officinalis-Lycium barbarumcoupled-herbs against oligoasthenozoospermia. Sci Rep 2021;11:2220. [CrossRef]
49. Yang J, Kim CS, Tu TH, Kim MS, Goto T, Kawada T, et al. Quercetin protects obesity-induced hypothalamic inflammation by reducing microglia-mediated inflammatory responses via HO-1 induction. Nutrients 2017;9:650. [CrossRef]
50. Morselli E, de Souza Santos R, Gao S, Ávalos Y, Criollo A, Palmer BF, et al. Impact of estrogens and estrogen receptor-a in brain lipid metabolism. Am J Physiol Endocrinol Metab 2018;315:E7-14. [CrossRef]
51. Dachev M, BryndováJ, Jakubek M, Moucka Z, Urban M. The effects of conjugated linoleic acids on cancer. Processes 2021;9:454. [CrossRef]
52. Münzberg H, Morrison CD. Structure, production and signaling of leptin. Metabolism 2015;64:13-23. [CrossRef]
53. Getrude NO, Albert NE, Gabriel G, Finbarrs-Bello E, Elizabeth C, Austin OI. Beta-carotene:Positive effect on oxidative stress, lipid peroxidation, insulin and leptin resistance induced by dietary fat consumption. J Adv Med Med Res 2018;27:1-7. [CrossRef]
54. Rahimifard M, Sadeghi F, Asadi-Samani M, Nejati-Koshki K. Effect of quercetin on secretion and gene expression of leptin in breast cancer. J Tradit Chin Med 2017;37:321-5. [CrossRef]
55. Muni Swamy G, Ramesh G, Devi Prasad R, Meriga B. Astragalin, (3-O-glucoside of kaempferol), isolated from Moringa oleifera leaves modulates leptin, adiponectin secretion and inhibits adipogenesis in 3T3-L1 adipocytes. Arch Physiol Biochem 2020;128:938-44. [CrossRef]
56. Simpson ER. Aromatase:Biologic relevance of tissue-specific expression. Semin Reprod Med 2004;22:11-23. [CrossRef]
57. Sanderson JT, Hordijk J, Denison MS, Springsteel MF, Nantz MH, van den Berg M. Induction and inhibition of aromatase (CYP19) activity by natural and synthetic flavonoid compounds in H295R human adrenocortical carcinoma cells. Toxicol Sci 2004;82:70-9. [CrossRef]
58. Santini SE, Basini G, Bussolati S, Grasselli F. The phytoestrogen quercetin impairs steroidogenesis and angiogenesis in swine granulosa cells in vitro. J Biomed Biotechnol 2009;2009:419891. [CrossRef]
59. Khan SI, Zhao J, Khan IA, Walker LA, Dasmahapatra AK. Potential utility of natural products as regulators of breast cancer-associated aromatase promoters. Reprod Biol Endocrinol 2011;9:91. [CrossRef]
60. Panche AN, Diwan AD, Chandra SR. Flavonoids:An overview. J Nutr Sci 2016;5:e47. [CrossRef]
61. Park YJ, Choo WH, Kim HR, Chung KH, Oh SM. Inhibitory aromatase effects of flavonoids from Ginkgo bilobaextracts on estrogen biosynthesis. Asian Pac J Cancer Prev 2015;16:6317-25. [CrossRef]
62. Obakan-Yerlikaya P, Arisan ED, Coker-Gurkan A, Palavan-Unsal N. Breast cancer and flavonoids as treatment strategy. In:Breast Cancer:From Biology to Medicine. United Kingdom:IntechOpen;2017. 305. [CrossRef]
63. Tipsmark CK, Strom CN, Bailey ST, Borski RJ. Leptin stimulates pituitary prolactin release through an extracellular signal-regulated kinase-dependent pathway. J Endocrinol 2008;196:275-81. [CrossRef]
64. Denver RJ, Bonett RM, Boorse GC. Evolution of leptin structure and function. Neuroendocrinology 2011;94:21-38. [CrossRef]
65. Frühbeck G. Intracellular signalling pathways activated by leptin. Biochem J 2006;393:7-20. [CrossRef]