1. INTRODUCTION
Dengue is a mosquito-borne viral disease in tropical and subtropical countries worldwide. Dengue disease is caused by four structurally similar but serologically distinct dengue virus serotypes (DENV1~4) [1-3]. Humans are the only known hosts of DENVs and the dengue disease spectra range from undifferentiated fever to severe hemorrhagic fever. Although the vast majority of people infected with DENVs remain asymptomatic, dengue is preliminary characterized by typical dengue fever, and in severe cases, DENV infection may result dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) [4]. The severe dengue, DHF, and DSS include leakage of vascular fluids, hemorrhagic manifestation, and shock accompanied by thrombocytopenia and abundant complement activation [5], and under such conditions, the patients need immediate hospitalization.
The RNA genome of DENVs encodes only 10 proteins: The capsid protein (C-protein), pre-membrane protein (prM-protein), envelope protein (E-protein), and seven non-structural proteins (NS-protein) [6]. The E-protein is an external outer layer of mature DENVs and is composed of three domains, domain 1 (ED1), domain 2 (ED2), and domain 3 (ED3) [7]. Among the three domains, the ED3 is the major immunogenic domain and most likely to be involved DENV serotype differentiation [8-10]. Moreover, despite being very similar in sequences and structures among the four serotypes, the ED3 alone could define DENV serospecificity [11]. Therefore, the ED3 may be of interest for detail understanding of DENVs and may have far impact on future dengue disease management.
Although numerous attempts have been made to explore the DENV-specific residue determinants at atomic level, the identification of the DENV serotype-specific residue determinants still remains elusive. This is because of huge variations in sequences, dengue pathogenesis, generation of neutralizing and sub-neutralizing anti-DENV antibodies in natural DENV infection, and immunogenicity of different DENV serotypes in different geographic locations [12]. In addition, the dengue research community is still lacking any model system for investigation of DENV infection and its management in details. Therefore, development of a model system is of worth interest to explore DENV-serospecificity. As well as, dengue management can be aided through formulating a tetravalent ED3-based vaccine candidate in future [13]. Therefore, this review article deals with much information on dengue infection, DENV seroprevalence, and dengue severity, mainly, focusing on DHF and DSS. We also focused on dengue diagnosis using traditional methods, challenges in dengue diagnosis and dengue disease management through formulating, and ED3-based dengue vaccine through rational consideration of differences in the immunogenicity against four ED3s and antigen interference of four ED3s in a multivalent ED3 formulation.
2. DENGUE DISEASE AND DENGUE VIRUS SEROTYPE
Dengue, a mosquito-borne viral disease caused by structurally similar but antigenically distinct four dengue virus serotypes (DENV1~4), is one of the world’s emerging viral disease threats [1-2]. Despite numerous attempts over the past three decades, the “dengue” is still a mystery [14]; the world is still lacking any dengue-specific treatment and/or vaccine [13]. This is perhaps originated from the inherent sequences of very similar but distinctive four DENV serotypes [15]. The typical clinical signs and symptoms of dengue disease include undifferentiated high fever, vomiting, musculoskeletal pain, myalgia, re-o-pain, rash, nausea, and headache which were observed in almost all cases of DENV infections [16] [Table 1]. A detail analysis indicated that DENV1 and DENV2 result very similar patterns of clinical symptoms which were quite different from those caused by DENV3 and DENV4, for example, ache-pain and diarrhea were absent in DENV3 and DENV4 infections, respectively [16]. However, the number of samples analyzed in each category was very limited, therefore, vast number samples from different geolocations are worth considering for developing rationale for infecting DENV serotypes with clinical signs and symptoms of dengue infection.
Table 1: The association of infecting DENV serotypes with clinical signs and symptoms. Subjects included were hospitalized dengue cases in Chittagong, Bangladesh. The table is reproduced from Roy et al., 2022 [16].
| Serotype (number, n) | Fever (%) | Vomiting (%) | M-pain (%) | Ache and Pain (%) | Myalgia (%) | Re-o-pain (%) | Rash% | Diarrhea (%) | Nausea (%) | Headache (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| DENV1 (n=7) | 100 | 42 | 42 | 42 | 42 | 28 | 14 | 14 | 28 | 28 |
| DENV2 (n=8) | 100 | 25 | 25 | 12 | 50 | 37 | 25 | 12 | 25 | 62 |
| DENV3 (n=4) | 100 | 25 | 50 | -- | 100 | 50 | 50 | 25 | 25 | 100 |
| DENV4 (n=13) | 100 | 30 | 23 | 38 | 84 | 30 | 15 | -- | 53 | 100 |
| DENV1,4 (n=4) | 100 | 66 | 33 | 100 | 66.66 | 66 | - | - | 100 | 100 |
| DENV1,3 (n=4) | 100 | 66 | 16 | 66 | 33 | 16 | 16 | 83 | 100 | |
| DENV1,3,4 (n=1) | 100 | 100 | 100 | 100 | 100 |
There are several controversies over the association of infecting DENV serotype and dengue disease severity. For example, the DENV2 was found as the most prevalent serotype with the maximum severity in Northwest India [17], but in Tamil Nadu, DENV2 and DENV4 constituted 20.6% and 20.0%, respectively, of total severe dengue cases [17]. Furthermore, all four DENV serotypes were present in the dengue history (1956–2017) in Tamil Nadu, India [18]. The phylogenetic analysis of DENV isolates collected from several dengue endemic regions concluded that both the Asian and American/African DENV1 genotype cocirculated in Madurai, Tamil Nadu, the DENV2 might be established as a cosmopolitan genotype and the DENV3 genotypes were Asian strains than Indian strains [18]. On the other hand, the DENV4 has been the most dominant serotype since 1968 in Tamil Nadu with a shift in Clade C to D due to genetic distinction [18]. Interestingly, in the past, DENV3 was the most prevailed serotype in Bangladesh [19], Cameroon [20], and India [21], however, the 2016–2019 dengue outbreaks in Bangladesh had DENV4 as the foremost prevailed serotype, followed by DENV2, DENV1, and DENV3 serotypes [16]. Although any explanation for such DENV serotype replacement is yet to be available, such DENV serotype variation in recurrent dengue outbreaks might have impact on dengue incidence and dengue disease severity [22]. Moreover, not only the sero-conversion but also concurrent multiple DENV infections have been reported in many dengue endemic countries worldwide [23-27]. For example, 2018–2019 dengue outbreak in Chittagong, Bangladesh, had 24% concurrently multiple DENV infections (concurrently infected with more than 1 DENV serotype) [16]. Altogether, these observations clearly suggested that DENV serotyping based on clinical signs and symptoms and establishing association of infecting DENV serotypes and dengue severity are seemed more difficult that previously though [16].
The mosquito Aedes aegypti carrying dengue virus was originally found in tropical and subtropical zones, however, currently has spread to the majority of the continents [27]. However, disease severity varied a lot in different geolocations [28], very little is known about the association serotype and disease severity and the reasons for different disease severity, even against the same serotype in different location and in the same location but in different dengue endemics [28]. One possible origin could be random mutations in the DENV viral RNA genome over time in dengue endemic regions. Being a RNA virus, the RNA-dependent RNA polymerase makes errors during replication and generates sequence variation in DENV genome, and consequently, dengue viruses exist as population of genetic variants, also called “quasispecies” [29,30]. Furthermore, successive mutations throughout the DENV genome may arise from random mutations during the viral cycle, in part due to experimental deficits and, consequently, an environment prone to the error of the RNA polymerase [31].
3. DENV INFECTION
DENV infection results various degrees of disease spectra and symptoms depending on age and repeated/recurrent DENV encounter. For example, children <15 years of age are very vulnerable with high fatality/mortality following DENV infections [32], however, disease severity decreased with ages with almost complete DENV clearance within 5 days of infection [33,34]. However, shock or death may occur due to progress of the disease beyond the acute febrile stage to a plasma leakage stage [35] along with leukopenia and thrombocytopenia [36,37] in elderly people as well [38,39]. However, recent DENV outbreaks showed that dengue infection has been becoming a serious threat to older people as well as observed in Thailand and China [38,39].
In the past two decades, DENVs have been spreading worldwide very aggressively both as single serotype and concurrent multiple DENV infections. For example, in the 80s, DENV1 and DENV4 were mostly restricted in North Brazil [40]. However, DENV1 soon prevailed nationwide, and in 2009, the DENV1 again re-emerged inflicting an extensive epidemic in almost all areas of Brazil [41]. All four DENV serotypes cocirculated during 2000–2009, however, DENV2 was the most prevailed DENV type in Bangladesh followed by DENV1 and DENV3 during the same in 2000–2009 dengue outbreaks [42]. Interestingly, the DENV2 serotype was closely related to those reported from Southeast Asia. These observations suggested that DENVs in Bangladesh have increased in genotypic range and DENV genotypic mutations found in various Asian countries were also present in Bangladesh. In addition, the current spread of DENV4 originated in Southeast Asia [43], spread to the Indian subcontinent. However, there was no uniform distribution pattern among the genotypes, but a significant risk factor was for dengue viruses found in Vietnam [44]. Dengue viruses have spread unexpectedly internationally in the past few decades, after the outbreak of dengue virus in many countries including Europe and the USA [44]. It was also observed that cocirculation of DENV serotypes increases the risk of concurrent infections [45].
Although the reasons for such increase in dengue incidence and dengue disease severity are mostly unknown, the recurrent single serotype, concurrent multiple serotype infections may be worsening the dengue threats worldwide, in addition to being contributed from climate change (global worming) [46,47], frequent air transportation, increased population density [47,48], extreme poverty, and inadequate sanitation [49].
4. SEVERE DENGUE MANIFESTATION: DHF AND DSS
A primary single DENV serotype infection may be self-limiting. This is because the natural anti-DENV antibody developed against the infecting DENV serotypes may neutralize the specific DENV serotype following concurrent secondary infection [4]. On the other hand, a secondary subsequent heterotypic and/or concurrent multiple DENV infection might elicit severe syndrome [5,23] through antibody-dependent enhancement (ADE) being promoted by the primary sub-neutralizing anti-dengue antibody developed during primary DENV infection [42,50]. Although the precise mechanism by which DHF and DSS develop remains undefined, three principal theories have been postulated predominantly as the major contributors to the pathogenesis of DHF and DSS. These theories include the ADE theory, the virulent virus theory, and the T-lymphocyte activation theory [51]. However, these theories altogether cannot fully explain the complex dengue disease etiology in details. Moreover, the residues responsible for DENV-specific (serotype-specific) and DENV-cross-reactive (sero-cross-specificity) recognition of anti-DENV antibodies still remain elusive. This is because of huge variations in sequences, severity, and immunogenicity of four DENV serotypes in different geographic locations [52,53]. In addition, different studies focused on different targets and reported different residues as DENV-serospecificity determinants [54,55]. The risk of developing severe dengue can be increased due to subsequent infection (secondary heterologous infection) by another serotype [Figure 1] [56-58].
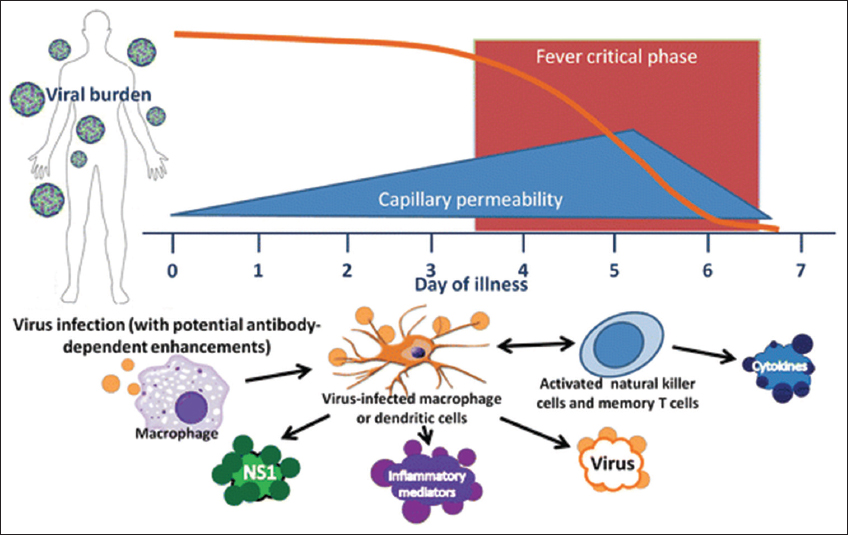 | Figure 1: Severe dengue manifestation. The DENVs infect monocyte-macrophage-dendritic cell lineage. Cytokines released from infected cells may result high fever, increased capillary permeability, rash, joint pain, and elevations of hepatic transferase enzymes which may lead severe dengue manifestation. The [Click here to view] |
5. DENGUE DIAGNOSIS
The precise diagnosis of a disease is the key to start treatment. As stated in the previous section based on clinical signs and symptoms, diagnosis of dengue is far more complicated than previously thought as the dengue disease spectra overlap with other febrile diseases [16]. Moreover, over 50% of DENV-infected patients may remain asymptomatic [60]. Therefore, determination of the virus, virus components, and anti-DENV antibodies are inevitable for confirmation of DENV infections. However, DENV diagnosis is in a state of dependence and general dengue diagnostic tools suffer from a very bad prediction. For example, many laboratory-based dengue diagnostic methods used for detecting dengue virus, viral RNA [61], viral antigens [62], and anti-dengue antibodies [63] have been reported which require specialized laboratory setup. The NS1-based diagnosis and MAC-ELISA are also quite commonly used [64]. However, the NS1 is very short-lived in infected human blood and it suffers from high false-negative prediction [64,65]. To some extent, some other routine tests, namely, full blood cell count, coagulation tests, routine biochemical, and lipid profile, are also prescribed. However, all these tests are far from being absolute confirmatory [66]. For example, it has been reported that anti-DENV IgM antibodies may be directly related to primary dengue infection and dengue disease severity [67]. However, only about 22% and 9% of patients with anti-DENV IgM and anti-DENV IgG antibodies, respectively, had adverse effects and asymptomatic DENV infection. Similarly, moreover, reverse transcriptase polymerase chain reaction is considered as gold standard in DENV serotyping for confirming the presence of viral RNA in the infected sera [68]. However, RNA is very unstable [69,70], therefore, it may suffer from high false-negative prediction [71]. Therefore, quantitative and qualitative diagnoses with high accuracy indeed needed to ensure timely exploitation of dengue disease in neglected communities.
6. ED3 IN DENGUE RESEARCH AND DENGUE MANAGEMENT
The E-protein is the outer layer of mature DENVs which is composed of three domains (ED1~ED3) [7,72,73]. Despite having very similar sequences and structures [7], the ED3s of DENV1-4 could define DENV serospecificity [9,74], and monoclonal antibodies developed against ED3s alone could restrict DENV infection in in vitro cell culture studies and in natural dengue infections [75,76]. Furthermore, all four ED3s could generate long-lasting immune response in mice model [77,78]. Altogether, these observations suggested that the domain ED3s of DENV1-4 may serve as a simplified model for investigating the natural immune response against DENV infection and may provide an worth considering tool for future DENV vaccine design [79]. It is quite well documented that the ED3 domain contains major determinants DENV serospecificity and sero-cross-specificity and virus-specific neutralization sites [80,81]. Interestingly, ED3s of DENVs exhibit the lowest sequence similarities with other flaviviruses cocirculating in the same endemic areas [82], and therefore, ED3s can be used as alternative to the existing DENV diagnosis tool for both qualitative and quantitative evaluations of natural dengue infection. For examples, detection of anti-DENV IgG and anti-DENV-IgM antibodies using ED3s may help differentiating acute and convalescent phases of dengue disease [83,84]. Altogether, these observations clearly suggested that artificially synthesized ED3s of DENV1-4 can be representatives of real DENVs, for both investigation of dengue infection and future dengue disease management, from diagnosis to vaccine design.
7. CHALLENGES RESTRICTING HUMAN EFFORTS AGAINST DENGUE
Dengue is still an unsolved mystery despite human efforts for over the past three decades and it is still one of the major public health threats [85]. The very first challenge is the proper understanding of the distinct antigenic determinants of DENV1-4 serotypes, each serotype being capable of causing diseases with different severity, and hence hinders the development of a common antigenic determinants conferring serospecificity against multiple DENV serotypes [86]. The second challenge is the ambiguous and structural context-dependent reports on serospecific and sero-cross-reactive immunological responses against DENV infections [87]. The third is the accumulation of random mutations during virus life cycle and variations found in sequence information in the successive dengue endemics. In addition, there is no established animal model system for dengue research to enable rational preclinical vaccine development [85]. Furthermore, very little is known about the mechanisms of dengue pathogenesis [85]. This is because all four DENVs are very similar in sequences and structures, they are very heterogeneous from immunological viewpoint [88], that is, DENV2, DENV3, and in some cases, DENV4 infections reported to downregulate chemokine responses while DENV1 increased IL-15, IL-6, and MCP-1 titers significantly. These findings suggested that profiles of “protective” or “pathologic” cytokines might contribute to subsequent subclinical or clinically apparent secondary DENV infection [89,90]. Furthermore, the dengue sickness might be a consequence of a complicated interplay of viral residences and host immune responses [91]. In addition, the association dengue pathogenesis with infecting DENV serotypes including single serotype primary and secondary DENV infection, secondary heterotypic, and concurrent multiple DENV infections is mostly unknown. For example, natural DENV infections not only reported to generate serospecific anti-DENV antibodies but also resulted sub-neutralizing sero-cross-reacting anti-DENV antibodies. Such heterogeneous immunogenicity of four DENVs, the sero-cross-talks of anti-DENV antibodies and immune interference among four different DENVs in several dengue vaccines initiatives might be restricting the success of several dengue vaccines in different stages of trials [92,93].
8. NATURAL IMMUNE RESPONSES AGAINST DENGUE INFECTION
It has been reported that a primary DENV infection may develop a neutralizing polyclonal antibody response against the infecting DENV serotype. However, such polyclonal anti-DENV antibody response may be sub-neutralizing making the subject susceptible to a secondary heterotypic DENV infection [91]. A primary anti-DENV IgM antibodies appear within 4–5 days
of infection (onset of fever) and could sustain for up to 3 months [94]. The primary anti-DENV IgG antibodies first appear about a week after onset of fever, sustain several weeks to months, and start declining to lower level which may persist for months to years [95]. Both the structural proteins (E-protein, PrM-protein, and C-protein) and non-structural proteins (NS1, NS3, and NS5) could be the prime targets of primary anti-DENV antibodies [96,97]. However, it is not known well antibodies developed against which components of DENVs are better protective and neutralizing. Moreover, the effects of primary anti-DENV antibodies on the outcomes of secondary heterotypic DENV and concurrent multiple DENV infections are mostly unknown. Therefore, it is worth investing the effects of primary anti-DENV antibodies on secondary heterotypic DENV infections in recurrent DENV infections in dengue endemic regions and concurrent multiple DENV infections in a single dengue session [98]. A detail analysis of anti-DENV antibodies from both serospecificity and sero-cross-reactivity is inevitable for formulating future tetravalent dengue vaccine resulting neutralizing anti-DENV antibodies against all four DENV serotypes.
9. CURRENT DENGUE RESEARCH STATUS
Despite numerous attempts over the past 30 years of dengue research, the success is still at the bottleneck. This is because of the mysterious inherent immunogenicity of four different DENVs, complex sero-cross-talks of anti-DENV antibodies, differential seroprevalence, and serotype-dependent dengue severity [99]. Previous attempts to control the mosquito vector had very limited success, making vaccines the most effective choice to combat this debilitating disease [13]. Although there is no dengue vaccine currently available, many promising vaccine candidates are in mid to late stages of development [100]. There are several difficulties; however, the generation of DENV-specific antibodies neutralizing all four DENV serotypes in a single dengue vaccine formulation is the most difficult one [101]. The two most promising vaccines, namely, Dengvaxia developed by Sanofi Pasteur [102] showed controversial efficacy at different geolocations and in different dengue-endemic regions [102]. For example, it could not generate type-specific antibodies against all four DENV serotypes at similar level, rather induced a skew anti-DENV antibody response against DENV1 serotype [102,103]. On the other hand, the TV003/005 developed by the National Institute of Allergy and Infectious Diseases [104] and reported to induce high levels of anti-DENV antibodies against all four DENV serotypes, however, much details on its efficacy in different ethnic groups in different dengue-endemic regions are yet to be available [105].
The domain 3 of dengue envelop protein (ED3) has been reported to mediate host-virus interactions and monoclonal antibodies developed against ED3s could neutralize DENVs almost completely both under in vivo and in vitro conditions [76]. Therefore, the fragmented DENV-specific ED3s would be a promising dengue vaccine candidate [106]. This is because, despite being a fragmented domain of the whole envelop protein, ED3s of DENVs were immunogenic and could retain the serospecificity of DENVs [Figure 2] [8,107]. Furthermore, the recombinant ED3s could the natively folded into stable native-like structure. Altogether, the ED3s may, therefore, serve as a simple model system for dengue research [107] and, accordingly, may serve as a template for future dengue vaccine instead of using live-attenuated DENVs [108].
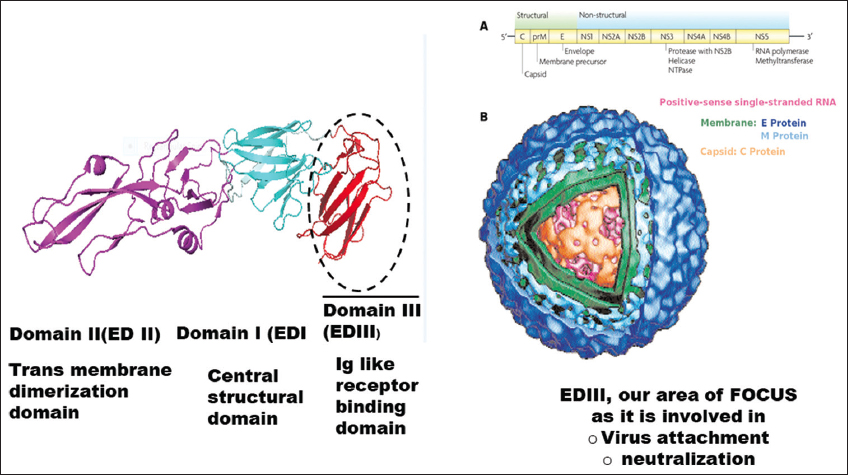 | Figure 2: The genomic organization (A) and structure (B) of DENV. The DENV genome encodes 10 proteins where the envelop protein constitutes the outermost surface and is consisted of three domains. The figure is reprinted from Elahi et al., 2014 [74]. [Click here to view] |
10. CONCLUSION
Despite extensive approaches over the past 30 years of dengue research, the world community is still lacking any dengue-specific treatment strategy and/or vaccines. The extensive literature review and discussions done in this review clearly suggested that the understanding of dengue disease etiology is still at its infancy. This is might be due to the very complex immune interference phenomenon including heterogeneous immunogenicity of DENVs, immune prevalence, immune tolerance, and mysterious cross-talks of different anti-DENV antibodies against several tetravalent dengue vaccine formulations. In addition, the generation of protective anti-DENV antibodies neutralizing all four serotypes seemed very much challenging. Furthermore, there is no model system for systematic dengue research, however, the domain 3 of dengue envelop protein (ED3) that we discussed here may serve as a very simplified model system for investigating natural anti-DENV responses against primary, secondary, secondary heterotypic, and concurrent multiple DENV infections and may, therefore, pave a way for designing an ED3-based tetravalent dengue vaccines ensuring neutralizing antibodies against all DENV serotypes in a single vaccine formulation.
11. ACKNOWLEDGMENT
MMI thanks GIR-MOE and Research and Publication Cell of Chittagong University, Bangladesh, for funding. Very special thank goes to Dr. Kuroda’s research group at Tokyo University of Agriculture and Technology, Tokyo, Japan, who is extensively doing dengue research-based DENV-specific ED3s.
12. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
13. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
14. ETHICAL APPROVALS
Ethical approvals not required for the review articles.
15. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
16. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Stanaway JD, Shepard DS, Undurraga EA, Halasa YA, Coffeng LE, Brady OJ,
2. World Health Organization. WHO, Dengue and Severe Dengue. Fact Sheet No. 117. Geneva:World Health Organization;2016.
3. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL,
4. Soo KM, Khalid B, Ching SM, Chee HY. Meta-analysis of dengue severity during infection by different dengue virus serotypes in primary and secondary infections. PLoS One 2016;11:e0154760. [CrossRef]
5. Guzman MG, Alvarez M, Halstead SB. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome:An historical perspective and role of antibody-dependent enhancement of infection. Arch Virol 2013;158:1445-59. [CrossRef]
6. Holmes EC, Twiddy SS. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol 2003;3:19-28. [CrossRef]
7. Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature 2004;427:313-9. [CrossRef]
8. Kulkarni MR, Islam MM, Numoto N, Elahi M, Mahib MR, Ito N,
9. Hussain M, Idrees M, Afzal S. Development of global consensus of dengue virus envelope glycoprotein for epitopes based vaccine design. Curr Comput Aided Drug Des 2015;11:84-97. [CrossRef]
10. Sultana M, Hasan N, Mahib MR, Islam MM. Systematic mutational analysis of epitope-grafted ED3 immunogenicity reveals a DENV3-DENV4 bi-serospecific ED3 mutant. Asian Pac J Trop Med 2022;15:63-70. [CrossRef]
11. Roehrig JT, Johnson AJ, Hunt AR, Bolin RA, Chu MC. Antibodies to dengue 2 virus E-glycoprotein synthetic peptides identify antigenic conformation. Virology 1990;177:668-75. [CrossRef]
12. Kleef EV, Bambrick H, Hales S. The Geographic Distribution of Dengue Fever and the Potential Influence of Global Climate Change TropIKA.net, 2010;1-22.
13. Thisyakorn U, Thisyakorn C. Latest developments and future directions in dengue vaccines. Ther Adv Vaccines 2014;2:3-9. [CrossRef]
14. Akshatha HS, Pujar GV, Sethu AK, Bhagyalalitha M, Singh M. Dengue structural proteins as antiviral drug targets:Current status in the drug discovery &development. Eur J Med Chem 2021;221:113527. [CrossRef]
15. Tamura T, Zhang J, Madan V, Biswas A, Schwoerer MP, Cafiero TR. Generation and characterization of genetically and antigenically diverse infectious clones of dengue virus serotypes 1-4. Emerg Microb Infect 2022;11:227-39. [CrossRef]
16. Roy MG, Uddin K, Islam D, Singh A, Islam MM. All four dengue virus serotypes co-circulate in concurrent dengue infections in a single dengue session in Chittagong, Bangladesh. Biores Commun 2022;8:1042-8. [CrossRef]
17. Gupta A, Rijhwani P, Pahadia MR, Kalia A, Choudhary S, Bansal DP,
18. Sankar SG, Sundari TM, Anand AA. Emergence of dengue 4 as dominant serotype during 2017 outbreak in South India and associated cytokine expression profile. Front Cell Infect Microbiol 2021;11:681937. [CrossRef]
19. Shirin T, Muraduzzaman AK, Alam AN, Sultana S, Siddiqua M, Khan MH,
20. Tchetgna HS, Yousseu FS, Kamgang B, Tedjou A, McCall PJ, Wondji CS. Concurrent circulation of dengue serotype 1, 2 and 3 among acute febrile patients in Cameroon. PLoS Negl Trop Dis 2021;15:e0009860. [CrossRef]
21. Mehta TK, Shah PD. Identification of prevalent dengue serotypes by reverse transcriptase polymerase chain reaction and correlation with severity of dengue as per the recent world health organization classification (2009). Indian J Med Microbiol 2018;36:273-8. [CrossRef]
22. Ainle MO, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborío S, Nuñez A,
23. Masyeni S, Yohan B, Sasmono RT. Concurrent infections of dengue virus serotypes in Bali, Indonesia. BMC Res Notes 2019;12:129. [CrossRef]
24. Tazeen A, Afreen N, Abdullah M, Deeba F, Haider SH, Kazim SN,
25. Utama IM, Lukman N, Sukmawati DD, Alisjahbana B, Alam A, Murniati D,
26. Reddy MN, Dungdung R, Valliyott L, Pilankatta R. Occurrence of concurrent infections with multiple serotypes of dengue viruses during 2013-2015 in northern Kerala, India. Peer J 2017;5:e2970. [CrossRef]
27. Dhanoa A, Hassan SS, Ngim CF, Lau CF, Chan TS, Adnan NA,
28. Morales I, Salje H, Saha S, Gurley ES. Seasonal distribution and climatic correlates of dengue disease in Dhaka, Bangladesh. Am J Trop Med Hyg 2016;94:1359-61. [CrossRef]
29. Freeman MC, Coyne CB, Green M, Williams JV, Silva LA. Emerging arboviruses and implications for pediatric transplantation:A review. Pediatr Transpl 2019;23:e13303. [CrossRef]
30. Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microiol 1997;51:151-78. [CrossRef]
31. Craig S, Thu MM, Lowry K, Wang XF, Holmes EC, Aaskov J. Diverse dengue Type 2 virus population contain recombination and both parental viruses in a single mosquito host. J Virol 2003;77:4463-7. [CrossRef]
32. Simmons JJ, John JH, Reynolds FH. Experimental studies of dengue. Philippine J Sci 1931;44:1-251.
33. Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg 1988;38:172-80. [CrossRef]
34. Thomas SJ, Strickman D, Vaughn DW. Dengue epidemiology:Virus epidemiology, ecology, and emergence. Adv Virus Res 2003;61:235-89. [CrossRef]
35. Rigau-Perez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vorndam AV. Dengue and dengue hemorrhagic fever. Lancet 1998;352:971-7. [CrossRef]
36. Isarangkura PB, Pongpanich B, Pintadit P, Phanichyakarn P, Valyasevi A. Hemostatic derangement in dengue hemorrhagic fever. Southeast Asian J Trop Med Public Health 1987;18:331-9.
37. Ashburn PM, Craig CF. Experimental investigations regarding the etiology of dengue fever. J Infect Dis 1907;4:440-75. [CrossRef]
38. Kalayanarooj S, Vaughn DW, Nimmannitya S, Green S, Suntayakorn S, Kunentrasai N,
39. Chhong LN, Poovorawan K, Hanboonkunupakarn B, Phumratanaprapin W, Soonthornworasiri N, Kittitrakul C. Prevalence and clinical manifestations of dengue in older patients in Bangkok Hospital for Tropical Diseases, Thailand. Trans R Soc Trop Med Hyg 2020;114:674-81. [CrossRef]
40. Kain D, Jechel DA, Melvin RG, Jazuli F, Klowak M. Hematologic parameters of acute dengue fever versus other febrile illnesses in ambulatory returned travelers. Curr Infect Dis Reports 2021;23:1-7. [CrossRef]
41. Nogueira FB, Faria NR, Nunes PC, Nogueira RM, Filippis AM, Santos FB. First detection and molecular characterization of a DENV- 1/DENV-4 co-infection during an epidemic in Rio de Janeiro, Brazil. Clin Case Rep 2018;6:2075-80. [CrossRef]
42. Wikramaratna PS, Simmons CP, Gupta S, Recker M. The effects of tertiary and quaternary infections on the epidemiology of dengue. PLoS One 2010;5:e12347. [CrossRef]
43. Suzuki K, Dungsombat J, Nakayama EE, Saito A, Egawa A, Sato T, Rahim R,
44. Sang S, Helmersson JL, Quam MB, Zhou H, Guo X, Wu H,
45. Guzman MG, Harris E. Dengue. Lancet 2015;385:453-65. [CrossRef]
46. Hsieh CC, Cia CT, Lee JC, Sung JM, Lee NY, Chen PL,
47. Wang WH, Chen HJ, Lin CY, Assavalapsakul W, Wang SF. Imported dengue fever and climatic variation are important determinants facilitating dengue epidemics in Southern Taiwan. J Infect 2020;80:121-42. [CrossRef]
48. Mahmood S, Irshad A, Nasir JM, Sharif F, Farooqi SH. Spatiotemporal analysis of dengue outbreaks in Samanabad town, Lahore metropolitan area, using geospatial techniques. Environ Monit Assess 2019;191:55. [CrossRef]
49. Costa SD, Branco MD, Junior JA, Rodrigues ZM, Queiroz RC, Araujo AS,
50. Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 1998;11:480-96. [CrossRef]
51. Srikiatkhachorn A, Wichit S, Gibbons RV, Green S, Libraty DH, Endy TP,
52. Sasmal SK, Takeuchi Y, Nakaoka S. T-Cell mediated adaptive immunity and antibody-dependent enhancement in secondary dengue infection. J Theor Biol 2019;470:50-63. [CrossRef]
53. Kurosu T, Chaichana P, Phanthanawiboon S, Khamlert C, Yamashita A, A-Nuegoonpipat A,
54. Rodriguez-Roche R, Blanc H, Bordería AV, Díaz G, Henningsson R, Gonzalez D,
55. Fleith RC, Lobo FP, Santos PF, Rocha MM, Bordignon J, Strottmann DM, Patricio DO,
56. Messer WB, Alwis RD, Yount BL, Royal SR, Huynh JP, Smith SA,
57. Halstead, S. Recent advances in understanding dengue. Natl Lib Med 2019;8:F1000 Faculty Rev-1279. [CrossRef]
58. Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T,
59. Lyday B, Chen T, Kesari S, Minev B. Overcoming tumor immune evasion with an unique arbovirus. J Transl Med 2015;13:3. [CrossRef]
60. Simon-Lorière E, Duong V, Tawfik A, Ung S, Ly S, Casadémont I,
61. Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol 1992;30:545-51. [CrossRef]
62. Matheus S, Deparis X, Labeau B, Lelarge J, Morvan J, Dussart P. Use of four dengue virus antigens for determination of dengue immune status by enzyme-linked immunosorbent assay of immunoglobulin G avidity. J Clin Microbiol 2005;43:5784-6. [CrossRef]
63. Kuno G, Gómez I, Gubler DJ. An ELISA procedure for the diagnosis of dengue infections. J Virol Methods 1991;33:101-13. [CrossRef]
64. Pok KY, Lai YL, Sng J, Ng LC. Evaluation of nonstructural 1 antigen assays for the diagnosis and surveillance of dengue in Singapore. Vector Borne Zoonotic Dis 2010;10:1009-16. [CrossRef]
65. Acosta PO, Granja F, Meneses CA, Nascimento IA, Sousa DD, Júnior WP,
66. Banerjee A, Paul UK, Bandyopadhyay A. Diagnosis of dengue fever:Roles of different laboratory test methods. Int J Am Med 2018;5:395-399. [CrossRef]
67. Liu L, Huang J, Zhong M, Yuan K, Chen Y. Seroprevalence of dengue virus among pregnant women in Guangdong.China. Viral Immunol 2020;33:48-53. [CrossRef]
68. Bhatnagar J, Blau DM, Shieh WJ, Paddock CD, Drew C, Liu L,
69. Anwar A, Wan G, Chua KB, August JT, Too HP. Evaluation of pre-analytical variables in the quantification of dengue virus by real-time polymerase chain reaction. J Mol Diagn 2009;11:537-42. [CrossRef]
70. Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med 2006;27:126-39. [CrossRef]
71. Koo C, Kaur S, Teh ZY, Xu H, Nasir A, Lai YL,
72. Mondotte JA, Lozach PY, Amara A, Gamarnik AV. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J Virol 2007;81:7136-48. [CrossRef]
73. Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A 2003;100:6986-91. [CrossRef]
74. Elahi M, Islam MM, Noguchi K, Yohda M, Toh H, Kuroda Y. Computational prediction and experimental characterization of a “size switch type repacking“during the evolution of dengue envelope protein domain III (ED3). Biochim Biophys Acta 2014;1844:585-92. [CrossRef]
75. Gromowski GD, Barrett ND, Barrett AD. Characterization of dengue virus complex-specific neutralizing epitopes on envelope protein domain III of dengue 2 Virus. J Virol 2008;82:8828-37. [CrossRef]
76. Matsui K, Gromowski GD, Li L, Schuh AJ, Lee JC, Barrett AD. Characterization of dengue complex-reactive epitopes on dengue 3 virus envelope protein domain III. Virology 2009;384:16-20. [CrossRef]
77. Apt D, Raviprakash K, Brinkman A, Semyonov A, Yang S, Skinner C,
78. Etemad B, Batra G, Raut R, Dahiya S, Khanam S, Swaminathan S,
79. Islam MM, Miura S, Hasan MN, Rahman N, Kuroda Y. Anti-dengue ED3 long-term immune response with T-cell memory generated using solubility controlling peptide tags. Front Immunol 2020;11:333. [CrossRef]
80. Gromowski GD, Barrett AD. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology 2007;366:349-60. [CrossRef]
81. Petty SS, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ,
82. Combe M, Lacoux X, Martinez J, Méjan O, Luciani F, Daniel S. Expression, refolding and bio-structural analysis of a tetravalent recombinant dengue envelope domain III protein for serological diagnosis. Protein Exp Purif 2017;133:57-65. [CrossRef]
83. Emmerich P, Mika A, Schmitz H. Detection of serotype-specific antibodies to the four dengue viruses using an immune complex binding (ICB) ELISA. PLoS Negl Trop Dis 2013;7(12):e2580. [CrossRef]
84. Batra G, Nemani SK, Tyagi P, Swaminathan S, Khann N. Evaluation of envelope domain III-based single chimeric tetravalent antigen and monovalent antigen mixtures for the detection of anti-dengue antibodies in human sera. BMC Infect Dis 2011;11:64. [CrossRef]
85. Friberg H, Beaumier CM, Park S, Pazoles P, Endy TP, Mathew A,
86. Murugesan A, Manoharan M. Dengue virus. Emerging and reemerging viral pathogens. 2020;16:281-359. [CrossRef]
87. Reich NG, Shrestha S, King AA, Rohani P, Lessler J, Kalayanarooj S,
88. Sigera PC, Amarasekara R, Rodrigo C, Rajapakse S, Weeratunga P, Silva NL,
89. Friberg H, Bashyam H, Maeda TT, Potts JA, Greenough T, Kalayanarooj S,
90. Woda M, Friberg H, Currier JR, Srikiatkhachorn A, Macareo LR, Green S. Dynamics of dengue virus (DENV)-Specific B cells in the response to DENV serotype 1 infections, using flow cytometry with labeled virions. J Infect Dis 2016;214:1001-9. [CrossRef]
91. Endy TP. Dengue human infection model performance parameters. J Infect Dis 2014;209:S56-60. [CrossRef]
92. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S,
93. Godói IP, Lemos LL, Araújo VE, Bonoto BC, Godman B, Júnior AA. CYD-TDV dengue vaccine:Systematic review and meta-analysis of efficacy, immunogenicity and safety. J Comp Eff Res 2017;6:165-80. [CrossRef]
94. St John AL, Rathore AP. Adaptive immune responses to primary and secondary dengue virus infections. Nat Rev Immunol 2019;19:218-30. [CrossRef]
95. Koraka P, Murgue B, Deparis X, Setiati TE, Suharti C, Gorp EC,
96. Olán LL, Sánchez GM, Cordero JG, Gutiérrez AE, Argumedo LS, Castañeda BG,
97. Alwis RD, Beltramello M, Messer WB, Petty SS, Wahala WM, Kraus A,
98. Halstead S. Recent advances in understanding dengue. F1000Res 2019;8:1279. [CrossRef]
99. Bosch I, Reddy A, Puig H, Ludert JE, Perdomo-Celis F, Narváez CF,
100. Khetarpal N, Khanna I. Dengue fever:Causes, complications, and vaccine strategies. J Immunol Res 2016;2016:6803098. [CrossRef]
101. Swaminathan S, Khanna N. Dengue vaccine development:Global and Indian scenarios. Int J Infect Dis 2019;84:80-6. [CrossRef]
102. Thomas SJ, Yoon IK. A review of Dengvaxia®:development to deployment. Hum Vaccin Immunother 2019;15:22952314. [CrossRef]
103. White MP, Elliott LR, Grellier J, Economou T, Bell S, Bratman GN,
104. Whitehead SS. Development of TV003/TV005, a single dose, highly immunogenic live attenuated dengue vaccine;What makes this vaccine different from the Sanofi-Pasteur CYD™vaccine?Expert Rev Vaccines 2016;15:509-17. [CrossRef]
105. Nivarthi UK, Swanstrom J, Delacruz MJ, Patel B, Anna P, Durbin AP,
106. Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ,
107. Elahi M, Islam MM, Noguchi K, Yohda M, Kuroda Y. High resolution crystal structure of dengue-3 envelope protein domain III suggests possible molecular mechanisms for serospecific antibody recognition. Proteins 2013;81:1090-5. [CrossRef]
108. Chávez JH, Silva JR, Amarilla AA, Figueiredo LT. Domain III peptides from flavivirus envelope protein are useful antigens for serologic diagnosis and targets for immunization. Biologicals 2010;38:613-8. https://doi.org/10.1016/j.biologicals.2010.07.004