β-glucanases are industrially significant glycosyl hydrolases which hydrolyze the glycosidic bonds in insoluble or partially soluble β-glucan molecules by acting as exo- and endo-hydrolases. Molecular characterization and structural elucidation are vital for uncovering the structural and functional relationship of β-glucan degrading enzymes. In the present study, full-length gene sequences of exo-β-1,4-glucanase (EXO-14) and endo-β-1,3-glucanase (ENDO-13), isolated from Streptomyces althioticus TBG-MR17 and Streptomyces cinereoruber subsp. cinereoruber TBG-AL13, respectively, were obtained. The primary sequence analysis and sequence based structural analysis were performed in silico. The obtained 1737 bp EXO14 gene and 1293 bp ENDO13 gene encoded 578 and 438 putative amino acids, respectively. The in silico functional domain predictions showed a multi-domain architecture of both enzymes and showed diverse physiochemical properties. The substrate binding analysis through molecular docking delivered the possible architecture of active sites, the tunnel type in EXO-14 prominently cleaves external β-1,4- linkages and open cleft like structure in ENDO-13 has more propensity to cut internal bonds. Overall, the results generate valuable information for continuing the understanding of β-glucanase enzymes especially from Streptomyces spp., and provide significance for upgrading their catalytic properties through enzyme engineering.
Edison LK, Reji SR, Pradeep NS. In silico perceptions in structural elucidation of Exo-β-1,4-glucanase and Endo-β-1,3-glucanase from Streptomyces spp. J App Biol Biotech. 2020;8(6):67-81. https://doi.org/10.7324/JABB.2020.80612
Genes encoding b-glucanase enzymes have been widely explored in various organisms because of enormous importance in industrial applications. b-glucanase gained commercial importance for its role in beer fermentation and poultry feed formulation [1,2], the enzyme was also weighted for its ecological significance [3,4]. Considering commercial applications, molecular redesigning of the enzyme may enhance its properties at industrial scale. This can be achieved by understanding the enzyme at proteomic and genomic level, which can guide the manipulation of the enzyme to suit industrial process.
Protein motifs of b-glucanases have been annotated in the carbohydrate-active enzymes (CAZy) modules as sequence related families [5] and the activities were annotated according to sequence similarities, protein structural folding, and recognized enzyme activities [6]. Understanding molecular structure of enzymes is essential to unfold the functional properties. The molecular structure predominantly controls the chemical and physical properties of enzymes which retains their particular roles in nature and biological systems [7].
Bioinformatics aid in understanding the functional role of proteins based on available sequence at the genomic or proteomic level. Computational biology can help in constructing the three dimensional structure of a protein, that further helps to predict functional roles and these predictions are helpful in outlining required modifications for activity enhancement [8]. Structural and functional characterization of several b-glucanase in the past has helped in defining the functional and structural – relationship of this protein [9,10]. Molecular docking is another bioinformatic tool that serves to identify binding affinity and strength of association between two molecules, especially in drug designing. In enzyme studies, the tool recognizes catalytic domains for binding of substrate molecules [11].
Many molecular in silico characterization reports are available in regard to b-glucanases from prokaryotes and eukaryotes [12], among this source of fungi and bacteria are more prominent; however, studies in Streptomyces are rarely reported [13,14]. In this report, we have isolated complete sequence of exo-b-1,4-glucanase (EXO14) and endo-b-1,3-glucanase (ENDO-13) genes from Streptomyces althioticus TBG-MR17 and Streptomyces cinereoruber subsp. cinereoruber TBG-AL13, respectively. The potent b-glucanase producers were isolated from Western Ghats of Kerala and detailed molecular characterization studies were conducted in silico. With an aim to understand the enzyme substrate interaction, homology modeling and docking analysis of predicted EXO14 and ENDO13 proteins were deliberated.
S. althioticus TBG-MR17 and S. cinereoruber subsp. Cinereoruber TBG-AL13 were isolated from Western Ghats regions in Kerala, India. The strains were grown and maintained in International Streptomyces Project 4 agar at 28°C. Escherichia coli strain DH5a was purchased from Invitrogen™, (USA) and grown in Luria Bertini (LB) agar (HiMedia, India) at 37°C. GoTaq® G2 Hot Start Green PCR Master Mix and pGEM®-T Easy Vector were purchased from Promega (USA). The primers were synthesized in Eurofin Genomics, Bengaluru. DNA sequencing reaction was done in ABI PRISM 3730xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using BigDye® Terminator v3.1 Cycle Sequencing Kit from Applied Biosystems based on Sanger’s dideoxy chain termination method.
High purity genomic DNAs were isolated from the mycelium of Streptomyces strains using the method of Murray and Thompson [15] with adequate modifications. PCR amplification of EXO-14 and ENDO-13 genes was carried out using the primers EX3f/EX3r (EX3f-AACCCGCACAATGAGTCGCACCA; EX3r-GTCAGCCGGTCACGACACCGG) and EN3f/EN3r (EN3f-CACCCGACGGAAGAGAGAGYCATGMCC; EN3r-CAGCGTGTCACCGTCCACTTCTGG), respectively. The primers designed based on the conserved regions of EXO-14 and ENDO-13 genes retrieved respectively from the phylogenetic neighboring species of S. althioticus TBG-MR17 and S. cinereoruber subsp. Cinereoruber TBG-AL13 available in the NCBI database. PCR reaction mixtures (25 mL) contained approximately 25 ng of DNA, 10 ng of each primers and 12.5 mL GoTaq® G2 Hot Start Green PCR Master Mix. The thermal program included an initial denaturation at 95°C for 2 min followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 66°C for 30 sec, and extension at 72°C for 1.30 sec, and a final extension at 72°C for 5 min. The amplicons were purified using Wizard® SV Gel and PCR clean-up system (Promega, USA), cloned into pGEM®-T Easy Vector and transformed to DH5a E. coli cells. The purified plasmids from positive colonies were sequenced using primers TvectF/TvectR (TvectF-GGGTTTTCCCAGTCACGACGT; TvectR- CGCCAAGCTATTTAGGTGACAC).
Forward and reverse contig sequences of EXO14 and ENDO13 genes were assembled using CAP contig program of BIOEDIT v7.2.5 [16]. Vector sequences were separated from the both assembled gene sequences using VecScreen tool from NCBI (https://www.ncbi.nlm.nih.gov/tools/vecscreen/). Open reading frames (ORF) were determined and the nucleotide sequences were translated to protein, using ExPASy Translate tool (http://web.expasy.org/translate/). Homology searches of protein sequences were carried out in NCBI GenBank database using BLASTp algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with default settings. Sequence alignments were performed with MAFFT v7 (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/) with the E-INS-I algorithm [17]. The aligned sequences were viewed using ESPript 3.0 [18]. Physicochemical parameters such as molecular weight, theoretical isoelectric point (pI), half-life, instability index, number of positive (+R) and negative residues (-R), aliphatic index, extinction coefficient, and grand average of hydropathicity (GRAVY) values were determined by the EXPASY tool ProtParam (http://web.expasy.org/protparam/) [19]. The phylogenetic trees were constructed using neighbor-joining algorithm in MEGA X [20]. Protein functional domains and motifs were identified using conserved domain database (CDD) search in NCBI [21]. The CAZyme families and carbohydrate binding modules of predicted putative protein were identified using the database for CAZy ANnotation (dbCAN) (http://csbl.bmb.uga.edu/dbCAN/index.php) [22]. Signal peptide cleavage sites were predicted by SignalP v4.1 (http://www.cbs.dtu.dk/services/SignalP/). Protein topology or secondary structure predictions were performed using the PSIPRED v3.3 [23].
Three dimensional models of EXO-14 and ENDO-13 protein were generated using I-TASSER webserver of University of Michigan, USA maintained by Zhang Lab [24]. It used multiple threading approaches for selecting the templates and predicted full-length models of EXO-14 and ENDO-13 proteins by iterative structure assembly. Based on Z-score, one template was selected from each threading program to predict the target model of each protein. The program selected ten best structural analogs from protein data bank based on closest structural similarity and generated five models by clustering the stimulated structural confirmations based on pair-wise structural similarity with the template. The three dimensional models were evaluated based on TM-score (template modeling Score), C-score (confidence score), and RMSD (Root-mean-square deviation). The confidence of models was measured quantitatively by C-score, normally in range of −5–2. The higher C-score value signified a high confidence model. The RMSD and TM-score calculated mainly based on the C-score. Finally using the TM-align structural alignment program I-TASSER aligned the best model to all available PDB structures. The models were visualized by the Maestro 11.5 element tool of Schrödinger Suites 2018-1 (Schrödinger, LLC, New York, NY, 2018) and finally validated by assessing the model stereochemical quality using BioLuminate module of Schrödinger Suites 2018-1. Superimposition of models with corresponding templates was performed using Chimera 1.12.
Protein structures in pdb file format were directly imported to molecular docking software (Glide, Schrödinger Suites 2018-1). Protein structural minimizations and optimizations were carried out accordingly the Protein Preparation Wizard (PrepWizard) in SchrödingerSuites 2018-1. The preparation protocol built loops and side chains with missing atoms, added hydrogen molecules, optimized H-bonding network and finally a restrained minimization was done to get a refined protein structure for docking studies. Active sites (ligand binding sites) of proteins were predicted using SiteMap application of Schrödinger Suites 2018-1 [25]. The binding site within the catalytic domain was considered for docking study.
Two dimensional structures of ligand molecules in sdf format were retrieved from PubChem compound database (https://pubchem.ncbi.nlm.nih.gov/) of NCBI. The ligands were prepared by adding hydrogen, eliminate the discrepancies between bond length and angle and finally converted to the 3D structure using the LigPrep module of Schrödinger Suites 2018-1.
Receptor grid of 15Å × 15Å × 15Å grid points and spacing 0.25 was generated at the center of predicted active site, using the Grid Generation module of Schrödinger Suites 2018-1. The prepared grid was selected and docked with a single best conformation of the ligands using Glide dock-XP mode in Schrödinger Suites 2018-1. Subsequently, the best gliding pose of ligand to protein with maximum glide score (G score) and glide energy (G energy) was generated [26].
The PCR amplification of ENDO-13 and EXO-14 genes yielded ~1400 bp and ~2000 bp corresponding intense amplicons [Supplementary Figure 1a]. Then, the amplicons were eluted and cloned into pGEMT vector [Supplementary Figure 1b]. The recombinant colonies with pGEMT/ENDO13 and pGEMT/EXO14 constructs were selected by colony PCR [Supplementary Figure 1c]. Validation of the presence of gene of interest was done by isolating the plasmids from positive colonies and subjected to PCR amplification using gene specific (EN3f/EN3r and EX3f/EX3r) and vector specific primers (TvectF and TvectR) [Supplementary Figure 1d and e]. The sequencing with TvectF/TvectR primers obtained full length protein coding sequences from both genes. The ORF of EXO-14 gene was 1737 bp and coding 578 putative amino acids [Supplementary Figure 2] and ENDO-13 gene was 1293 bp coding 438 putative amino acids [Supplementary Figure 3].
BLASTp search revealed that both proteins exhibited sequence similarity and identity to specific b-glucanases of Streptomyces spp. The amino acid sequence of EXO-14 showed 99% of sequence identity with Streptomyces sp. 4F cellulose 1,4-b-cellobiosidase (GenBank Accession No. WP058917325) and Streptomyces griseorubens cellulose 1,4-b-cellobiosidase (GenBank Accession No. WP037642617). The ENDO-13 protein amino acid sequence exhibited 82% identity with Streptomyces globisporus ENDO13 (GenBank Accession No. WP030690772) and Streptomyces sp. TLI-105 laminarinase like protein (GenBank Accession No. WP093872583). Multiple sequence alignments of EXO-14 protein with homologous sequence revealed a consensus sequence VIYNLPGRDCAALASNG, which is the key characteristics consensus pattern, V-x-Y-x(2)-P-x-R-D-C-[GSAF]-x(2)-[GSA](2)-x-G, of all other EXO14 (cellulose 1,4-b-cellobiosidase) protein [Figure 1]. This confirmed that the catalytic domain of EXO14 resembled that of cellulose 1,4-b-cellobiosidase (EXO14). MSA of ENDO-13 protein showed the presence of consensus sequence EIDVMENVGFEP, similar to the characteristic consensus pattern, E-[LIV]-D-[LIVF]-x(0,1)-E-x(2)-[GQ]-[KRNF]-x-[PSTA], of all other ENDO13 (laminarinase) proteins [Figure 2]. The computed physicochemical analysis of the predicted EXO-14 and ENDO-13 proteins is shown in Table 1. The EXO14 has high number of negative amino acid residues than ENDO13. The instability index <40 is predicted as stable, the values obtained showed that both EXO-14 and ENDO-13 proteins are considered as stable. Low GRAVY values of EXO-14 (−0.446) and ENDO-13 (−0.424) indicated both have the property to interact with water. The nucleotide sequence for EXO14 and ENDO13 was deposited in GenBank database under the accession number of MG983485 and MH719000 respectively.
.png) | Figure 1: Multiple sequence alignment of EXO14 protein and other EXO-b-1,4-glucanase. The selected sequences from NCBI database are as follows: Streptomyces sp. 4F (WP058917325), Streptomyces griseorubens (WP037642617), Streptomyces vinaceus (WP086699320), Streptomyces pseudogriseolus (WP086681553), Streptomyces viridosporus (WP081241171), Streptomyces toyocaensis (WP037926726), S. griseoflavus (WP004922770), Streptomyces antibioticus (WP059195250), Streptomyces mirabilis (WP037706533) and Streptomyces canus (WP028807940). Conserved residues are shown in dark shaded boxes and light shaded boxes indicated similar residues. Conserved active site region of EXO-b-1,4-glucanase protein are specified in green box. The catalytic amino acids residues are indicated by arrows. [Click here to view] |
.png) | Figure 2: Multiple sequence alignment of ENDO13 protein and other ENDO-b-1,3-glucanase. The selected sequences from NCBI database are as follows: Streptomyces globisporus (WP030690772), Streptomyces venezuelae (WP041663301), Streptomyces pristinaespiralis (WP037772968), Streptomyces bikiniensis (WP030222470), Streptomyces flavochromogenes (WP030317051), Streptomyces sp. TLI (WP093872583), Streptomyces sp. CB02009 (WP073905603), Streptomyces antioxidants (WP079343793), Streptomyces griseus (WP069173959) and Streptomyces bobili (WP086771346). Conserved residues are shown in dark shaded boxes and light shaded boxes indicated similar residues. Conserved active site region of endo-b-1,3-glucanase protein are specified in green box. The catalytic amino acids residues are indicated by arrows. [Click here to view] |
Table 1: Physicochemical properties of proteins EXO-14 and ENDO-13.
| Parameters | EXO14 | ENDO13 |
|---|---|---|
| Number of amino acid residues | 578 | 430 |
| Molecular weight | 61152.67 | 44360.76 |
| Chemical formula | C2661H4046N742O892S14 | C1918H2956N582O615S11 |
| Theoretical PI | 4.34 | 8.11 |
| +R residues | 38 | 33 |
| −R residues | 76 | 31 |
| Extinction coefficient | 111270–111645 | 79410–79910 |
| Instability index | 24.65 | 29.37 |
| Aliphatic index | 66.26 | 59.35 |
| Estimated half-life in Escherichia coli | >10 h | >10 h |
| GRAVY | −0.446 | −0.424 |
The phylogenetic tree constructed based on the BLASTp revealed that the EXO-14 protein grouped into a clade with Streptomyces sp. 4F cellulose 1,4-b-cellobiosidase and Streptomyces griseorubens cellulose 1,4-b-cellobiosidase. The sequence showed more close similarity to the Streptomyces sp. 4F cellulose 1,4-b-cellobiosidase [Figure 3a]. Constructed neighbor-joining phylogenetic tree of ENDO-13 protein identified that it was grouped in a first clade and showed more similarity to S. globisporus ENDO13 [Figure 3b]. However, it displayed a prominent discrepancy from other sequences, this evidently confirmed the ENDO-13 protein from S. cinereoruber subsp. cinereoruber TBG-AL13 is a novel one.
.png) | Figure 3: Phylogenetic analysis (a) evolutionary history of EXO14 protein. The optimal tree with the sum of branch length = 0.51628067 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches, (b) the evolutionary history of ENDO13 protein. The optimal tree with the sum of branch length = 0.78494038 is shown. The evolutionary history of both proteins were inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. Each analysis involved 11 amino acid sequences. [Click here to view] |
CDD search of EXO-14 protein in concise mode showed that protein comprises two main functional domains, N-terminal carbohydrate binding module 2 (CBM2) family, and C-terminal glycosyl hydrolase family 6 (GH6). dbCAN domain search of EXO-14 protein done through Hidden Markov models for each CAZy families confirmed the protein belongs to GH6 CAZyme family with N-terminal CBM2 domain (residues 37 to 136) and C-terminal GH6 catalytic domain (residues 181 to 522) [Supplementary Figure 4]. The amino acid sequence of ENDO-13 protein perfectly matched with domain of glycosyl hydrolase 16 families (GH16) in CDD search. The active site containing GH16 catalytic domain was located at the N-terminal region and in the C-terminal, a Ricin-B-lectin domain was identified, which were very typical for ENDO13 enzymes. dbCAN annotation represented Ricin-B-lectin domain as structurally identical to carbohydrate binding module 13 (CBM13) in the CAZy database. Further, the study confirmed the length of N-terminal GH16 catalytic domain (residues 50–296) and C-terminal CBM13 domain (residues 312–429) [Supplementary Figure 5].The signal peptide prediction showed that both EXO-14 and ENDO-13 proteins were secretory in nature, with signal peptide of 32 and 35 amino acids, respectively. The cleavage site of EXO-14 existed in between the amino acid positions of Ala-32 and Ala-33 and ENDO-13 cleavage occurred in between Ala-35 and Ala-36 amino acid positions [Supplementary Figures 6 and 7]. Based on the results of signal analysis, CDD search and dbCAN analysis a schematic representation of EXO14 and ENDO13 proteins are shown in Figure 4. Secondary structures of EXO-14 and ENDO-13 proteins were predicted with high confidence using the PSIPREDv3.3 based on local sequence similarity. Both proteins have varied secondary structures containing several alpha-helices and beta-sheets with loop structural intervals, while both have a predicted N-terminal helical structure which indicated N-terminal leader peptide [Figure 5].
.png) | Figure 4: Schematic representation of structural and functional domains of (a) EXO-14 and (b) ENDO-13 proteins based on signal peptide analysis, CDD search and dbCAN analysis. [Click here to view] |
.png) | Figure 5: Predicted secondary structure of EXO-14 and ENDO-13 by PSIPRED v3.3 (a) predicted secondary structure of EXO-14 protein have 11 helixes and 15 sheets (b) predicted secondary structure of ENDO-13 have only 2 helixes and 24 sheets. a-helixes indicated in orange and b-sheets in blue. [Click here to view] |
The best 3D structural model of EXO14 predicted by I-TASSER using protein 4b4fA as the protein model target showed C-score = −2.07, estimated TM-Score = 0.47 ± 0.15, and RMSD = 12.7 ± 4.3 Å [Figure 6a]. I-TASSER also identified the enzyme commission classification of EXO-14 protein EC3.2.1.91 using structure based function annotation algorithm of COFACTOR. The ENDO-13 structural model predicted using 3dgtA as template protein, showed C-score = −1.01, estimated TM-Score = 0.59 ± 0.14, and RMSD = 9.3 ± 4.6 Å [Figure 6b]. The identified EC classification of ENDO-13 by I-TASSER was EC 3.2.1.39. The predicted structures were further validated using the superimposition of their corresponding templates. Superimposed structures of EXO-14 and ENDO-13 with its corresponding templates are shown in Figure 6c and d, respectively.
.png) | Figure 6: Molecular modeling of EXO-14 and ENDO-13 proteins and validation through Ramachandran plot (RM plot) (a) predicted 3D model of EXO-14 protein showed a tunnel like catalytic pocket, (b) 3D model of predicted ENDO-13 protein showed an open cleft like catalytic pocket, (c) superimposition of EXO-14 (beige color) and 4b4fA (blue color) (d) Superimposition of ENDO13 (beige color) and 3dgtA (blue color). [Click here to view] |
The molecular docking of prepared EXO-14 protein was done with a ligand cellotriose (PubChem CID: 5287993), a smallest unit of long chain b-(1,4)-D-glucan. Among the predicted five different active sites of EXO-14 protein, the active site (site score 1.008, size 102, Dscore 1.052, volume 407.484) with a groove containing catalytic residues performed a favorable docking interaction with cellotriose, which produced better docking score of -7.393 and Emodel energy −79.575. The docking confirmed that the catalytic pockets are encompasses within the tunnel and the cellotriose molecule showed desirable hydrophobic and H-bonding interactions within the binding cavity. The key amino acid residues involved in the binding of cellotriose were found to be Tyr239, Asp245, Ala247, Ala248, Ser251, Asp292, Asn296, Thr299, Asn300, Thr306, His345, Gly347, Trp348, Trp351, Asn354, Thr383, Ala384, Asn385, Asn479, Thr480, Gly481, Trp483, Met533, Pro542, Arg543, and Asn544 [Figure 7a]. Among these, only three residues (Asp245, His345, and Arg543) are involved in H-bonding interactions.
 | Figure 7: Ligand interaction diagrams (a) interaction of cellotriose molecule with EXO14 protein active site amino acid residues (b) interaction of laminaritriose molecule with ENDO13 protein active site amino acid residues. [Click here to view] |
The ENDO-13 docking was executed with ligand laminaritriose (PubChem CID: 11477625), a smallest unit of b-(1,3)-D-glucan long chain. The active site is open cleft like with site score of 0.952, size 994, Dscore 1.070, and volume 749.455 produced a positive interaction at better energy level with docking score of -11.026 and Emodel energy -78.365. The detected amino acid residues of ENDO-13 protein involved in the laminaritriose binding were Thr69, Tyr108, Tyr112, Tyr117, Thr118, Ser119, Ala155, Ser157, Leu159, Leu160, Asn162, Ser168, Trp170, Ser173, Gly174, Glu175, Asn177, Pro196, Ile264, Asn266, and Tyr272. Six amino acid residues, namely, Thr118, Ser157, Glu175, Ser173, Asn162, and Trp170 were actively involved in eight hydrogen bonding interactions for grasping the ligand molecule within the catalytic pocket [Figure 7b].
Streptomyces sp. from soil has been a reservoir of compounds and enzymes that gained scientific, medical, and industrial importance. Streptomyces isolates are acknowledged as chief producers of extracellular enzymes. However, current understanding of the glucanolytic enzymes from Streptomyces is very limited. As per our studies, two soil isolates S. althioticus TBG-MR17 and S. cinereoruber subsp. cinereoruber TBG-AL13 were potent producers of extracellular EXO14 and ENDO13, respectively. In this study, we have isolated the complete gene sequence of EXO-14 from S. althioticus TBG-MR17 and ENDO-13 from S. cinereoruber subsp. cinereoruber TBG-AL13, and scrutinized the biological structure and interaction topographies with particular substrates using in silico approach.
PCR amplification provided 1737 bp complete gene sequence of EXO-14 which encoded a putative protein of 578 amino acids. BLASTp analysis and phylogenetic tree construction confirmed that EXO-14 protein has very close resemblance to the Streptomyces sp. 4F cellulose 1,4-b-cellobiosidase. The functional domain and motif architectures were identified using CDD search and dbCAN annotation, this was vital for determining the molecular functions of proteins. The EXO-14 protein has N-terminal CBM2 domains and C-terminal GH6 domain, the characteristics functional domains of enzyme EXO14 (cellulose 1,4-b-cellobiosidase, or cellobiohydrolase) [27]. According to Tomotsune et al., [28] cellulose producing Streptomyces have been assigned to the protein families of GH5, GH6, GH9, GH12, and GH48 in CAZy database. Annotation of complete genome sequence of a pine-boring wood wasp associated Streptomyces sp. SirexAA-E (ActE) predicted 1,4-b-cellobiohydrolase with GH6 and CBM2 domains, but structural information was not elucidated [29]. In fungi and plants, GH modules are accompanied by CBM modules [30,31]. Mostly, CBM2 are fused on the C-terminal side of the catalytic domain in all glycoside hydrolase families, whereas in GH5 and GH6 domain families it is located on the N-terminal end of domain architecture [32]. Both GH and CBM domains are separated by a linker sequence, rich with Pro/Thr residues [33,34]. In CBM2/GH6, the average length of linker is 35 amino acids [35]. In our case, linker with 45 amino acids was found in between these two domains. As per report of Gilkes et al. [36], the catalytic site of EXO14 (cellulose 1,4-b-cellobiosidase) has a key characteristics consensus pattern, of V-x-Y-x(2)-P-x-R-D-C-[GSAF]-x(2)-[GSA](2)-x-G. The EXO-14 protein alignment also showed similar catalytic core pattern, VIYNLPGRDCAALASNG, in between amino acid 236 and 254.
The complete sequence of ENDO13 has 1293 bp that encodes a putative protein of 438 amino acids. Based on the multiple sequence alignment, phylogenetic analysis and domain annotations the ENDO-13 protein came under GH16 family of ENDO13 and showed more similarity to S. globisporus ENDO13. However, it displayed a prominent divergence from other sequences, which evidently confirmed the ENDO-13 protein a novel one. Functional annotation of ENDO13 protein confirmed that it has a N-terminal Ricin-B-lectin such as GH16 and C-terminal CBM13 domains which is typical for ENDO13 enzymes. Both domains are separated by a 16 amino acid long linker sequence. GH16 is one of the largest CAZyme family, including endoglucanases involved in glucan degradation [37]. Endo-1,3-b-glucanase from Cellulosimicrobium cellulans reported a N-terminal GH16 catalytic domain and C-terminal CBM13 regions separated by a Gly/Ser-rich linker [38]. Catalytic region of ENDO-13 protein showed a consensus sequence WPNSGEIDVMENVGFEP, similar to the characteristic consensus pattern, W-P-x(2)-G-E-[LIV]-D-[LIVF]-x(0,1)-E-x(2)-[GQ]-[KRNF]-x-[PSTA], found in all other ENDO13 (laminarinase) proteins. The two glutamic acid residues, Glu-175 and Glu-180, within this conserved region are thought to be involved in catalysis [39], though the aspartic acid residue (Asp-177) is significant in preserving the relative position of above catalytic amino acids [40]. In addition, minor differences were revealed in active site sequences, WPKCGEIDIMELLG, of Clostridium thermocellum ENDO13, Lic16A [41]. Our Streptomyces ENDO-13 also showed such some differences in the catalytic site regions when comparing with Laminarinase genes of Rhodothermus marinus, Thermotoga neapolitana, Bacillus circulans, and Strongylocentrotus purpuratus [42].
SignalP program detected the EXO-14 protein has a TAT signal peptide sequence of 32 amino acid in length. It helps to cleave the mature protein in between the amino acids Ala-32 and Ala-33. TAT pathway mostly secretes proteins in native folded state, found in both Streptomyces and E. coli [43,44]. 35 amino acids long N-terminus TAT family signal peptide domain was also found in ENDO-13 protein. It cleaves the mature protein in between Ala-35 and Ala-36 positions.
The analysis of physicochemical property showed that the isoelectric point (pI) of EXO14 was 4.34; this detailed the need to develop buffer system for pH based purification [45]. At pI, proteins are compact and stable [46]. Estimated half-life of EXO-14 in E. coli was more than 10 h and the designated instability index value of 24.65; this predicted that the protein was stable. The high aliphatic value of 66.26 showed that the protein was stable at varying range of temperatures since the aliphatic index is directly related with thermostability of protein [47]. Very low GRAVY value −0.446 concludes that the protein makes good interaction with water molecules. Secondary structure of EXO14 protein has 11 helixes and 15 sheets. Physicochemical properties deduces that the ENDO-13 has a pI value of 8.1 (pI>7.0), this reveals the basic nature of protein. Estimated half-life in E. coli was more than 10 h and the protein predicted as stable with instability index of 29.37. The aliphatic value of 59.35 showed that the ENDO-13 was less thermostable than EXO-14. Low GRAVY value, −0.424 revealed the good hydrophilic interaction. Predicted secondary structure has only 2 helixes and 24 sheets. Greater b-sheet content above other consistent secondary structures is a collective feature of b-1,3 glucanases [45].
Predicted three-dimensional structure of EXO-14 and ENDO-13 proteins using I-TASSER proved it as good quality with proper fold confirmation. Structural modeling in I-TASSER is based on the secondary structure enhanced profile-profile threading alignment [48]. Initially, it predicts secondary structure and then goes to tertiary structure. In the current study, for EXO-14 protein modeling, 4b4fA (Thermobifida fusca Cel6B) [49] was selected as best template with Z-score 2.46 and model 1 was selected as top ranked model based on high C-score (−2.07), signified the model was good. This was additionally validated by highest TM-Score (0.57 ± 0.15) and RMSD (12.7 ± 4.3 Å). For the molecular modeling of ENDO-13 protein, the template 3dgtA with Z-score= 8.44 (endo-1,3-b-glucanase from Streptomyces sioyaensis) [50] was selected as a best template and the best model was built with high C-score value 1.01, estimated TM-Score = 0.59 ± 0.14 and estimated RMSD = 9.3 ± 4.6 Å. The superimposed models of EXO-14 and ENDO-13 confirmed that the active sites of both enzymes are completely overlapped with corresponding templates. These two models were selected for further molecular docking studies to understand the enzyme substrate interaction.
The EXO-14 docking studies used cellotriose as ligand. It forms a favorable interaction with active site groove (site score 1.008, size 102, Dscore 1.052, volume 407.484) of protein and produced better docking score of −7.393 and Emodel energy −79.575. As mentioned by Wu et al. [51], we also found that a tunnel-type active site within the protein structure allows the polysaccharide chain to enter the active site from one entrance of tunnel in an “exo” fashion and release the products from other side while firmly clamping the chain. The key amino acid residues involved in the binding of cellotriose were found to be Tyr239, Asp245, Ala247, Ala248, Ser251, Asp292, Asn296, Thr299, Asn300, Thr306, His345, Gly347, Trp348, Trp351, Asn354, Thr383, Ala384, Asn385, Asn479, Thr480, Gly481, Trp483, Met533, Pro542, Arg543, and Asn544. The three residues involved in H-bonding interactions are Asp245, His345, and Arg543. By correlating the multiple sequence alignment of EXO-14 protein with other Streptomyces cellobiohydrolase, all these residues are conserved in all sequences. According to Chinnathambi et al. [52], cbhII putative catalytic site residues are conserved in different Trichoderma sp.
The molecular docking studies of ENDO13 protein were executed with ligand laminaritriose. The active site prepared by Site Map consisted site score of 0.952, size 994, Dscore 1.070, and volume 749.455 was used for the molecular docking studies of ENDO-13 protein. Here, the active site is an open cleft like structure, more propensities to cut internal bonds [51]. The docking results produced a positive interaction at better energy level with docking score of -11.026 and Emodel energy −78.365. The detected amino acid residues involved in catalysis binding were Thr69, Tyr108, Tyr112, Tyr117, Thr118, Ser119, Ala155, Ser157, Leu159, Leu160, Asn162, Ser168, Trp170, Ser173, Gly174, Glu175, Asn177, Pro196, Ile264, Asn266, and Tyr272. 6 amino acid residues, namely, Thr118, Ser157, Glu175, Ser173, Asn162, and Trp170 were actively involved in eight hydrogen bonding interactions.
In short, the study enlighten the theoretical overview of EXO14 and ENDO13 encoding genes and the corresponding predicted putative proteins from Western Ghats isolates S. althioticus TBG-MR17 and S. cinereoruber subsp. cinereoruber TBG-AL13, respectively. It delivers an overall idea about protein structural domain architecture that eventually governs their stability, function, and interaction with particular substrates. The study of structure–function relationships exclusively uncovers the complete potential of both enzymes. Overall, it provides new information into the Streptomyces b-glucanases and moreover delivers a basis for enhancing the expression studies and protein stability toward industrial applications.
Authors would like to acknowledge Woman Scientist Division, Kerala State Council for Science, Technology and Environment, Kerala for financial support.
Authors declared that they do not have any conflicts of interest.
1. Wettstein DV, Mikhaylenko G, Froseth JA, Kannangara CG. Improved barley broiler feed with transgenic malt containing heat-stable (1,3-1,4)-b-glucanase. Proc Natl Acad Sci USA 2000;97:13512-7. [CrossRef]
2. Lamp AE, Evans AM, Moritz JS. The effects of pelleting and glucanase supplementation in hulled barley based diets on feed manufacture, broiler performance, and digesta viscosity. J Appl Poult Res 2015;24:295-303. [CrossRef]
3. Bull AT, Chesters CG. The biochemistry of laminarin and the nature of laminarinase. Adv Enzymol 1963;28:325. [CrossRef]
4. Mann JW, Jeffries TW, Macmillan JD. Production and ecological significance of yeast cell wall degrading enzymes from Oerskovia. Appl Environ Microbiol 1978;36:594. [CrossRef]
5. Blackman LM, Cullerne DP, Hardham AR. Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genomics 2014;15:785. [CrossRef]
6. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res 2009;37:233-8. [CrossRef]
7. Bielecki S, Galas E. Microbial beta-glucanases different from cellulases. Crit Rev Biotechnol 1991;10:275-304. [CrossRef]
8. Bayat A. Science, medicine, and the future: Bioinformatics. BMJ 2002;324:1018-22. [CrossRef]
9. McGregor N, Morar M, Fenger TH, Stogios P, Lenfant N, Yin V, et al. Structure-function analysis of a mixed-linkage β-glucanase/xyloglucanase from the Key ruminal Bacteroidetes Prevotella bryantii B(1)4. J Biol Chem 2016;291:1175-97. [CrossRef]
10. Lim J, Lee C, Dhakshnamoorthy V, Park JS, Hong S. Molecular characterization of Streptomyces coelicolor A(3) SCO6548 as a cellulose 1,4-b-cellobiosidase. FEMS Microbiol Lett 2016;363:245. [CrossRef]
11. Lanka S, Talluri VR, Ganesh J, Latha NL. Homology modeling, molecular dynamic simulations and docking studies of a new cold active extracellular lipase, EnL a from Emericella nidulans NFCCI 3643. Trends Bioinform 2015;8:37-51. [CrossRef]
12. Hrmova M, Fincher GB. Structure-function relationships of b-D-glucan endo-and exohydrolases from higher plants. Plant Mol Biol 2001;47:73-91. [CrossRef]
13. George J, Arunachalam R, Paulkumar K, Wesely EG, Shiburaj S, Annadurai G. Characterization and phylogenetic analysis of cellulase producing Streptomyces noboritoensis SPKC1. Interdiscip Sci 2010;2:205-12. [CrossRef]
14. da Vinha FN, Gravina-Oliveira MP, Franco MN. Cellulase production by Streptomyces viridobrunneus SCPE-09 using lignocellulosic biomass as inducer substrate. Appl Biochem Biotechnol 2011;164:256-67. [CrossRef]
15. Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 1980;8:4321-5. [CrossRef]
16. Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95-8.
17. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 2013;30:772-80. [CrossRef]
18. Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 2014;42:W320-4. [CrossRef]
19. Gasteiger E, Hoogland C, Gattiker A. Protein identification and analysis tools on the ExPASy server. In: John M, editor. Walker, the Proteomics Protocols Handbook. United States: Humana Press; 2005. p. 571-607. [CrossRef]
20. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9. [CrossRef]
21. Marchler-Bauer A. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res 2013;41:384-52. [CrossRef]
22. Yin Y, Mao X, Yang J, Chen X, Mao F, Ying X. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 2012;40:445-51. [CrossRef]
23. McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics 2000;16:404-5. [CrossRef]
24. Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc 2010;5:725-38. [CrossRef]
25. Halgren T. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 2009;49:377-89. [CrossRef]
26. Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 2006;49:6177-96. [CrossRef]
27. Leo VV, Asem D, Zothanpuia, Singh BP. Actinobacteria: A highly potent source for holocellulose degrading enzymes. In: Singh JS, Singh DP, editors. Actinobacteria, New and Future Developments in Microbial Biotechnology and Bioengineering. Amsterdam: Elsevier; 2018. p. 191-205. [CrossRef]
28. Tomotsune K, Kasuga K, Tsuchida M, Shimura Y, Kobayashi M, Agematsu H, et al. Cloning and sequence analysis of the cellulase genes isolated from two cellulolytic streptomycetes and their heterologous expression in Streptomyces lividans. Int J Soc Mater Eng Resour 2014;20:213-8. [CrossRef]
29. Takasuka TE, Book AJ, Lewin GR, Currie CR, Fox BG. Aerobic deconstruction of cellulosic biomass by an insect associated Streptomyces. Science 2013;3:1-10. [CrossRef]
30. Lopez-Casado G, Urbanowicz BR, Damasceno CM, Rose JK. Plant glycosyl hydrolases and biofuels: A natural marriage. Curr Opin Plant Biol 2008;11:329-37. [CrossRef]
31. Hakkinen M, Arvas M, Oja M, Aro N, Penttila M, Saloheimo M, et al. Reannotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb Cell Fact 2012;11:134. [CrossRef]
32. Teegardin KA, James S, Barabote RD. Bioinformatic analysis of glycoside hydrolases in the proteomes of mesophilic and thermophilic Actinobacteria. MOJ Proteomics Bioinform 2017;5:75-81. [CrossRef]
33. Tomme P, Diane P, Emily DA, Robert AC, Miller JR, Antony R, et al. Comparison of a fungal (Family I) and bacterial (Family II) cellulose-binding domain. J Bacteriol 1995;117:4356-63. [CrossRef]
34. Jensen MS, Fredriksen L, MacKenzie AK, Pope PB, Leiros I, Chylenski P. Discovery and characterization of a thermostable two-domain GH6 endoglucanase from a compost metagenome. PLoS One 2018;13:e0197862. [CrossRef]
35. Sammond DW, Payne CM, Brunecky R, Himmel ME, Crowley MF, Gregg T, et al. Cellulase linkers are optimized based on domain type and function: Insights from sequence analysis, biophysical measurements, and molecular simulation. PLoS One 2012;7:e48615. [CrossRef]
36. Gilkes NR, Claeyssens M, Aebersold R, Henrissat B, Meinke A, Morrison HD, et al. Structural and functional relationships in two families of beta-1, 4-glycanases. Eur J Biochem 1991;202:367-77. [CrossRef]
37. Temuujina U, Chia W, Chang YB, Honga S. Identification and biochemical characterization of SCO3487 from Streptomyces coelicolor A3(2), an exo-and endo-type-agarase producing neoagarobiose. J Bacteriol 2012;194:142-9. [CrossRef]
38. Tanabe Y, Pang Z, Oda M. Cloning and sequencing of endo-1,3-b-glucanase from Cellulosimicrobium cellulans. J Biol Macromol 2008;8:60-3.
39. Juncosa M, Pons J, Dot T, Querol E, Planas A. Identification of active site carboxylic residues in Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase by site-directed mutagenesis. J Biol Chem 1994;269:14530-5.
40. Michel G, Chantalat L, Duee E, Barbeyron T, Henrissat B, Kloareg B, et al. The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: A novel insight in the evolution of clan-B glycoside hydrolases. Structure 2001;9:513-25. [CrossRef]
41. Fuchs KP, Zverlov VV, Velikodvorskaya GA, Lottspeich F, Schwarz WH. Lic16A of Clostridium thermocellum, a non-cellulosomal, highly complex endo-b-1,3-glucanase bound to the outer cell surface. Microbiology 2003;149:1021-31. [CrossRef]
42. Krah M, Misselwitz R, Politz O, Thomsen KK, Welfle H, Borriss R. The laminarinase from thermophilic eubacterium Rhodothermus marinus: Conformation, stability, and identification of active site carboxylic residues by site-directed mutagenesis. Eur J Biochem 2003;257:2101-11. [CrossRef]
43. Ize B, Stanley NR, Buchanan G, Palmer T. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol 2003;48:1183-93. [CrossRef]
44. Anne J, Vrancken K, Mellaert LV, Impe JV, Bernaerts K. Protein secretion biotechnology in gram-positive bacteria with special emphasis on Streptomyces lividans. Biochim Biophys Acta 2014;1843:1750-61. [CrossRef]
45. Abraham A, Narayanan SP, Philip S, Nair DG, Chandrasekharan A, Kochupurackal J. In silico characterization of a novel b-1,3-glucanase gene from Bacillus amyloliquefaciens-a bacterial endophyte of Hevea brasiliensis antagonistic to Phytophthora meadii. J Mol Model 2013;19:999-1007. [CrossRef]
46. Sahay A, Shakya M. In silico analysis and homology modelling of antioxidant proteins of spinach. J Proteomics Bioinform 2010;3:148-54. [CrossRef]
47. Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem 1980;88:1895-8.
48. Deshmukh RA, Jagtap S, Mandal MK, Mandal SK. Purification, biochemical characterization and structural modelling of alkali-stable b-1,4-xylan xylanohydrolase from Aspergillus fumigatus R1 isolated from soil. BMC Biotechnol 2016;16:11. [CrossRef]
49. Sandgren M, Wu M, Karkehabadi S, Mitchinson C, Kelemen BR, Larenas EA, et al. The structure of a bacterial cellobiohydrolase: The catalytic core of the Thermobifida fusca family GH6 cellobiohydrolase Cel6B. J Mol Biol 2013;425:622-35. [CrossRef]
50. Hong TY, Hsiao YY, Meng M, Li TT. The 1.5 a structure of endo-1, 3-beta-glucanase from Streptomyces sioyaensis: Evolution of the active-site structure for 1, 3-beta-glucan-binding specificity and hydrolysis. Acta Crystallogr D 2008;64:964-70. [CrossRef]
51. Wu M, Nerinckx W, Piens K, Ishida T, Hansson H, Sandgren M, et al. Rational design, synthesis, evaluation and enzyme-substrate structures of improved fluorogenic substrates for family 6 glycoside hydrolases. FEBS J 2013;80:184-98. [CrossRef]
52. Chinnathambi V, Balasubramanium M, Gurusamy R, Paramasamy G. Molecular cloning and expression of a family 6 cellobiohydrolase gene cbhII from Penicillium funiculosum NCL1. Adv Biosci Biotechnol 2015;6:213-22. [CrossRef]
 | Supplementary Figure 1: Cloning of ENDO13 and EXO14 genes in pGEM®-T Easy Vector (a) isolated Genomic DNA, Lane 1: Streptomyces althioticus TBG-MR17, Lane 2: Streptomyces cinereoruber subsp. cinereoruber TBG-AL13 (b) the PCR amplicons of ENDO13 (~1400 bp) and EXO14 (~2000 bp) genes (c) construction of pGEMT/EXO14 and pGEMT/ENDO13 plasmid (d) screening of positive colonies by colony PCR. Lane 1-5: Positive colonies with ENDO13 gene, Lane M: 1 kb DNA ladder, Lane 6-10: Positive colonies with EXO14 gene (e) confirmation of the presence of gene of interest in isolated plasmids. Lane 1: PCR amplification of pGEMT/ENDO13 with gene specific primers, Lane 2: PCR amplification of pGEMT/EXO14 with gene specific primers, Lane 3: pGEMT/ENDO13 plasmid, Lane 4: pGEMT/EXO13 plasmid, Lane 5: PCR amplification of pGEMT/ENDO13 with Tvect primers, Lane 6: PCR amplification of pGEMT/EXO14 with Tvect primers [Click here to view] |
 | Supplementary Figure 2: Aligned gene sequences obtained after sequencing and frame analysis. (a) EXO-14 sequence, 5’3’frame 1 showed 1737 bp full gene sequence. (b) ENDO-13 sequence, 5’3’frame 3 showed 1293 bp full gene sequence. Yellow color sequence denotes the pGEMT vector sequences, green, and red codons represents start and stop codons, respectively [Click here to view] |
 | Supplementary Figure 3: The ORF of EXO-14 gene with corresponding putative amino acids [Click here to view] |
.png) | Supplementary Figure 4: The ORF of ENDO-13 gene with corresponding putative amino acids [Click here to view] |
 | Supplementary Figure 5: Conserved domain search of EXO-14 protein. (a) NCBI conserved domain search based on sequence alignment recognized EXO-14 has linker which separates N-terminal CBM2 family and C-terminal GH6 family domains. (b) dbCAN domain search also confirmed the above domain architecture and the protein belongs to GH6 CAZyme family [Click here to view] |
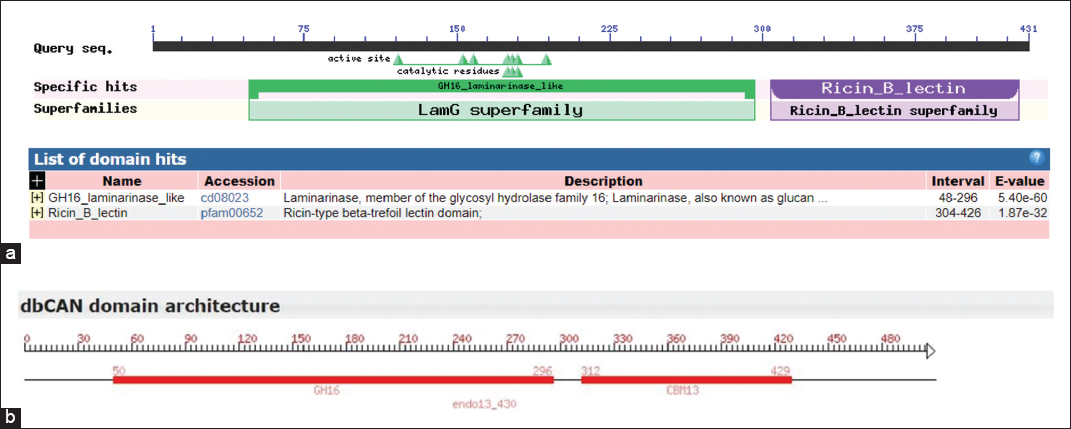 | Supplementary Figure 6: Conserved domain search of ENDO13 protein. (a) NCBI conserved domain search based on sequence alignment recognized ENDO-13 has N-terminal GH16-laminarinase-like domain and C-terminal Ricin-B-lectin family domains, both are separated by a linker sequence. (b) dbCAN domain architecture of ENDO-13 confirmed, the protein belongs to GH16 CAZyme family and Ricin-B-lectin domain was a carbohydrate binding module 13 (CBM13) family of CAZy database [Click here to view] |
 | Supplementary Figure 7: Signal peptide prediction of EXO-14 and ENDO-13 proteins. (a) Predicted signal sequence of EXO-14 protein showed cleavage site in between A-32 and A-33 with cutoff value 0.450. (b) Predicted signal sequence of EXO-14 protein showed a cleavage site in between A-35 and A-36 residues with 0.450 cutoff [Click here to view] |
1. Wettstein DV, Mikhaylenko G, Froseth JA, Kannangara CG. Improved barley broiler feed with transgenic malt containing heat-stable (1,3-1,4)-β-glucanase. Proc Natl Acad Sci USA 2000;97:13512-7. https://doi.org/10.1073/pnas.97.25.13512 | |
2. Lamp AE, Evans AM, Moritz JS. The effects of pelleting and glucanase supplementation in hulled barley based diets on feed manufacture, broiler performance, and digesta viscosity. J Appl Poult Res 2015;24:295-303. https://doi.org/10.3382/japr/pfv028 | |
3. Bull AT, Chesters CG. The biochemistry of laminarin and the nature of laminarinase. Adv Enzymol 1963;28:325. https://doi.org/10.1042/bj0860028 | |
4. Mann JW, Jeffries TW, Macmillan JD. Production and ecological significance of yeast cell wall degrading enzymes from Oerskovia. Appl Environ Microbiol 1978;36:594. https://doi.org/10.1128/AEM.36.4.594-605.1978 | |
5. Blackman LM, Cullerne DP, Hardham AR. Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genomics 2014;15:785. https://doi.org/10.1186/1471-2164-15-785 | |
6. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res 2009;37:233-8. https://doi.org/10.1093/nar/gkn663 | |
7. Bielecki S, Galas E. Microbial beta-glucanases different from cellulases. Crit Rev Biotechnol 1991;10:275-304. https://doi.org/10.3109/07388559109038212 | |
8. Bayat A. Science, medicine, and the future: Bioinformatics. BMJ 2002;324:1018-22. https://doi.org/10.1136/bmj.324.7344.1018 | |
9. McGregor N, Morar M, Fenger TH, Stogios P, Lenfant N, Yin V, et al. Structure-function analysis of a mixed-linkage β-glucanase/ xyloglucanase from the Key ruminal Bacteroidetes Prevotella bryantii B(1)4. J Biol Chem 2016;291:1175-97. https://doi.org/10.1074/jbc.M115.691659 | |
10. Lim J, Lee C, Dhakshnamoorthy V, Park JS, Hong S. Molecular characterization of Streptomyces coelicolor A(3) SCO6548 as a cellulose 1,4-β-cellobiosidase. FEMS Microbiol Lett 2016;363:245. https://doi.org/10.1093/femsle/fnv245 | |
11. Lanka S, Talluri VR, Ganesh J, Latha NL. Homology modeling, molecular dynamic simulations and docking studies of a new cold active extracellular lipase, EnL a from Emericella nidulans NFCCI 3643. Trends Bioinform 2015;8:37-51. https://doi.org/10.3923/tb.2015.37.51 | |
12. Hrmova M, Fincher GB. Structure-function relationships of β-D-glucan endo-and exohydrolases from higher plants. Plant Mol Biol 2001;47:73-91. https://doi.org/10.1023/A:1010619128894 | |
13. George J, Arunachalam R, Paulkumar K, Wesely EG, Shiburaj S, Annadurai G. Characterization and phylogenetic analysis of cellulase producing Streptomyces noboritoensis SPKC1. Interdiscip Sci 2010;2:205-12. https://doi.org/10.1007/s12539-010-0069-y | |
14. da Vinha FN, Gravina-Oliveira MP, Franco MN. Cellulase production by Streptomyces viridobrunneus SCPE-09 using lignocellulosic biomass as inducer substrate. Appl Biochem Biotechnol 2011;164:256-67. https://doi.org/10.1007/s12010-010-9132-8 | |
15. Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 1980;8:4321-5. https://doi.org/10.1093/nar/8.19.4321 | |
16. Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95-8. | |
17. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 2013;30:772-80. https://doi.org/10.1093/molbev/mst010 | |
18. Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 2014;42:W320-4. https://doi.org/10.1093/nar/gku316 | |
19. Gasteiger E, Hoogland C, Gattiker A. Protein identification and analysis tools on the ExPASy server. In: John M, editor. Walker, the Proteomics Protocols Handbook. United States: Humana Press; 2005. p. 571-607. https://doi.org/10.1385/1-59259-890-0:571 | |
20. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9. https://doi.org/10.1093/molbev/msy096 | |
21. Marchler-Bauer A. CDD: Conserved domains and protein three-dimensional structure. Nucleic Acids Res 2013;41:384-52. https://doi.org/10.1093/nar/gks1243 | |
22. Yin Y, Mao X, Yang J, Chen X, Mao F, Ying X. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 2012;40:445-51. https://doi.org/10.1093/nar/gks479 | |
23. McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics 2000;16:404-5. https://doi.org/10.1093/bioinformatics/16.4.404 | |
24. Roy A, Kucukural A, Zhang Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat Protoc 2010;5:725-38. https://doi.org/10.1038/nprot.2010.5 | |
25. Halgren T. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 2009;49:377-89. https://doi.org/10.1021/ci800324m | |
26. Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 2006;49:6177-96. https://doi.org/10.1021/jm051256o | |
27. Leo VV, Asem D, Zothanpuia, Singh BP. Actinobacteria: A highly potent source for holocellulose degrading enzymes. In: Singh JS, Singh DP, editors. Actinobacteria, New and Future Developments in Microbial Biotechnology and Bioengineering. Amsterdam: Elsevier; 2018. p. 191-205. https://doi.org/10.1016/B978-0-444-63994-3.00013-8 | |
28. Tomotsune K, Kasuga K, Tsuchida M, Shimura Y, Kobayashi M, Agematsu H, et al. Cloning and sequence analysis of the cellulase genes isolated from two cellulolytic streptomycetes and their heterologous expression in Streptomyces lividans. Int J Soc Mater Eng Resour 2014;20:213-8. https://doi.org/10.5188/ijsmer.20.213 | |
29. Takasuka TE, Book AJ, Lewin GR, Currie CR, Fox BG. Aerobic deconstruction of cellulosic biomass by an insect associated Streptomyces. Science 2013;3:1-10. https://doi.org/10.1038/srep01030 | |
30. Lopez-Casado G, Urbanowicz BR, Damasceno CM, Rose JK. Plant glycosyl hydrolases and biofuels: A natural marriage. Curr Opin Plant Biol 2008;11:329-37. https://doi.org/10.1016/j.pbi.2008.02.010 | |
31. Hakkinen M, Arvas M, Oja M, Aro N, Penttila M, Saloheimo M, et al. Reannotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb Cell Fact 2012;11:134. https://doi.org/10.1186/1475-2859-11-134 | |
32. Teegardin KA, James S, Barabote RD. Bioinformatic analysis of glycoside hydrolases in the proteomes of mesophilic and thermophilic Actinobacteria. MOJ Proteomics Bioinform 2017;5:75-81. https://doi.org/10.15406/mojpb.2017.05.00158 | |
33. Tomme P, Diane P, Emily DA, Robert AC, Miller JR, Antony R, et al. Comparison of a fungal (Family I) and bacterial (Family II) cellulose-binding domain. J Bacteriol 1995;117:4356-63. https://doi.org/10.1128/JB.177.15.4356-4363.1995 | |
34. Jensen MS, Fredriksen L, MacKenzie AK, Pope PB, Leiros I, Chylenski P. Discovery and characterization of a thermostable two-domain GH6 endoglucanase from a compost metagenome. PLoS One 2018;13:e0197862. https://doi.org/10.1371/journal.pone.0197862 | |
35. Sammond DW, Payne CM, Brunecky R, Himmel ME, Crowley MF, Gregg T, et al. Cellulase linkers are optimized based on domain type and function: Insights from sequence analysis, biophysical measurements, and molecular simulation. PLoS One 2012;7:e48615. https://doi.org/10.1371/journal.pone.0048615 | |
36. Gilkes NR, Claeyssens M, Aebersold R, Henrissat B, Meinke A, Morrison HD, et al. Structural and functional relationships in two families of beta-1, 4-glycanases. Eur J Biochem 1991;202:367-77. https://doi.org/10.1111/j.1432-1033.1991.tb16384.x | |
37. Temuujina U, Chia W, Chang YB, Honga S. Identification and biochemical characterization of SCO3487 from Streptomyces coelicolor A3(2), an exo-and endo-type-agarase producing neoagarobiose. J Bacteriol 2012;194:142-9. https://doi.org/10.1128/JB.05978-11 | |
38. Tanabe Y, Pang Z, Oda M. Cloning and sequencing of endo-1,3- β-glucanase from Cellulosimicrobium cellulans. J Biol Macromol 2008;8:60-3. | |
39. Juncosa M, Pons J, Dot T, Querol E, Planas A. Identification of active site carboxylic residues in Bacillus licheniformis 1,3-1,4-beta- D-glucan 4-glucanohydrolase by site-directed mutagenesis. J Biol Chem 1994;269:14530-5. | |
40. Michel G, Chantalat L, Duee E, Barbeyron T, Henrissat B, Kloareg B, et al. The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: A novel insight in the evolution of clan-B glycoside hydrolases. Structure 2001;9:513-25. https://doi.org/10.1016/S0969-2126(01)00612-8 | |
41. Fuchs KP, Zverlov VV, Velikodvorskaya GA, Lottspeich F, Schwarz WH. Lic16A of Clostridium thermocellum, a non-cellulosomal, highly complex endo-b-1,3-glucanase bound to the outer cell surface. Microbiology 2003;149:1021-31. https://doi.org/10.1099/mic.0.26153-0 | |
42. Krah M, Misselwitz R, Politz O, Thomsen KK, Welfle H, Borriss R. The laminarinase from thermophilic eubacterium Rhodothermus marinus: Conformation, stability, and identification of active site carboxylic residues by site-directed mutagenesis. Eur J Biochem 2003;257:2101-11. https://doi.org/10.1046/j.1432-1327.1998.2570101.x | |
43. Ize B, Stanley NR, Buchanan G, Palmer T. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol 2003;48:1183-93. https://doi.org/10.1046/j.1365-2958.2003.03504.x | |
44. Anne J, Vrancken K, Mellaert LV, Impe JV, Bernaerts K. Protein secretion biotechnology in gram-positive bacteria with special emphasis on Streptomyces lividans. Biochim Biophys Acta 2014;1843:1750-61. https://doi.org/10.1016/j.bbamcr.2013.12.023 | |
45. Abraham A, Narayanan SP, Philip S, Nair DG, Chandrasekharan A, Kochupurackal J. In silico characterization of a novel β-1,3-glucanase gene from Bacillus amyloliquefaciens-a bacterial endophyte of Hevea brasiliensis antagonistic to Phytophthora meadii. J Mol Model 2013;19:999-1007. https://doi.org/10.1007/s00894-012-1645-3 | |
46. Sahay A, Shakya M. In silico analysis and homology modelling of antioxidant proteins of spinach. J Proteomics Bioinform 2010;3:148-54. https://doi.org/10.4172/jpb.1000134 | |
47. Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem 1980;88:1895-8. | |
48. Deshmukh RA, Jagtap S, Mandal MK, Mandal SK. Purification, biochemical characterization and structural modelling of alkali-stable β-1,4-xylan xylanohydrolase from Aspergillus fumigatus R1 isolated from soil. BMC Biotechnol 2016;16:11. https://doi.org/10.1186/s12896-016-0242-4 | |
49. Sandgren M, Wu M, Karkehabadi S, Mitchinson C, Kelemen BR, Larenas EA, et al. The structure of a bacterial cellobiohydrolase: The catalytic core of the Thermobifida fusca family GH6 cellobiohydrolase Cel6B. J Mol Biol 2013;425:622-35. https://doi.org/10.1016/j.jmb.2012.11.039 | |
50. Hong TY, Hsiao YY, Meng M, Li TT. The 1.5 a structure of endo-1, 3-beta-glucanase from Streptomyces sioyaensis: Evolution of the active-site structure for 1, 3-beta-glucan-binding specificity and hydrolysis. Acta Crystallogr D 2008;64:964-70. https://doi.org/10.1107/S0907444908021550 | |
51. Wu M, Nerinckx W, Piens K, Ishida T, Hansson H, Sandgren M, et al. Rational design, synthesis, evaluation and enzyme-substrate structures of improved fluorogenic substrates for family 6 glycoside hydrolases. FEBS J 2013;80:184-98. https://doi.org/10.1111/febs.12060 | |
52. Chinnathambi V, Balasubramanium M, Gurusamy R, Paramasamy G. Molecular cloning and expression of a family 6 cellobiohydrolase gene cbhII from Penicillium funiculosum NCL1. Adv Biosci Biotechnol 2015;6:213-22. https://doi.org/10.4236/abb.2015.63021 | |
Year
Month