1. INTRODUCTION
Non-coding RNAs are class of functional regulatory non-protein coding RNAs and have potential role in different biomedical applications [1]. Scientific studies related to non-coding RNAs have expand impetus continuously from past to current time showing its ability in the area of gene expression modulation and control in cellular innate and adaptive immunity [2]. Micro-RNAs (mi-RNAs) are members of the small non-coding RNA family, have an important role in regulation of gene expression. They are non-coding regulatory RNA, small in size of approx ~22 nucleotides belongs to class of endogenous post-transcriptional regulators known to interact with expression of other genes, commonly identified as target genes [3]. Interaction of mi-RNA with target genes generally results in suppression of gene expression of target genes, by the means of translational repression, messenger RNA (mRNA) cleavage, and deadenylation [4]. Mi-RNAs and their associated proteins are found in abundance in the ribo-nucleoprotein complexes in the cells. As per the research report of different studies, it is proved that mi-RNAs are found in different biologically important body fluids such as serum, plasma, urine, peritoneal cavity fluid, cerebrospinal fluid, and fluid of follicles [5,6]. These mi-RNAs released in extracellular fluids and cells act as potential biomarkers for several diseases and cancer [7].
Now, it is evident from research result that mi-RNAs have role in the development of the several types of cancer and it is due to the dysregulation of the different oncogenic and tumor suppressor mi-RNAs. Each transcribed mi-RNA targets specific mRNA in the cytoplasm and performs regulatory mechanisms in a complex manner by formation of mi-RNAs-based RNA silencing complexes (RISCs) [Figure 1] [8]. The pre-transcribed mi-RNAs (pri-mi-RNAs) are dysregulated by unregulated expression of DROSHA/DGCR8 microprocessor proteins by either mutation or epigenetic factors [9]. The mi-RNAs may be upregulated or downregulated in HCC and perform transverse actions on tumor progression. The upregulated mi-RNAs act as onco-miRs, while the downregulated mi-RNAs act as a tumor suppressor.
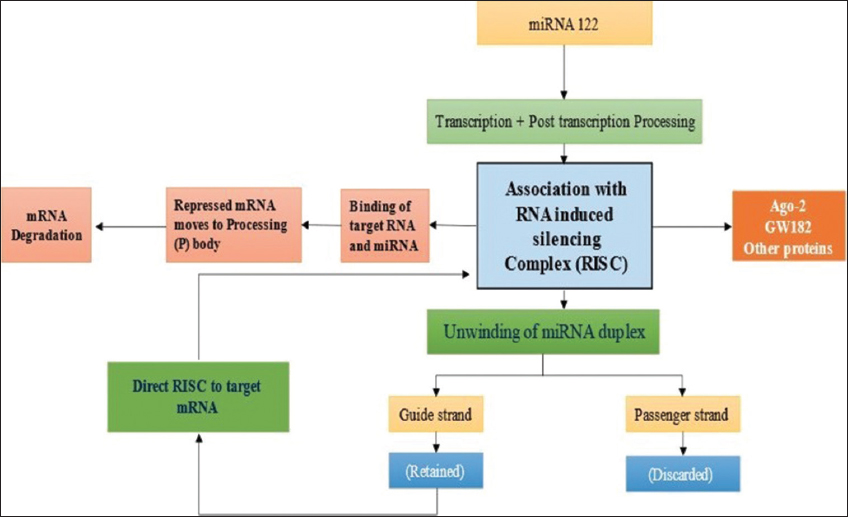 | Figure 1: Summary of miR-122 cellular expression and processing [8-10]. [Click here to view] |
The clinical significance of mi-RNA was identified in the therapy of several diseases. Mi-RNA antagonists and mi-RNA mimics are two approaches involved in utilization of mi-RNA molecule as therapeutic agents. Mi-RNA antagonists represent antisense RNA approach in which chemically modified mi-RNA are introduced to bind to target mi-RNA, that is, over expressed mi-RNAs in diseased cell type results in formation of irreversible mi-RNA duplex which is ultimately degraded. Mi-RNA antagonists find their application when target mi-RNA acquires a gain of function in diseased cell type. On the other hand, mi-RNA mimics come into play when target mi-RNA encounters loss of function. Mi-RNA mimics are introduced into diseased cell type to restore normal function. Advantages associated with utilization of mi-RNA as therapeutic molecules include their specific nature, tolerance in normal tissues, and few delivery issues as compared to protein encoding plasmid vector.
At present, several mi-RNAs have been discovered with relevance to the different important diseases. Here, focuses are made to the mi-RNAs having important role in the genesis and progression of liver cancer. In this regard, two main mi-RNAs selected are miR-122 and miR-21, details are as follows.
1.1. Mi-RNA 122 (miR-122)
Liver is considered as one of the most vital organ of mammalian body system. This organ has ability of self-regeneration of hepatic cells as well as it is actively involved in metabolic activities. Several mi-RNA has been accounted to its specific key role in biological activities of liver [Figure 2] [10]. Among liver related mi-RNAs, miR-122 is most abundant prominent biomarker for its biological activities and comprises about 52% of the total hepatic mi-RNAs. It is also among the highest expressed mi-RNA in liver cell with approximate 66,000 copies per hepatic cell. In humans, gene encoding miR-122 is specifically located on the chromosome number 18, has important role in several liver functions along with reported anti-inflammatory activity [11].
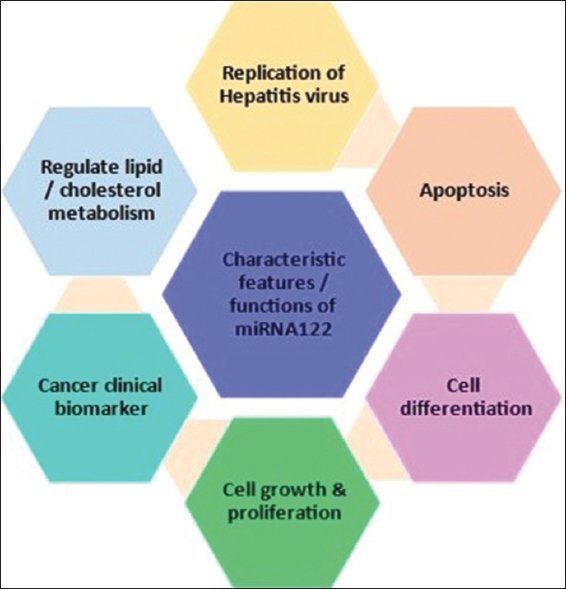 | Figure 2: Major cellular function of miR-122 [12,13]. [Click here to view] |
Studies suggested the multifunctional role of miR-122 in liver associated diseases and also involved in life cycle of pathogens infecting liver cells. It has been reported that down regulated miR-122 expression in liver fibrosis is due to prolonged chronic alcohol consumption. Regulation of inflammation, diet-induced obesity, cholesterol and iron metabolism, and fat metabolism are other clinical implications involving miR-122 [Figure 2]. However, now a days, most of the studies emphasized the crucial role of miR-122 in the development of liver cancer [12,13]. Figure 1 summarizes the outline of cellular processing of miR122.
1.2. Mi-RNA 21 (miR-21)
MiR-21 is the first time reported and discovered mi-RNA among all mi-RNAs identified, also known as has-miR21, miR-21 which is highly conserved and abundant micro-RNA. It is reported to be expressed in several cell types and is involved in several biologically important functions related to health and diseases [Figure 3]. The most promising application of miR-21 is in clinical diagnostics as a potential biomarker in early cancer diagnosis. The three main target of miR-21 are programmed cell death 4, tropomyosin 1 and phosphatase and tensin homolog (PTEN) [14-17]. The miR-21 is located in vacuole membrane protein-1 present on chromosome-7. High concentration of miR-21 supports its easy detection within different biological fluids named serum, urine, plasma, etc. The stability of miR-21 in body fluids supports utilization of miR-21 as a biomarker. The study conducted have reported miR-21 to act as oncogene and when over-expressed results in malignancy [18]. Mi-RNA upregulation by cytokines indicates their potential role as inflammatory response. It is found in various reports that miR-21 has involved in neoplastic as well as non-neoplastic diseases. Jenike and Halushka have provided a detailed review including expression of miR-21 in sixteen non-neoplastic diseases and 13 neoplastics [19]. Besides being a highly expressed mi-RNA, miR-21 is also expressed in large numbers of cell types with extremely high concentration in monocytes, macrophages, and dendritic cells. However, high level of expression of miR-21 in several cell types appears as a challenge for clinical application of miR-21 as a specific biomarker. Hence, relevant researches are essential requirements for the establishment of miR-21 as potential biomarker or as regulatory molecule.
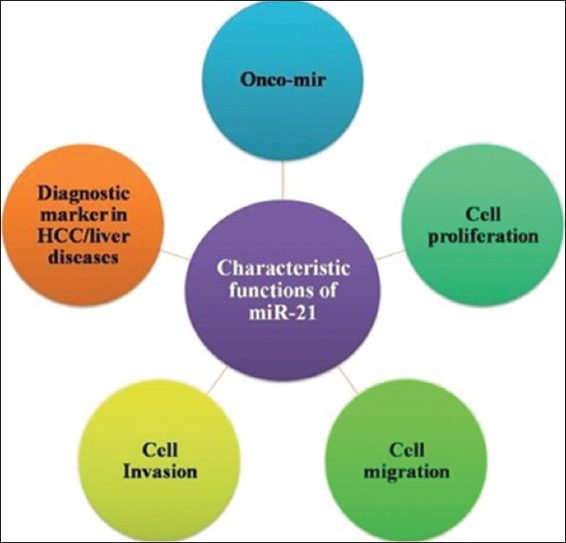 | Figure 3: Major cellular function of miR-21 [15,16,18,19]. [Click here to view] |
2. BIOMEDICAL APPLICATIONS
2.1. Mi-RNA-122 in Liver Cancer Diagnosis
Studies conducted have established miR-122 to exhibit potential to be utilized as biomarker in liver cancer diagnosis. The diagnostic potential application is attributed to differential gene expression of miR-122 in cancerous and non-cancerous cells. Dai et al., studied the applicability of miR-122 in the diagnosis as well as prognosis of cancer with specific reference to hepatocellular carcinoma (HCC). This study reported the elevated expression of miR-122 in the case of HCC in comparison to miR-122 expression in normal cells. As per the research result, it is also recommended that combination of miR-122 with alpha-feto protein can further enhance the diagnostic efficiency to utilize mi-RNA expression as biomarker for cancer diagnosis [20]. Another specifically studied clinical application of miR-122 is in Hepatitis C virus (HCV) infection. As HCV replication requires interaction of miRNA-122 with 5’ non-coding region of the virus, so decreased miR-122 expression has been directly related to reduction in replication of HCV [21]. The durable schematic representation regarding cell culture for HCV infection is limited only to the researcher using specific clones for HCV cell culture and cultured cells derived from human specific hepatoma cell line Huh7. Human specific hepatic cell lines specific to lentiviral vector showed a higher expression pattern of miR-122 comparable to endogenous expression pattern in Huh7 cell line. Cells of Hep3B cell line gradually corresponded miR-122 (Hep3B/miR122) among the studied cell lines showed a critical improvement of HCV in cell culture engendering [22].
Upregulation of an instant objective of miR-122 and arginine carrier SLC7A1 belongs to solute transporter family 7 involve in prolonged increment of arginine level inside cell. Arginine served as substrate for enzyme nitric oxide synthetase and this enzyme levels were extended in HCC cells with silenced miR-122 expression inside cell by extended safety by use of a kinase inhibitor anti-cancer drug sorafenib [23]. The most notable mi-RNA reported in the development of human liver cancer is miR-122, which is thought to be a prospective marker linked to the damage of liver tissue [24]. Due to the existence of the HCV core, miR204-HPCAL1-lncRNAHOTTIP and miR-122-TGFBRAP1 pathways were reasonably connected with the progression of cancer. Mi-RNAs such as miR-122 and miR-204 over-expression may control the progression of HCC by down regulating the expression of TGFBRAP1 and HOTTIP [25]. The transcriptional studies reported implicated that hypoxia-inducible factor 1a (HIF1a) in the enlistment of miR-122 and recognized the oxygen-detecting prolyl hydroxylase space 1 (PHD1) as a miR-122 target. Further investigations showed that HIF1a-subordinate acceptance of miR122 took part in a feed-forward pathway for liver security by means of the improvement of hepatic HIF responses through PHD1 repression [26]. Increased expression of GRHL2 in mice livers and liver of ALD patients inhibits the transcription of miR-122. Anti-miR-122 articulation reduced the extent of liver damage in mice after ethanol ingestion. By reducing HIF1 levels, miR-122 appears to protect the liver from damage brought on by ethanol. To diminish the harshness of ALD in patients, these cycles may be managed [27]. The capacity of miR-122 to advance separation of early stage and grown-up foundational microorganisms to hepatocytes in vitro suggests its expected outcome in dynamic hepatic separation program. As a key injury biomarker of liver, miR-122 has shown astonishing prospective for timely and susceptible in situ detection of DILI [28]. Glycyrrhetinic acid has positive impact on DILI, but its administration is challenged by long-term and/or high dose effect [29]. By raising Sirt1 and activating the AMPK pathway, Long et al. found that miR-122 suppression protects hepatic cells from lipid metabolic trouble such as non-alcoholic fatty liver disease (NAFLD) and suppresses lipogenesis [30]. ADAM10 and c-Met are directly targeted by both mi-RNAs miR-122 and miR144. Upregulated expression of mi-RNAs miR-122 and miR-144 prompted diminished articulation of ADAM10 and c-Met in the UM cell lines and impaired cell expansion, movement, cell cycle, and flaking of c-Met ecto-space [31]. Sorafenib is commonly utilized in the treatment of HCC; however, the drug dose not process to be uniformly beneficial for all patients [32]. A major issue associated with sorafenib is the acquired resistance [33]. Being a conserved liver specific mi-RNA, miR-122 has been reported to play a key role in liver metabolism as well as homeostasis. miR-122 is looked on a potential molecule to act as biomarker for detecting/diagnostics of liver damage [28]. Long et al. reported miR-122 to be a potential biomarker and also a suitable drug target for NAFLD [30]. In the study, both in vitro (free fatty acid treated Hep G2 and Huh7) and in vivo (mice fed with high fat diet) were analyzed with observations of excess lipid production and triglyceride secretion in presence of miR-122 upregulation which downregulates Sirt1. MiR-122 inhibition resulted in suppressed lipogenesis and reduced excess lipid production. Fauda et al. reported utilization of combination of four miRNA named as miR-299, miR-200, miR-335, and miR-21) to identify HCC [34]. Most of the studies conducted pertaining to application of miR-122 as biomarker is confined to liver related disorders.
2.2. HCC Development by Oncogenic mi-RNAs
Oncogenic mi-RNAs are frequently expressed during different cancer cases such as breast, lung cancer, lymphoma, prostate cancer, melanoma, colorectal cancer, and liver cancer. OncomiRs are those dysregulated mi-RNAs that are upregulated in HCC and other cancer types. Many onco-miRNA types that are specific to their targets undergo specific mechanisms in tumor cell growth and invasion, whereby some are mentioned in Table 1. Specifically, miR-21 expression increased in cells, serum, and other body fluids of HCC patients [4]. miR-21 in HCC cases has oncogenic properties in most estimations [35]. Other miRNAs such as miR-21/-221/-451 suppress the activity of PTEN deleted on chromosome 10) tumor suppressor gene, a key control gene in cancer pathway [36,37].
Table 1: Upregulated mi-RNAs functioning as oncogenic mi-RNAs for HCC
| miRNA | Targets | Mechanism | References |
|---|---|---|---|
| miR-21 | Inhibit Kruppel like factor 5. miR-21 upregulation activates HCC tumor cell invasion, development, and migration Target programmed cell death 4 factor | Cell growth, migration, and invasion | [38] |
| miR-25 | Target on TNF-related apoptosis-inducing ligand | Apoptotic cell death | [39] |
| miR-96 | Target and control SMAD signalling pathway | Promotes schistosomiasis-related hepatic fibrosis Increase collagen expression | [40] |
| miR-451 | Target interleukin-6R-STAT3 pathway | Angiogenesis | [41] |
| miR-221 | TRIAL resistance regulation and tumorigenicity enhancer through PTEN and tissue inhibitor of metalloproteinase | Cell death/apoptosis | [42,43] |
HCC: Hepatocellular carcinoma
2.3. HCC Development by Tumor Suppressor mi-RNAs
Several mi-RNAs expression is reported to be downregulated during HCC development that serves as tumor suppressor by targeting immune cells and proteins for activation and silence oncogenes by targeting specific mRNAs employed tumor cell growth and invasion [44]. The downregulated mi-RNAs may have the capacity to silence the oncogenic gene activity and modify oncogenic proteins by targeting their mRNA sequences. The mi-RNAs of the let-7 family were named as tumor-suppressors since they target RAS-associated protein families [45]. These mi-RNAs have control over genes related to hepatic fibrosis, lowering tumorigenicity. Details are given in Table 2.
Table 2: Downregulated mi-RNAs functioning as a tumor suppressor.
| miRNA | Targets | Mechanism | References |
|---|---|---|---|
| miR-122a | Modulate cyclin G1 expression | Control cell cycle | [46] |
| miR-486-3p | Target fibroblast growth factor receptor 4 and estimated glomerular filtration rate | Apoptosis the potent target for HCC treatment | [46] |
| miR-3622a-3p | Downregulate spalt-like transcription factor 4 | Inhibit cancer cell proliferation and metastasis | [47] |
| miR-29b | Promote expression of tumor-suppressor genes | Repress DNA methyltransferase, Immune cell malignancy | [48] |
HCC: Hepatocellular carcinoma
Gramantieri et al. reported in their study that miR-122a is abundant in HCC cases among which nearly 60–70% was down-regulated, and hence confirms the hepatocarcinogenesis potential of the down-regulated miR-122a [48]. The expression of five miRNAs, that is, miR-671-3p, miR-486-3p, miR-328-3p, miR-378a-3p, and miR-378a-5p was identified in another study and it was reported to be in a significantly reduced state in HCC cells compared to normal cells in HCC patients [46]. The expression level of onco-miRs has been correlated with tumorigenesis and/or tumor suppression. The miR-29 overexpression significantly reduces or inhibits tumorigenesis in an in vivo research study by promoting apoptosis and directly targeting antiapoptotic proteins, for example, BCL2 and MCL1 [49,50]. With the investigations and detailed research on dysregulated mi-RNAs in HCC cases, extracellular mi-RNAs can be applied as candidate biomarkers for HCC detection.
2.4. mi-RNAs as a Biomarker for HCC Associated with Various Diseases
The developmental stages of HCC in most patients are silent at early stages, that is, no initial symptoms have been shown by the patient. Many diagnostic techniques, imaging devices and conventional tumor markers such as α-fetoprotein, have been developed and adopted for diagnosing HCC and screening out tumor cells in the liver [51]. However, diagnosis with such conventional markers provides unsatisfactory results and increases the necessity for predictive biomarkers. The mi-RNAs possess potential application as an effective non-invasive biomarker for HCC prediction for their key role in fibrosis and liver regeneration [52]. The important mi-RNAs investigated till date to use as biomarkers for HCC and liver disease include miR-21, miR-121, miR-16, and miR-221 [53]. The HCC developmental stages are common in patients with liver cirrhosis, hepatitis B virus infection, etc. The mi-RNAs (upregulated/downregulated), circulating in the biological fluids of HCC patients, exist as apoptotic bodies or encapsulated exosomes or bound to serum proteins, lipids, and membranes [54]. The ability to circulate mi-RNAs, for example, miR-122 and miR-15b (upregulated miRNAs) in the serum of HCC patients for earlier diagnosis of HCC in association with hepatitis B virus was reported [55]. A research study constructed a mi-RNA combination composed of has-miR-221 and has-miR-29c using step-wise logistic regression analysis and confirmed the combination as a promising tool for HCC diagnosis. Patients with HBV-associated HCC have significantly lower serum miR-101 expression compared to other cells in HBV-associated cirrhosis and hepatitis, suggesting that miR-101 has biomarker potential for detecting and identifying HBV-associated HCC [56]. The miR-122 proves to be a valuable diagnostic tool (as biomarker) in HCC caused by HCV. MiR-122, miR-885-5p, and miR-22 were all found to have diagnostic efficiency and accuracy for advanced HCC detection in patients with liver cirrhosis [57]. Table 3 summarizes specificity and selectivity of selected mi-RNAs.
Table 3: Sensitivity and specificity of selected mi-RNAs as biomarkers in early-stage HCC [43,58,59].
| mi-RNA | Sample | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| miR-21 | Serum/plasma | 84–89.5 | 71.2–92.0 |
| miR-122 | Serum | 70.7–81.6 | 69.1–83.3 |
| miR-29b | Serum | 75.4 | 87.5–89.6 |
| miR-15b | Serum | 98.3 | 15.3 |
| miR-215 | Serum | 78.7-80 | 91 |
| miR-122a | Plasma | 70.6 | 67.1 |
| miR-223 | Serum | 80 | 76.5 |
| miR-18a | Serum | 86.1 | 75.0 |
| miR-130b | Serum | 87.7 | 81.4 |
| miR-483-5p | Plasma | 55.7 | 85.7 |
HCC: Hepatocellular carcinoma
3. THERAPEUTIC POTENTIAL OF Mi-RNAs FOR HCC
Applications of mi-RNAs for the development of therapeutic targets for practical implementation are entice by the regulation of numerous targets in cascades related to HCC pathway and enhancement of mi-RNA based cancer therapy. As already mentioned, the downregulated mi-RNAs act as tumor suppressors and activate tumor suppression proteins. The downregulated mi-RNAs can be restored in liver cancer patients as tumor treatment. In some cases, the upregulated mi-RNAs that act as onco-miRNAs were knocked down in HCC cases using the CRISPR/Cas9 system to silence the oncogenes activation [46]. Figure 4 illustrates an outline of down regulation of miR-122 and upregulation of miR-21 involved in tumor formation in liver cells. Table 4 summarizes research studied conducted to assess the potential role of different mi-RNA in HCC and related liver disorders.
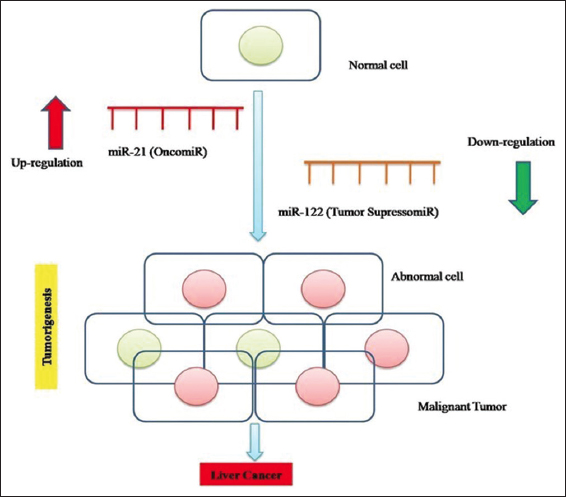 | Figure 4: Illustration of liver cancer development mediated by miR-21 (oncomir) and miR-122 (tumor suppressor-miR). [Click here to view] |
Table 4: Summary of clinical studies conducted to diagnose/analyze liver disorders thorough miRNA as biomarker.
| mi-RNA | Clinical implication/Disease | Target cell/Molecule | Critical finding | References |
|---|---|---|---|---|
| miR-122 | DILI | Glycyrrhetinicacid | Clinical diagnosis and treatment of DILI | [29] |
| miR-122 | mi-RNA as potential candidate to enhance hepatic ischemia tolerance | Murine modes of hepatic IR injury | Study depicted potential role of miR-122 in enhancing hepatic ischemia tolerance | [26] |
| miR-122 | Identical of miR-122 to negatively affect expression of cytotoxic gene in λ AAV vector | miR-122 enriched Huh7 cell line | • The study reported HEK293 miR-122 cells with 122T sequence to effectively attenuate cytotoxic transgene expression | [60] |
| miR-122 | Implication in NAFLD pathogenesis | HepG2 and Huh-7 cells | miR-122 inhibition protects liver cells from lipid metabolic disorders like NAFLD along with suppression of lipogenesis | [30] |
| miR-21 | HepG2.2.15 and Huh7-1.3 cells | miR-21 higher expression in HCC group and miR-122 lower expression inhibited proliferation and invasion of HepG2.2.15 and Huh7-1.3 cells. Luciferase reporter gene along with RNA immunoprecipitation assay illustrated smad family 7 to be target gene of miR-21 | [39] | |
| miR-21 | Diagnosis of HCC | PDK1/AKT | miRNA-21 could alter HSCs to CAFs by downregulation of its descendent target PTEN to activate signaling pathway PDK1/AKT. In extension, CAFs advanced cancer enhancement by angiogenesis through secretion of interleukins IL-6 and IL-8. Further crosstalk between HSCs and cancer cells explicate the molecular mechanism involved in HCC invasion | [61] |
| has- miR-122 | Diagnosis of HCC | The study reported down regulation of has-miR-122 in HCC patients which further acts as biomarker for diagnosis of HCC in blood sample | [24] | |
| Inc-RNAs, mRNA, Circ-RNAs | Diagnosis of HCC | Huh7 cell line | Over expressed miR-122, miR-204 was reported to be linked to inhibition of HCC progression through down regulation of TGFBRAPI and HOTTIP | [25] |
| miR-122 | miR-122 expression in ethanol induced liver disease | C57BI/6 mice | GRHL2 inhibits miR-122 expression resulting in low level of miR-122 | [27] |
| mi-RNA | Diagnosis of HCC | Epithelial-mesenchymal transition (EMT), HCC | mi-RNAs (122,148a, 1246) raised in serum exosomes in HCC patients contrasted with liver cirrhosis (LC) and normal control (NC) individuals | [62] |
| mi-RNA | Development of inflammatory bowel diseases (IBDs) | Potential role of miR (miR -122, 196A2, 124A) polymorphism in clinical phenotype modulation among IBD like Cronn’s disease and ulcerative colitis | [63] | |
| miR-122 | miR-122 regulation by free fatty acids and impact of miR-122 on triglyceride synthesis | Huh7 BNL-IME HEK293 | Histological studies of different tissues from mice exhibited FFA to increase hepatic expression secretion of miR-122 | [64] |
| miR-21 | miR-21 dysregulates phosphatase and tengin homolog (PTEN) resulting inhibition of At R | HCC cells | miR-21 is potential therapeutic agent to overcome cellular resistance against sorafenib | [65] |
| miR-193a | HCC, Negative regulator of UPA | HCC cell lines | • miR-193a transfected 00HA22T/VGH HCC cells exhibited anti proliferative potential | [32] |
| miR-122 | Establish cell line for HCV propagation | HCV cc Huh7 | New permissive cell line for HCV cc can be potential for research related to HCV and other therapeutic methods | [22] |
| miR-122 | miR122 target gene expression in liver disease | HCC, HCV | miR-122 was recently shown to modulate miRNA and target gene articulation in the liver and result in the deficiency of HCV with negligible poison levels in a non-human primate | [66] |
HCC: Hepatocellular carcinoma, HCV: Hepatitis C virus, DILI: Drug induced liver injury
4. FUTURE PROSPECTS AND CHALLENGES
Scientific studies on mi-RNA identification and continuous investigation of their biogenesis, regulatory mechanisms, and mi-RNA mediated gene modulation advances the utility of miRNAs for disease diagnosis. Mi-RNAs have been recognized as effective molecular tool for identification of candidate genes for various diseases and as biomarker are particularly under study for cancer diagnosis and development of treatment targets. Analysis of published literature on modulation of the mi-RNAs expression in various tumor affected cells, depict the potential role of mi-RNAs as biomarkers for liver diseases and HCC development. With specific reference to cancer biology, several scientific studies established the sound role of mi-RNA to be utilized as biomarker for cancer diagnosis. Up-regulation and down-regulation of specific mi-RNA are directly related to increase/decrease in cancer cell proliferation through inflexing expression of genes metabolic pathways involved in cancer cell biology. Mi-RNAs effective approach in combination or with other markers and imaging techniques can bean successful early diagnosis of liver cancer (HCC). Development of biological database along with other computer software programs specifically of mi-RNA study and analysis can confer valuable support analyzing diagnostic potential of different mi-RNA and their respective therapeutic potential. Gene level modification is another area which can be explored to optimize the application of mi-RNAs in healthcare and medicine. Development of process protocol and diagnostic kits is core potential target of mi-RNAs-based studies. Such objectives and goal-oriented research can change the perspective of diagnostics on global scale with mi-RNA’s to be among key players.
5. CONCLUSION
The present review indicates that the biological aspects of mi-RNAs mainly miR-122 and miR-21 can be potentially employed as a biomarker and diagnostic tool for HCC diagnosis. There are existing challenges which needs to be addressed to explore complete potential of mi-RNA in the field of molecular diagnostics. Manipulation of pathways and biogenesis in immune cells could lead to new treatment methods for cancer, infectious illness, autoimmune diseases, and other immunological malignancies on a practical level. The combinations of mi-RNAs in place of targeting single mi-RNA could be utilized as effective early stage prognostic biomarker which provide high diagnostic accuracy and efficiency. Finally, it could be concluded that the molecular expression pattern of miR-122 and miR-21 provide molecular information for the development and progression of HCC.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
Data available on request.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Arraiano CM. Regulatory noncoding RNAs:Functions and applications in health and disease. FEBS J 2021;288:6308-9. [CrossRef]
2. Peltier DC, Roberts A, Reddy P. LNCing RNA to immunity. Trends Immunol 2022;43:478-95. [CrossRef]
3. Bartel DP. MicroRNAs:Genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [CrossRef]
4. O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. [CrossRef]
5. Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis 2008;14:27-41. [CrossRef]
6. Da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins:A possible new form of cell communication within the ovarian follicle. Biol Reprod 2012;86:71. [CrossRef]
7. Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, Kahn CR. Extracellular miRNAs:From biomarkers to mediators of physiology and disease. Cell Metab 2019;30:656-73. [CrossRef]
8. Zhang R, Jing Y, Zhang H, Niu Y, Liu, Wang J, et al. Comprehensive evolutionary analysis of the major RNA-induced silencing complex members. Sci Rep 2018;8:14189. [CrossRef]
9. Gurtner A, Falcone E, Garibaldi F, Piaggio G. Dysregulation of microRNA biogenesis in cancer:The impact of mutant p53 on Drosha complex activity. J Exp Clin Cancer Res 2016;35:45. [CrossRef]
10. Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122-a key factor and therapeutic target in liver disease. J Hepatol 2015;62:448-57. [CrossRef]
11. Zeinali F, Zarch SM, Jahan-Mihan A, Kalantar SM, Mehrjardi MY, Fallahzadeh H, et al. Circulating microRNA-122, microRNA-126-3p and microRNA-146a are associated with inflammation in patients with pre-diabetes and Type 2 diabetes mellitus:A case control study. PLoS One 2021;16:e0251697. [CrossRef]
12. Chun KH. Molecular targets and signaling pathways of microRNA-122 in hepatocellular carcinoma. Pharmaceutics 2022;14:1380. [CrossRef]
13. Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009;28:3526-36. [CrossRef]
14. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007;133:647-58. [CrossRef]
15. Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 2010;411:846-52. [CrossRef]
16. Zhu Q, Wang Z, Hu Y, Li J, Li X, Zhou, L, et al. miR-21 promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma. Oncol Rep 2012;27:1660-8.
17. Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 2007;282:14328-36. [CrossRef]
18. Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010;467:86-90. [CrossRef]
19. Jenike AE, Halushka MK. miR-21:A non-specific biomarker of all maladies. Biomark Res 2021;9:18. [CrossRef]
20. Dai M, Li L, Qin X. Clinical value of miRNA-122 in the diagnosis and prognosis of various types of cancer. Oncol Lett 2019;17:3919-29. [CrossRef]
21. Menon KM, Menon B, Gulappa T. Regulation of luteinizing hormone receptor mRNA expression in the ovary:The role of miR-122. Vitam Horm 2018;107:67-87. [CrossRef]
22. Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, et al. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. J Virol 2012;86:1382-93. [CrossRef]
23. Kishikawa H, Kimura K, Takarabe S, Kaida S, Nishida J. Helicobacter pylori antibody titer and gastric cancer screening. Dis Mark 2015;2015:156719. [CrossRef]
24. Quoc NB, Phuong ND, Ngan TK, Linh NT, Cuong PH, Chau NN. Expression of plasma hsa-miR122 in HBV-related hepatocellular carcinoma (HCC) in Vietnamese patients. MicroRNA 2018;7:92-9. [CrossRef]
25. Wang X, Peng J, Wang J, Li M, Wu D, Wu S, et al. Hepatitis C virus core impacts expression of miR122 and miR204 involved in carcinogenic progression via regulation of TGFBRAP1 and HOTTIP expression. Onco Targets Ther 2018;11:1173-82. [CrossRef]
26. Ju C, Wang M, Tak E, Kim B, Emontzpohl C, Yang Y, et al. Hypoxia-inducible factor-1a-dependent induction of miR122 enhances hepatic ischemia tolerance. J Clin Invest 2021;131:e140300. [CrossRef]
27. Satishchandran A, Ambade A, Rao S, Hsueh YC, Iracheta-Vellve A, Tornai D, et al. MicroRNA 122, regulated by GRLH2, protects livers of mice and patients from ethanol-induced liver disease. Gastroenterology 2018;154:238-52.e7. [CrossRef]
28. Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther 2015;15:142-50. [CrossRef]
29. Meng L, Wang Q, Wang L, Zhao Z, Xin GZ, Zheng Z, et al. miR122-controlled all-in-one nanoplatform for in situ theranostic of drug-induced liver injury by visualization imaging guided on-demand drug release. Mater Today Bio 2021;12:100157. [CrossRef]
30. Long JK, Dai W, Zheng YW, Zhao SP. miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol Med 2019;25:26. [CrossRef]
31. Amaro A, Croce M, Ferrini S, Barisione G, Gualco M, Perri P, et al. Potential onco-suppressive role of miR122 and miR144 in uveal melanoma through ADAM10 and C-Met inhibition. Cancers (Basel) 2020;12:1468. [CrossRef]
32. Salvi A, Conde I, Abeni E, Arici B, Grossi I, Specchia C, et al. Effects of miR-193a and sorafenib on hepatocellular carcinoma cells. Mol Cancer 2013;12:162. [CrossRef]
33. He M, Qin H, Poon TC, Sze SC, Ding X, Co NN, et al. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis 2015;36:1008-18. [CrossRef]
34. Fouda MS, Omran MM, Tarek G, Wahab AH. Development of a novel panel based on micro-RNAs (21, 29a, 200 and 335) and alpha-fetoprotein as diagnostic biomarkers for hepatocellular carcinoma associated with hepatitis C infection. Arab J Gastroenterol 2021;22:28-33. [CrossRef]
35. Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med 2016;14:143. [CrossRef]
36. Sartorius K, Sartorius B, Winkler C, Chuturgoon A, Makarova J. The biological and diagnostic role of miRNA's in hepatocellular carcinoma. Front Biosci (Landmark Ed) 2018;23:1701-20. [CrossRef]
37. Wagenaar TR, Zabludoff S, Ahn SM, Allerson C, Arlt H, Baffa R, et al. Anti-miR-21 suppresses hepatocellular carcinoma growth via broad transcriptional network deregulation. Mol Cancer Res 2015;13:1009-21. [CrossRef]
38. Wang Y, Zhang P, Yuan M, Li X. Overexpression of miRNA-21 promotes the proliferation and invasion in hepatocellular carcinoma cells via suppressing SMAD7. Technol Cancer Res Treat 2019;18. [CrossRef]
39. Feng X, Jiang J, Shi S, Xie H, Zhou L, Zheng S. Knockdown of miR-25 increases the sensitivity of liver cancer stem cells to TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signalling pathway. Int J Oncol 2016;49:2600-10. [CrossRef]
40. Luo X, Zhang D, Xie J, Su Q, He X, Bai R, et al. MicroRNA-96 promotes schistosomiasis hepatic fibrosis in mice by suppressing Smad7. Mol Ther Methods Clin Dev 2018;11:73-82. [CrossRef]
41. Liu X, Zhang A, Xiang J, Lv Y, Zhang X. miR-451 acts as a suppressor of angiogenesis in hepatocellular carcinoma by targeting the IL-6R-STAT3 pathway. Oncol Rep 2016;36:1385-92. [CrossRef]
42. Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009;16:498-509. [CrossRef]
43. Santhekadur PK, Das SK, Gredler R, Chen D, Srivastava J, Robertson C, et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kB and miR-221. J Biol Chem 2012;287:13952-8. [CrossRef]
44. Chang S, Sun G, Zhang D, Li Q, Qian H. MiR-3622a-3p acts as a tumor suppressor in colorectal cancer by reducing stemness features and EMT through targeting spalt-like transcription factor 4. Cell Death Dis 2020;11:592. [CrossRef]
45. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell 2019;179:1033-55. [CrossRef]
46. Ji L, Lin Z, Wan Z, Xia S, Jiang S, Cen D, et al. miR-486-3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis 2020;11:250. [CrossRef]
47. Lee HM, Nguyen DT, Lu LF. Progress and challenge of microRNA research in immunity. Front Genet 2014;5:178. [CrossRef]
48. Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, LiuCG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007;67:6092-9. [CrossRef]
49. Kwon JJ, Factora TD, Dey S, Kota J. A systematic review of miR-29 in cancer. Mol Ther Oncolytics 2019;12:173-94. [CrossRef]
50. Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 2010;51:836-45. [CrossRef]
51. Oura K, Morishita A, Masaki GT. Molecular and functional roles of microRNAs in the progression of hepatocellular carcinoma-a review. Int J Mol Sci 2020;21:8362. [CrossRef]
52. Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol 2015;21:10573-83. [CrossRef]
53. Abdel-Al A, El-Ahwany E, Zoheiry M, Hassan M, Ouf A, Abu-Taleb H, et al. miRNA-221 and miRNA-222 are promising biomarkers for progression of liver fibrosis in HCV Egyptian patients. Virus Res 2018;253:135-9. [CrossRef]
54. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [CrossRef]
55. Hung CH, Hu TH, Lu SN, Kuo FY, Chen CH, Wang JH, et al. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int J Cancer 2016;138:714-20. [CrossRef]
56. Xie Y, Yao Q, Butt AM, Guo J, Tian Z, Bao X, et al. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol Ther 2014;15:1248-55. [CrossRef]
57. Zekri AN, Youssef AS, El-Desouky ED, Ahmed OS, Lotfy MM, Nassar AA, et al. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol 2016;37:12273-86. [CrossRef]
58. Huang JT, Liu SM, Ma H, Yang Y, Zhang X, Sun H, et al. Systematic review and meta-analysis:Circulating miRNAs for diagnosis of hepatocellular carcinoma. J Cell Physiol 2016;231:328-35. [CrossRef]
59. Jiang Y, He J, Li Y, Guo Y, Tao H. The diagnostic value of MicroRNAs as a biomarker for hepatocellular carcinoma:A meta-analysis. Biomed Res Int 2019;2019:5179048. [CrossRef]
60. Ran G, Feng XL, Xie YL, Zheng QY, Guo PP, Yang M, et al. The use of miR122 and its target sequence in adeno-associated virus-mediated trichosanthin gene therapy. J Integr Med 2021;19:515-25. [CrossRef]
61. Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res 2018;37:324. [CrossRef]
62. Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, et al. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med 2018;7:1670-9. [CrossRef]
63. Ciccacci C, Politi C, Biancone L, Latini A, Novelli G, Calabrese E, et al. Polymorphisms inMIR122, MIR196A2, andMIR124Agenes are associated with clinical phenotypes in inflammatory bowel diseases. Mol Diagn Ther 2017;21:107-14. [CrossRef]
64. Chai C, Rivkin M, Berkovits L, Simerzin A, Zorde-Khvalevsky E, Rosenberg N, et al. Metabolic circuit involving free fatty acids, microRNA 122, and triglyceride synthesis in liver and muscle tissues. Gastroenterology 2017;153:1404-15. [CrossRef]
65. He C, Dong X, Zhai B, Jiang X, Dong D, Li B, et al. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 2015;6:28867-81. [CrossRef]
66. Braconi C, Henry JC, Kogure T, Schmittgen T, Patel T. The role of MicroRNAs in human liver cancers. Semin Oncol 2011;38:752-63. [CrossRef]