1. INTRODUCTION
Tomato (Solanum lycopersicum) producing area in Vietnam is over 23,000 hectares [1]. The production of tomato is constrained by both biotic and abiotic factors. Among the biotic constraints, bacterial wilt caused by Pseudomonas solanacearum (or Ralstonia solanacearum), is regarded as the most destructive. The bacterium is well adapted to soil conditions and can survive in the rhizosphere. It possesses a wide variety of virulence factors, enters the roots through artificial or natural openings, and colonizes vascular tissues, causing internal and external symptoms that result in the plant’s death [2]. Moreover, P. solanacearum infects about 200 plant species belonging to over 50 botanical families [3]. It is ranked second on the list of the top 10 most destructive plant pathogenic bacteria [4]. The current tomato disease control strategies include crop rotation, the breeding of resistant varieties, and the application of chemical pesticides [5]. In Vietnam, chemical pesticides have been the most used way for controlling tomato illnesses. However, improper use of chemical pesticides can have a significant detrimental impact on human health and the long-term viability of tomato farming due to their unfavorable biological influence on the farming environment [6]. Because of these negative effects, biological management is an alternative method in agriculture because it is sustainable, efficient, and less harmful to the environment [7]. Nevertheless, bacteria and fungi, which are frequently utilized in biological management, are frequently ineffectual against rhizosphere bacterial infections [2].
Bacteriophage (phage) therapy is proving efficient in controlling rhizosphere bacterial targets due to its specificity, safety, and cost-effectiveness [2]. It has been demonstrated that lytic phages are effective and safe when employed to manage bacterial infections in crops, while also being harmless to animals and the environment [8]. Numerous earlier research has shown that bacteriophages can effectively reduce tomato bacterial wilt [2,8,9]. In this study, three P. solanacearum-specific phages were isolated. The potential of the phages against P. solanacearum was evaluated based on their morphology, latent period, burst size, lytic activity, and genomic information.
2. MATERIALS AND METHODS
2.1. Phage Isolation
Tomato plant samples were collected from tomato fields in Duc Trong district, Lam Dong province, Vietnam, and transported to the laboratory for phage isolation under cold conditions. Ten grams of the sample was ground and mixed with 10 mL of distilled water and 10% (w/v) chloroform. The mixture was then vortexed for 5 min before being centrifuged at 4000 rpm (Tomy MX-100, Japan) for 10 min at room temperature to obtain the supernatant and discard the plant residue and chloroform layers. To remove the remaining chloroform, the supernatant was centrifuged at 10,000 rpm for 5 min at 4°C. The resultant supernatant was filtered using a 0.45-μm pore size filter (Minisart® NY25 17846, Sartorius, Germany), and the filtrate was used for phage enrichment. P. solanacearum PS025 which caused a disease rate of 93.3% in tomato in a 7-day pathogenicity experiment at a dose of 108 CFU.mL-1 (data not shown) was used as a host. In the initial step of phage enrichment, the filtrate was fed to a 100 μL log-phase P. solanacearum PS025 culture in a falcon tube containing 9 mL of TSB medium. The culture was centrifuged for 5 min at a temperature of 4°C after shaking at 150 rpm (LSI-3016R, Daihan Labtech, Korea) for 24 h at 30°C. The resultant supernatant was run through a 0.22-μm pore size filter (Minisart® NY25 17845, Sartorius, Germany), and the filtrate was subjected to a plaque assay. A mixture of 100 μL filtrate and 200 μL log-phase P. solanacearum PS025 culture was poured over a 1.5% Luria-Bertani (LB) agar plate. After incubating at 30°C overnight, a single transparent plaque was removed from the plate, suspended in SM buffer, incubated at 4°C overnight, and passed through a 0.22-μm filter. To purify the phage, the filtrate was subjected to the above protocol three times in a row.
2.2. Transmission Electron Microscopy Examination
A high titer (approximately 1010 PFU/mL) phage suspension sample was generated and then negatively stained with 5% uranyl acetate. Bacteriophage morphologies were examined at the Vietnam National Institute of Hygiene and Epidemiology using a transmission electron microscope (JEOL JEM-1010, Japan) operating at 80 kV voltage and an instrumental magnification of 25,000–30,000.
2.3. One-step Growth Curve
Based on previous research, the one-step growth curves of three phages were determined with minor modifications [10]. The P. solanacearum PS021 culture was incubated at 30 °C, 150 rpm until its OD600 reached 0.1 (approximately 107 CFU/mL). The phage was added at a multiplicity of infection (MOI) of 0.01. (phage: host). This mixture was incubated for 10 min at 30°C and 150 rpm before being centrifuged for 5 min at 10,000 rpm and 4°C. The pellet was resuspended in the same volume of TSB and incubated as before. Every 5 min, a portion of the volume was taken and diluted 100 fold in SM buffer on ice. The diluted samples were centrifuged at 10,000 rpm for 5 min at 4°C, and the supernatant was used for phage titration using the double agar-layer technique. The phage’s latent period and burst size were measured using a previously established method [11]. The experiment was repeated three times.
2.4. In Vitro Control of Phage against P. solanacearum
The host bacterial culture in TSB was shaken at 150 rpm and 30°C until it reached an OD600 of 0.1. Next, it was separated into four aliquots. Each of the three aliquots contained the phage at a MOI of 0.01, 0.1, or 1.0. The other aliquot contained no phage (the control). The four mixtures were shaken at 150 rpm and 30°C. The OD600 value was periodically determined. Each trial was performed three times.
2.5. Phage Genome Sequencing and Analysis
Phage genomic DNA was extracted according to the previous study by Andrews [12]. The phage genome was utilized as input for library preparation with the NEBnext Ultra II DNA Library Prep Kit, and sequencing was done on an Illumina (NextSeq550, USA) (150 bp paired end) at KTEST (Ho Chi Minh City, Vietnam). FASTQC v0.11.9 [13] was utilized to evaluate the sequence quality of read data in FASTQ format files before further analysis. Unicycler v0.4.8 in conservative mode was utilized to carry out de novo assembly [14]. QUAST version 5.0.2 was utilized to evaluate entire contig assemblies [15]. BLAST search against the NCBI database was used to identify viral origin [16]. The GenBank files generated by RAST were then manually inspected to validate identified open reading frames. Using Proksee, a circular representation of the phage genome was generated. Using ResFinder v4.1, the presence of possible antibiotic resistance factors was explored [17]. Using BACPHILP, it was determined if the phage was more likely to be virulent (lytic) or temperate (lysogenic) [18]. tRNA detection was conducted using tRNAscan-SE v2.0 [19].
2.6. Statistical Analysis
One-way analysis of variance was used to process the resulting data using the Statistical Package for the Social Sciences version 20 software for Mac. Duncan’s multiple range test demonstrated a statistically significant difference between treatments at P ≤ 0.05.
3. RESULTS AND DISCUSSION
3.1. Lytic Activity and Morphology of Phages
Three bacteriophages such as BHDT3, BHDT4, and BHDT8 were isolated from tomato plant samples obtained from Bac Hoi village, in Duc Trong district, Lam Dong province. Following purification, three phages exhibited clear plaques [Figure 1]. The phage morphologies were clarified based on transmission electron microscopy morphological analysis [Figure 1]. It was discovered that three phages have icosahedral heads with a diameter of approximately 87 nm (BHDT3), 54 nm (BHDT4), or 61 nm (BHDT8). Three phages belonged to the Podovirus family and were classified as Autographiviridae [20]. The one-step growth curves of three phages are described in Figure 2. The latent period and burst size of the BHDT3 are 205 (min) and 12 ± 1.7 (PFU/cell). The values of the BHDT4 are 235 (min) and 17 ± 2.7 (PFU/cell). The BHDT8 shows the lowest latent period of 160 (min) and the highest burst size of 45 ± 5.4 (PFU/cell). It indicated that the BHDT8 had the highest lytic activity in terms of latent period and burst size.
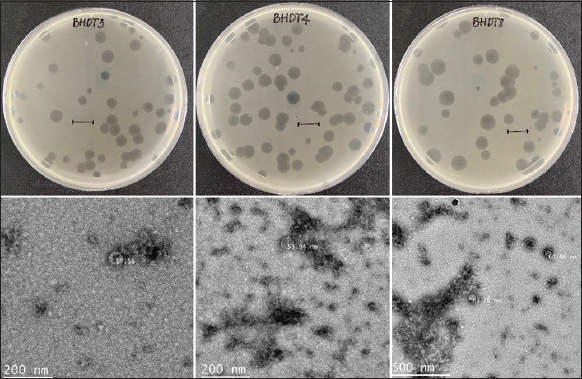 | Figure 1: Top agar overlay showing plaque morphology of three phages BHDT3, BHDT4, and BHDT8; the scale bar indicates 1 cm. Electron micrograph of three respective phages; the bar represents 200 nm or 500 nm. [Click here to view] |
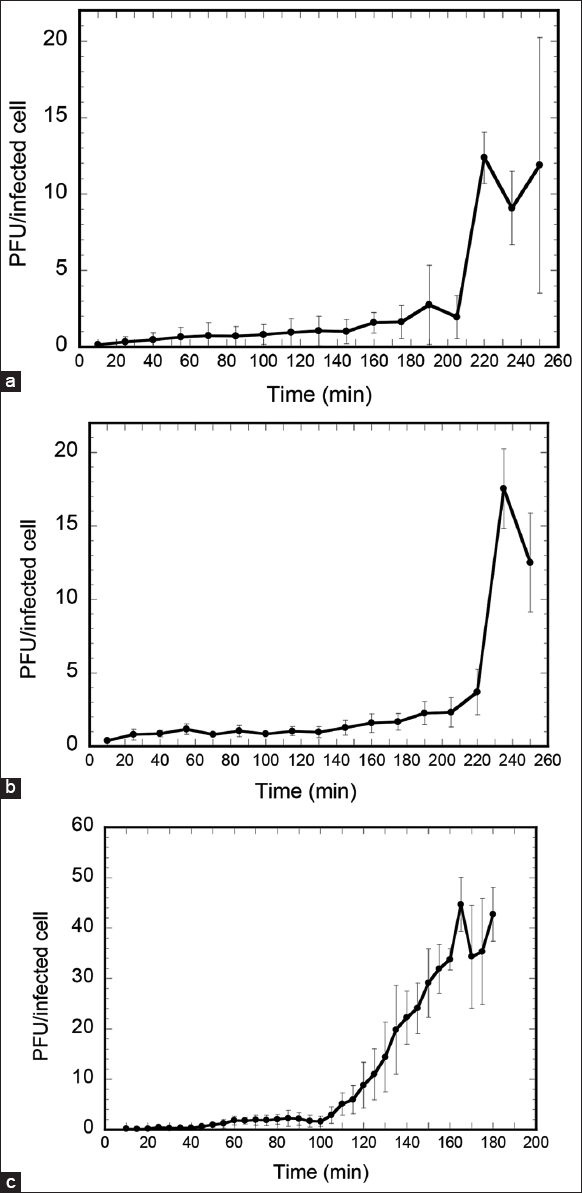 | Figure 2: The one-step growth curve of phage (a) BHDT3, (b) BHDT4, and (c) BHDT8. [Click here to view] |
3.2. Inactivation of P. solanacearum by Phages
The capacity of three phages to inhibit P. solanacearum growth at different MOI values in TSB was evaluated. As shown in Figure 3a, four treatments experienced an increase in OD600 during the first 2.0 h of incubation (with or without the phage). Nevertheless, from 2 h to 4 h of incubation, the OD600 of the bacterium-phage suspension at the MOI = 1.0 began to decrease, whereas that of the bacterium-phage suspension at the MOIs = 0.1, 0.01 and the control continued to rise. From 4 h to 6 h of incubation, the OD600 of the bacterium phage at three MOI values decreased, whereas the OD600 of the control dramatically increased. The sharp decrease in OD600 of the suspension was caused by the lysis of host cells by phages. The ability of BHDT3 to inactivate the host cells was demonstrated by the transparency of the bacterium-phage suspension until hour 34 (OD600 < 0.1), whereas the turbidity of the control was maintained throughout the experiment. The OD600 of the bacterium-phage solution increased after 34 h of incubation, indicating the growth of phage-resistant bacteria. Figure 3b and c depicted the capacity of phage BHDT4 and BHDT8 to inhibit P. solanacearum growth in TSB, respectively. The OD600 changes at the first 4 h were similar to that shown in Figure 3a. The ability of BHDT4 to inactivate the host cells was described by the transparency of the bacterium-phage suspension until the hour 36 (OD600 < 0.1). However, this ability of BHDT8 was much higher than that of the other two phages when the transparency of the bacterium-BHDT8 suspension was maintained until 52 h [Figure 3c]. It indicated a higher lytic activity of BHDT8 compared to BHDT3 and BHDT4. Therefore, phage BHDT8 was chosen to further analysis of its genome properties and safety for possible usage in tomato farming.
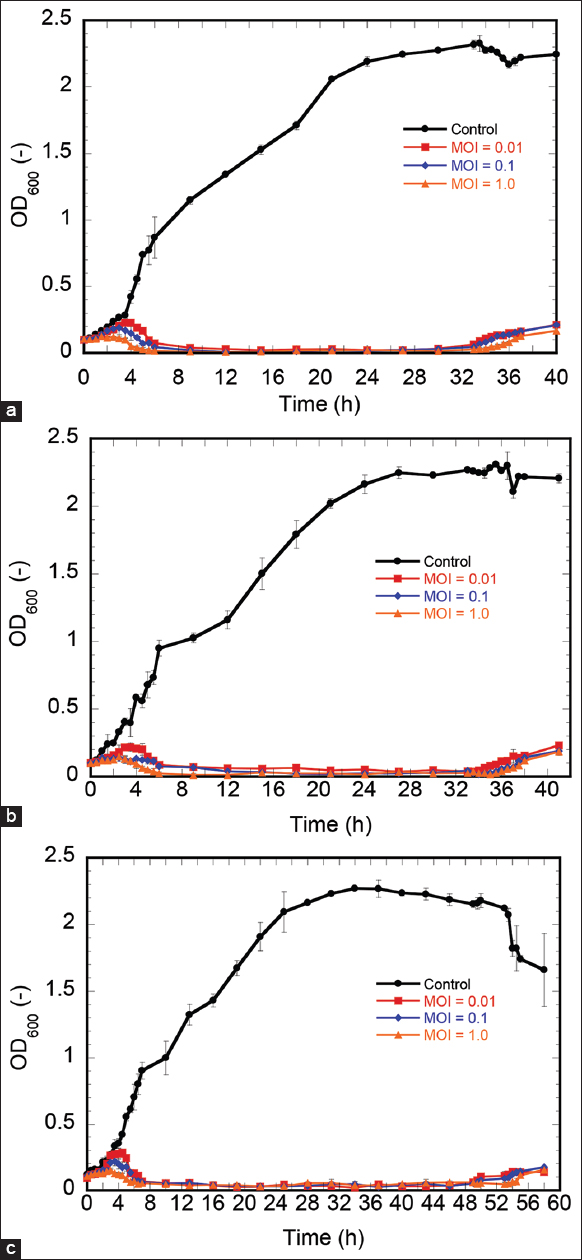 | Figure 3: Changes in optical density at 600 nm (OD600) during inactivation of P. solanacearum Ps021 by (a) BHDT3, (b) BHDT4, and (c) BHDT8 in TSB at 30°C at different multiplicities of infection values of 0.01 (square), 0.1 (diamond), and 1.0 (triangle). The negative control was without phage (circles). Error bars indicating 95% confidence intervals for the averaged values (n = 3) are not graphically detectable as the intervals are too narrow. [Click here to view] |
MOI value describes the number of phages against the number of host bacteria at the initial stage. In this study, the effect of three MOI values of 0.01, 0.1, and 1.0 on the inactivation of P. solanacearum by each phage was conducted and compared. There was no clear difference in the inactivation time among the three cases of MOI value. For instance, in Figure 3c, re-increase of OD600 value occurred at the same time (52 h) at MOI values of 0.01, 0.1, and 1.0. In the application of phages to control the pathogen, it prefers a smaller MOI value since it implies a smaller required amount of phage product.
3.3. Phage Genome Analysis
The nucleotide sequences for phage BHDT8 were submitted to NCBI under accession number OQ579022. The assembly generated one complete contig, with an estimated read depth of 2051x. The phage genome was 41,298 bp long with a total GC content of 62%, containing 47 predicted protein CDSs. The whole genome of phage BHDT8 encoding a variety of different functional proteins was annotated [Figure 4]. Genes related to DNA replication and modification enzymes were detected such as polymerase, exonuclease, endonuclease, helicase, primase, and peptidoglycan lytic exotransglycosylase. Furthermore, one lysozyme-encoding gene was also found in this genome. Genes encoding structural and packaging proteins were detected including capsid protein, head-to-tail connector, tail fiber, and tail tubular. The remaining predicted proteins were categorized as hypothetical [Supplementary Data from Table S1].
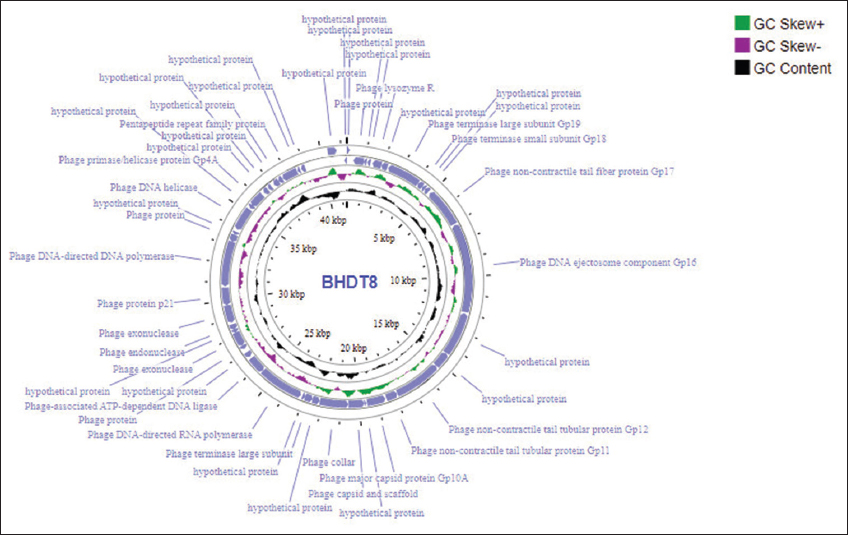 | Figure 4: Genome map of BHDT8. Relative orientations of open reading frames are indicated blue colors (sense and antisense). [Click here to view] |
The lifestyle of phage can be categorized into two states, lytic or lysogenic, with the former being essential for phage therapy candidates [21]. In its lysogenic state, a phage integrates its DNA into the host’s DNA to become a prophage, rendering itself dormant and suppressing the anti-bacterial properties typically displayed during its lytic state. The presence of a functional phage-encoded enzyme, integrase, is largely responsible for lysogeny’s mechanisms. Therefore, the presence of an integrase gene is not accepted for the therapy. Detecting any of the following would likely prevent a phage for therapeutic use: Genes encoding virulence factors, antimicrobial resistance, toxins, and transducing elements. Therefore, before using phages to control P. solanacearum infections in tomato, it is necessary to conduct a comprehensive genomic characterization of the phages. In this study, no known antibiotic resistant genes or virulence factors of pathogenic bacteria were detected in phage BHDT8. In addition, no known genes associated with lysogeny, such as integrase, were identified. The lifestyle prediction of the phage indicated that BHDT8 was likely to be virulent. The phage’s lytic nature suggested that it could serve as a potential agent against P. solanacearum, an infectious agent in Vietnam’s tomato.
4. CONCLUSION
The use of chemical pesticides has a significant impact on the viability of tomato farming and the health of the Vietnamese community. As a tool for preventing and treating bacterial diseases in plants, lytic bacteriophages have garnered considerable attention. In this study, three bacteriophages BHDT3, BHDT4, and BHDT8 specific to P. solanacearum causing bacterial wilt in tomato were isolated. Among the three phages, BHDT8 showed the highest capacity in the inactivation of P. solanacearum in vitro in broth. BHDT8 was also considered as a safe phage when there was no virulence or antibiotic resistance genes in its genome. Thus, it suggested that the phage would be a biotherapeutic agent against P. solanacearum in tomato.
5. ACKNOWLEDGMENTS
This research is funded by Vietnam National University Ho Chi Minh City, Vietnam (VNU-HCM) under grant number B2021-20-09.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
9. DATA AVAILABILITY
The nucleotide sequences for phage BHDT8 were submitted to NCBI under accession number OQ579022.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Chau MH, Chinh NX. Effect of plant density and fertilizer application rates on growth, fruit yield and quality of tomato (Solanum lycopersicum L.) in greenhouse condition. Asian Plant Res J 2021;8:22-31. [CrossRef]
2. Wang X, Wei Z, Yang K, Wang J, Jousset A, Xu Y, et al. Phage combination therapies for bacterial wilt disease in tomato. Nat Biotechnol 2019;37:1513-20. [CrossRef]
3. Hayward AC. Ralstonia solanacearum. In:Lederberg J, editor. Encyclopedia of microbiology. San Diego, CA:Academic Press;2000. 32-42.
4. Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 2012;13:614-29. [CrossRef]
5. Singh S, Gautam RK, Singh DR, Sharma TV, Sakthivel K, Roy SD. Genetic approaches for mitigating losses caused by bacterial wilt of tomato in tropical islands. Eur J Plant Pathol 2015;143:205-21. [CrossRef]
6. Nguyen TM, Le NT, Havukainen J, Hannaway DB. Pesticide use in vegetable production:A survey of Vietnamese farmers'knowledge. Plant Prot Sci 2018;54:203-14. [CrossRef]
7. Sharma P, Yadav M. Enhancing antibacterial properties of bacteriocins using combination therapy. J App Biol Biotech 2023;11:232-43. [CrossRef]
8. Wei C, Liu J, Maina AN, Mwaura FB, Yu J, Yan C, et al. Developing a bacteriophage cocktail for biocontrol of potato bacterial wilt. Virol Sin 2017;32:476-84. [CrossRef]
9. Thapa Magar R, Lee SY, Kim HJ, Lee SW. Biocontrol of bacterial wilt in tomato with a cocktail of lytic bacteriophages. Appl Microbiol Biotechnol 2022;106:3837-48. [CrossRef]
10. Hoang HA, Yen MH, Ngoan VT, Nga LP, Oanh DT. Virulent bacteriophage of Edwardsiella ictaluri isolated from kidney and liver of striped catfish Pangasianodon hypophthalmus in Vietnam. Dis Aquat Organ 2018;132:49-56. [CrossRef]
11. Pajunen M, Kiljunen S, Skurnik M. Bacteriophage fYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J Bacteriol 2000;182:5114-20. [CrossRef]
12. Tu VQ, Nguyen TT, Tran XT, Millard AD, Phan HT, Le NP, et al. Complete genome sequence of a novel lytic phage infecting Aeromonas hydrophila, an infectious agent in striped catfish (Pangasianodon hypophthalmus). Arch Virol 2020;165:2973-7. [CrossRef]
13. Andrews S. FastQC:A Quality Control Tool for High Throughput Sequence Data;2010. Available from:http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Last accessed on 2023 Mar 01].
14. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler:Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 2017;13:e1005595. [CrossRef]
15. Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST:Quality assessment tool for genome assemblies. Bioinformatics 2013;29:1072-5. [CrossRef]
16. Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST:A better web interface. Nucleic Acids Res 2008;36:W5-9. [CrossRef]
17. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 2020;75:3491-500. [CrossRef]
18. Hockenberry AJ, Wilke CO. BACPHLIP:Predicting bacteriophage lifestyle from conserved protein domains. PeerJ 2021;9:e11396. [CrossRef]
19. Lowe TM, Eddy SR. tRNAscan-SE:A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997;25:955-64. [CrossRef]
20. Turner D, Kropinski AM, Adriaenssens EM. A roadmap for genome-based phage taxonomy. Viruses 2021;13:506. [CrossRef]
21. Philipson CW, Voegtly LJ, Lueder MR, Long KA, Rice GK, Frey KG, et al. Characterizing phage genomes for therapeutic applications. Viruses 2018;10:188. [CrossRef]
Supplementary Data
Table S1: Results of BLASTP and predictive ORFs.
| CDS | Start (bp) | End (bp) | Length (a.a) | Blast protein | NCBI conserved domains | Final predict | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene product | Match (%) | Query cover (%) | E | Domain | E | |||||
| ORF-1 | 3 | 155 | 86 | Hypothetical protein | 105 | 100 | 6.0E−28 | Hypothetical protein | ||
| ORF-2 | 914 | 342 | 190 | Phage protein | 387 | 100 | 2.0E−135 | Phage protein | ||
| ORF-3 | 1069 | 911 | 52 | Hypothetical protein | 103 | 100 | 2.0E−27 | Hypothetical protein | ||
| ORF-4 | 1283 | 1101 | 99 | Hypothetical protein | 117 | 100 | 2.0E−32 | Hypothetical protein | ||
| ORF-5 | 1881 | 1384 | 165 | Putative lysozyme | 339 | 100 | 2.0E−117 | Endolysin_R21-like | 4.13E-43 | Phage lysozyme R |
| ORF-6 | 2228 | 1881 | 115 | Putative type II and type III secretion system protein precursor | 221 | 100 | 2.0E−72 | Hypothetical protein | ||
| ORF-7 | 4091 | 2274 | 605 | Large terminase subunit-like protein | 1257 | 100 | 0.0E+00 | Termin_lrg _T7 | 0 | Phage terminase large subunit Gp19 |
| ORF-8 | 4387 | 4088 | 99 | Putative terminase small subunit | 197 | 100 | 2.0E−63 | PHA01735 superfamily | 2.73E-08 | Phage terminase small subunit Gp18 |
| ORF-9 | 4572 | 4384 | 62 | Holin superfamily II protein | 128 | 100 | 3.0E−37 | Solute_trans_a superfamily | 6.74E-03 | Hypothetical protein |
| ORF-10 | 4816 | 4574 | 99 | Hypothetical protein | 160 | 100 | 5.0E−49 | M34_peptidase superfamily | 4.61E-04 | Hypothetical protein |
| ORF-11 | 7058 | 4923 | 711 | Putative non-contractile tail fiber protein | 1415 | 100 | 0 | Phage non-contractile tail fiber protein Gp17 | ||
| ORF-12 | 11,939 | 7122 | 1605 | Putative transglycosylase | 3265 | 100 | 0 | Phage DNA ejectosome component Gp16 | ||
| ORF-13 | 14,384 | 12,009 | 791 | Hypothetical protein | 1613 | 100 | 0 | Hypothetical protein | ||
| ORF-14 | 15,254 | 14,394 | 286 | Hypothetical protein | 560 | 100 | 0 | Hypothetical protein | ||
| ORF-15 | 17,850 | 15,265 | 861 | Tail tubular protein A | 1769 | 100 | 0 | Phage non-contractile tail tubular protein Gp12 | ||
| ORF-16 | 18,487 | 17,864 | 207 | Tail tubular protein A | 431 | 100 | 1.0E−152 | Phage non-contractile tail tubular protein Gp11 | ||
| ORF-17 | 19,555 | 18,545 | 336 | Major capsid-like protein | 682 | 100 | 0 | Phage major capsid protein Gp10A | ||
| ORF-18 | 19,699 | 19,586 | 37 | Hypothetical protein | 72 | 100 | 1.0E−15 | Hypothetical protein | ||
| ORF-19 | 20,571 | 19,699 | 290 | Scaffolding-like protein | 568 | 100 | 0 | Phage capsid and scaffold | ||
| ORF-20 | 22,116 | 20,572 | 514 | Head portal-like protein | 1058 | 100 | 0 | Phage collar, head-to-tail connector protein Gp8 | ||
| ORF-21 | 22,530 | 22,126 | 134 | Hypothetical protein | 251 | 100 | 8.0E−84 | Hypothetical protein | ||
| ORF-22 | 22,949 | 22,527 | 141 | Putative terminase large subunit | 289 | 100 | 2.0E−98 | Phage terminase large subunit | ||
| ORF-23 | 23,112 | 22,933 | 59 | Hypothetical protein | 117 | 100 | 1.0E−32 | Hypothetical protein | ||
| ORF-24 | 25,636 | 23,189 | 815 | RNA polymerase | 1708 | 100 | 0 | Phage DNA-directed RNA polymerase | ||
| ORF-25 | 26,571 | 25645 | 308 | ATP-dependent DNA ligase | 630 | 100 | 0 | Phage-associated ATP-dependent DNA ligase | ||
| ORF-26 | 26,974 | 26,672 | 100 | Hypothetical protein | 201 | 100 | 6.0E−65 | Hypothetical protein | ||
| ORF-27 | 27,406 | 27,185 | 73 | Putative capsid and scaffold protein | 142 | 100 | 1.0E−42 | Phage protein | ||
| ORF-28 | 28,197 | 27,403 | 264 | DNA polymerase exonuclease subunit | 552 | 100 | 0 | Phage exonuclease | ||
| ORF-29 | 28,384 | 28,184 | 66 | Hypothetical protein | 135 | 100 | 7.0E−40 | Hypothetical protein | ||
| ORF-30 | 28,673 | 28,368 | 101 | DNA endonuclease VII | 206 | 100 | 6.0E−67 | Phage endonuclease | ||
| ORF-31 | 29,734 | 28,763 | 323 | Exonuclease | 600 | 100 | 0 | Phage exonuclease | ||
| ORF-32 | 30,693 | 29,734 | 319 | Putative DNA-directed RNA polymerase | 645 | 100 | 0 | Phage protein p21 | ||
| ORF-33 | 33,215 | 30,762 | 817 | DNA polymerase | 1701 | 100 | 0 | Phage DNA-directed DNA polymerase | ||
| ORF-34 | 33,604 | 33,212 | 130 | Endonuclease | 269 | 100 | 5.0E−91 | Phage protein | ||
| ORF-35 | 33,789 | 33,604 | 61 | Hypothetical protein | 127 | 100 | 9.0E−37 | Hypothetical protein | ||
| ORF-36 | 35,192 | 33,915 | 425 | Replicative DNA helicase | 884 | 100 | 0 | Phage DNA helicase | ||
| ORF-37 | 35,466 | 35,260 | 68 | Hypothetical protein | 140 | 100 | 6.0E−42 | Hypothetical protein | ||
| ORF-38 | 36,248 | 35,445 | 267 | Putative DNA primase | 546 | 100 | 0 | Phage primase/helicase protein Gp4A | ||
| ORF-39 | 36,656 | 36,381 | 91 | Hypothetical protein | 172 | 100 | 1.0E−53 | Hypothetical protein | ||
| ORF-40 | 36,917 | 36,696 | 73 | Hypothetical protein | 152 | 100 | 2.0E−46 | Hypothetical protein | ||
| ORF-41 | 37,391 | 37,071 | 106 | Hypothetical protein | 213 | 100 | 2.0E−69 | Hypothetical protein | ||
| ORF-42 | 37,633 | 37,382 | 83 | Hypothetical protein | 167 | 100 | 5.0E−52 | Hypothetical protein | ||
| ORF-43 | 38,519 | 37,656 | 287 | Putative pentapeptide repeat protein | 554 | 100 | 0 | Pentapeptide repeat family protein | ||
| ORF-44 | 38706 | 38,545 | 53 | Hypothetical protein | 108 | 100 | 2.0E−29 | Hypothetical protein | ||
| ORF-45 | 39,002 | 38,760 | 80 | Hypothetical protein | 163 | 100 | 1.0E−50 | Hypothetical protein | ||
| ORF-46 | 40,334 | 40,795 | 153 | Hypothetical protein | 250 | 93 | 1.0E−81 | Hypothetical protein | ||
| ORF-47 | 41,296 | 41,159 | 45 | Hypothetical protein | ||||||
ORFs: Open reading frames