1. INTRODUCTION
Antimicrobial resistance (AMR) is a natural process; the public health emergency due to the uncontrolled spread of this phenomenon depends primarily on the overuse of antibiotics. About 700,000 people worldwide die each year due to the cause of AMR [1]. This concern is further compounded by the unregulated and often misuse of antibiotics, leading to the development of AMR and the rapid spread of these organisms, which poses a significant threat to the healthcare system and increases the risk of infectious diseases [2]. India, in particular, is vulnerable to the loss of antimicrobial efficacy due to the high treatment costs and limited access to antibiotics [3]. Urinary tract infections (UTIs) are among the most frequent bacterial infections, affecting both hospital patients and those in the community and are particularly common in women and children [4].
The cause of infection by Enterococcus faecium, Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa is major contributors to healthcare-acquired infections globally and is often caused by multidrug-resistant isolates, presenting significant challenge for clinical practitioners [5,6]. E. faecalis strains are known for their ability to thrive in various harsh environments, such as high temperatures, high salinity, and acidity [7], as well as their adaptation to the gastrointestinal tracts (GIT) of both humans and animals [8]. Furthermore, these bacteria are commonly found in soil, water, and the environment [9] and can be spread through poor hygiene practices and food contamination [10].
E. faecalis strains are known to cause severe pathogenic infections, such as gastrointestinal and UTIs, meningitis, bacteremia, and periodontitis, due to the presence of virulence factors and toxins [10]. Research has shown that these virulence factors and toxins can increase the severity of the disease caused by these bacteria [11,12]. Studies have identified specific virulence factors, such as enterococcal surface protein (esp), fsr regulator locus responsible for bacterial quorum sensing (encoded by fsrA, fsrB, and fsrC genes), structural pilin genes (ebpA, ebpB, and ebpC), cytolysin (cylL and cylS), and endocarditis-specific antigen (efa), that play a crucial role in the adherence, colonization, evasion, enzyme production, biofilm development, and pathogenicity of the bacterium, leading to more severe infections [11,13].
E. faecalis infections can also be challenging to treat with traditional antimicrobials, due to the bacteria’s high resistance rate to commonly used antimicrobials, particularly penicillins, tetracyclines, aminoglycosides, chloramphenicol, cephalosporin, and macrolides [14]. Therefore, analyzing the AMR of E. faecalis strains can aid in selecting the most appropriate antimicrobial agents for treatment [10,15]. Quorum sensing is a population density-dependent regulatory mechanism through which bacteria will communicate through signaling molecules called autoinducers. They are around 15 differential proteins which are expressed in Vancomycin-resistant E. faecalis and play a role in biofilm development like, Quorum sensing-related LuXs enzyme known as S-ribosylhomocysteine lyase highly secreted in multidrug-resistant bacteria. Extensive studies are required to explore the interaction of quorum-sensing proteins and enzymes of MDR bacteria [16]. Hence, the main objectives of our study were to assess the diversity of antibiotic-resistant E. faecalis in urine samples of children suffering from UTI in Kalaburagi City, Karnataka, India, and to perform the bioluminescence assay for the determination of quorum sensing among E. faecalis isolates.
2. MATERIALS AND METHODS
2.1. Collection of Urine samples
The urine samples for the study were collected from the UTI patients from various health-care centers, including Basaveshwara Teaching Hospital, Government Hospital, Pooja Diagnostic Centre, Nandan Diagnostic Centre, and Khaja Bande Nawaz Teaching Hospital (KBN) of Kalaburagi, Karnataka, India. The samples were transported to the laboratory under aseptic conditions, using sterile-labeled containers, and all standard microbiological precautions were taken during the analysis process.
2.2. Isolation and Identification of Enterococcus Species
All the urine samples were serially diluted and inoculated in 5 ml of brain heart infusion broth an enrichment media, incubated at 37°C for a period of 24 h, the samples were then streaked on Bile Esculin Azide agar media and the plates were incubated at 37°C for 24–48 h. The speciation of E. faecalis was carried out by Gram’s staining, colony morphology, culture characteristics of the colonies, and biochemical tests such as oxidase test, catalase test, gelatin hydrolysis test, sugar fermentation test, D-Fructose, D-Glucose, Lactose, Sorbitol, and Sucrose were used during the experiment.
2.3. Antimicrobial Susceptibility Test (AST)
The standard disc diffusion method was used for AST, performed according to guidelines of Clinical and Laboratory Standards Institute (CLSI) [17], on Mueller Hinton agar media (Hi-media, India) using antibiotics such as ceftriaxone, clindamycin, imipenem, linezolid, chloramphenicol, gentamycin, vancomycin, ceftazidime, tobramycin, ampicillin, oxacillin, and levofloxacin as mentioned in Table 1. The culture was spread over the plates using a sterile cotton swab and antibiotic discs were placed on the plates with pre-heated forceps. The plates were incubated at 37°C for 18–24 h and zones of inhibition were determined.
Table 1: Type of antibiotics with concentration used in antimicrobial susceptibility test.
| S. No. | Name of the antibiotics | Symbol | Concentration of antibiotics-disc (mg/ml) |
|---|---|---|---|
| 1. | Ceftriaxone | CTR | 30 |
| 2. | Clindamycin | CD | 30 |
| 3. | Imipenem | IPM | 30 |
| 4. | Linezolid | LZ | 30 |
| 5. | Chloramphenicol | C | 30 |
| 6. | Gentamycin | GEN | 10 |
| 7. | Vancomycin | VAN | 30 |
| 8. | Ceftazidime | CE | 30 |
| 9. | Tobramycin | TOB | 30 |
| 10 | Ampicillin | AMP | 30 |
| 11. | Oxacillin | OX | 30 |
| 12. | Levofloxacin | LEV | 30 |
2.4. Minimal Inhibitory Concentration (MICs) of Antibiotics
MIC was defined as the lowest antibiotic concentration point that killed visible bacteria. It was determined by AST according to the guidelines of CLSI. Gentamycin and vancomycin were dissolved in sterile distilled water, then diluted in the BHI broth to 2X the top concentration desired in the test (the maximum desired concentration is 128 μl/ml, diluted to 256 μg/ml). An overnight bacteria culture (E. faecalis) in BHI broth was diluted at 1:100 in fresh BHI and cultured at 37°C with shaking at 180 rpm to an optical density at 600 nm of 0.5. Then, 10 μl of 2 × 104 CFU was added into each well of a 96-well microtiter plate with 100 μl BHI of a series of 2-fold dilutions of an antibiotic. The plates were incubated at 37°C for 18–24 h. The optical density was determined at 600 nm using a microtiter plate reader [18].
2.5. Bioassay of Quorum Sensing
Quorum sensing assay was performed according to the protocol mentioned by Martín-Rodríguez and Fernández [19]; Da Silva Fernandes et al. [20] with slight modifications. A single colony of E. faecalis was added to the 3 ml of sterile autoinducer bioassay (AB) medium in a 15 ml sterile tube. The tubes were then incubated in an orbital shaker at 37°C, overnight at 40 rpm. The overnight incubated E. faecalis culture was diluted to 1:50 with fresh AB medium (approximately culture concentration will be 1–2 × 107 CFU/ml), 100 μl of diluted culture was then added to 96-well microplate. Water was added to one of the well as a negative control. The plate was then sealed with transparent sealing film. The microtiter plate was incubated for 18 h at 37°C for 150 rpm in an orbital shaker incubator. The luminescence reading was measured at OD600 nm, at intervals of 15 min for 5 h. All assays were performed in triplicate and the results compared to the bioluminescence were determined as follows:
 |
2.6. Isolation of Genomic DNA by Phenol-Chloroform Method
Genomic DNA was isolated from 40 E. faecalis isolates by phenol-chloroform method. A single isolated colony was inoculated in 5 ml of brain heart infusion broth and incubated at 37°C for 16–18 h in an incubator shaker at 160 rpm. After incubation, 2 ml of culture aliquot was taken in e-tubes and centrifuged at 10,000 rpm for 5 min at 4°C. The pellet was suspended in 400 μl of lysis buffer containing 100 μl of 10% of SDS and incubated at 65°C for 1 h. Next, 500 μl of saturated phenol was added and the tubes were centrifuged at 10,000 rpm for 10 min at 4°C, the supernatant was then mixed with an equal volume of chloroform: isoamyl alcohol (24:1) and 100 μl of sodium acetate. The mixture was centrifuged at 10,000 rpm for 10 min at 4°C and the supernatant was collected in new tubes a double volume of chilled alcohol was added, and then tubes were kept overnight for precipitation. After incubation, the tubes were centrifuged at 12,000 rpm for 15 min at 4°C, the supernatant was completely discarded and the pellet was washed twice with 70% alcohol, dried completely at room temperature, and the pellet was dissolved in 50 μl of 1 × TE buffer. The purity and concentration of DNA were measured by A260/A280 ratio ranging between 1.7 and 1.9 using Bio-spectrophotometer (Eppendorf Pvt., Ltd.) and confirmed by 1% of agarose gel electrophoresis.
2.7. Amplification Parameters
The 16sRNA gene in E. faecalis was amplified by polymerase chain reaction (PCR) assay. Two pairs of oligonucleotide primers used for the PCR reactions were 16s_F1 primer: forward, 5’-AGAGTTTGATCMTGGCTCAG-3’; reverse, 16S_R13primer 5’-TACGGYTACCTTGTT ACGACTT-3’. The PCR mixture for amplification reaction contains 10 μl of 2X master mix (Takara), including 0.4 μl of each primer, 0.5 μl of template DNA, and RNA/DNA free water was added to make up to 20 μl. DNA amplification was performed in the Alpha thermal cycler (Cole Parmar). The PCR conditions consisted of an initial denaturation at 95°C for 5 min; 35 cycles of denaturation at 95°C of 30 s of annealing at 58°C of 45 s and 30 s of extension at 72°C; ending with a final extension at 72°C for 10 min [21].
2.8. Detection and Documentation
DNA fragments were analyzed by electrophoresis on agarose gel (1 % wt/vol) using 1 μl of 6X DNA loading dye with SYBR safe for 5 μl of a template. The gel was photographed with Gel Chem doc (Biorad). 100 bp ladder was used as a molecular weight marker. PCR products were sequenced and identified by blasting against those available in the Gen-Bank database [http://blast.ncbi.nlm.nih.gov/Blast.cgi].
3. RESULTS
3.1. Isolation and Biochemical Characterization of E. faecalis
A total of 155 clinical urine samples were collected from five healthcare and diagnostic centers in Kalaburagi City. Among these 155 samples, 74 (47.74%) samples have shown positive for the presence of E. faecalis on bile esculin azide agar with black-greyish colonies formation as shown in Figure 1. Further, these 74 isolates were morphologically identified as Gram-positive, cocci-shaped, catalase-negative, and non-hemolytic. The isolates showed positive for fructose, glucose, lactose, sorbitol, and sucrose, respectively.
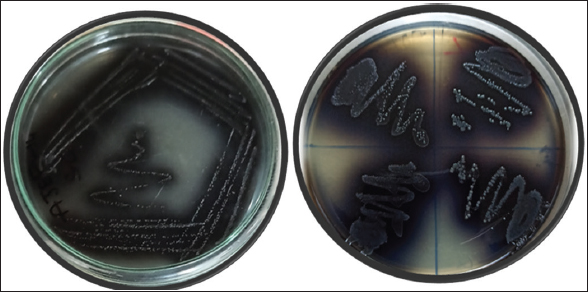 | Figure 1: Growth of Enterococcus faecalis isolates on bile esculin azide Agar (a) Single isolate of E. faecalis AJEF1; (b) E. faecalis Isolates of AJEF 2,3,4,5. [Click here to view] |
3.2. AST
The disc diffusion method was performed for all 74 isolates for different antimicrobial agents. Among these isolates, 40 isolates were resistant to both gentamycin and vancomycin with a percent of 54.05, respectively, as represented in Table 2 and Figure 2. The isolates were sensitive to oxacillin antibiotic at 89.18% and were intermediate to ceftazidime at 72.97% and they also intermediate to ampicillin, tobramycin, ceftriaxone, and clindamycin used antibiotics as mentioned in Table 2 and Figure 2. Twenty isolates were also found to be multi-drug resistant. The zone of inhibition was measured according to the standards established by the CLSI guidelines comparing with quality control strain E. faecalis NCIM 5025.
Table 2: Percentage of antibiotics resistance of E. faecalis isolates.
| S. No. | Name of the antibiotics | Sensitivity in percent | Intermediate in percent | Resistance in percent |
|---|---|---|---|---|
| 1. | Ceftriaxone | (11/74) 14.88 | (43/74) 58.10 | (20/74) 27.02 |
| 2. | Clindamycin | (31/74) 41.90 | (43/74) 58.10 | NIL |
| 3. | Imipenem | (32/74) 43.24 | (32/74) 43.24 | (10/74) 13.50 |
| 4. | Linezolid | (33/74) 44.59 | (41/74) 55.40 | NIL |
| 5. | Chloramphenicol | (26/74) 35.13 | (38/74) 51.35 | (10/74) 13.51 |
| 6. | Gentamycin | (22/74) 29.72 | (12/74) 16.21 | (40/74) 54.05 |
| 7. | Vancomycin | (16/74) 21.62 | (18/74) 24.32 | (40/74) 54.05 |
| 8. | Ceftazidime | (20/74) 27.02 | (54/74) 72.97 | NIL |
| 9. | Tobramycin | (28/74) 37.83 | (46/74) 62.16 | NIL |
| 10 | Ampicillin | (24/74) 32.43 | (50/74) 67.56 | NIL |
| 11. | Oxacillin | (66/74) 89.18 | (08/74) 10.81 | NIL |
| 12. | Levofloxacin | (44/74) 59.45 | (30/74) 40.54 | NIL |
NIL Indicates the absence of resistance in the percent of isolates. E. faecalis: Enterococcus faecalis
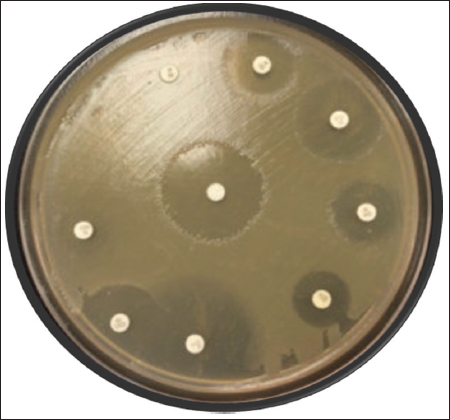 | Figure 2: Antimicrobial susceptibility test of Enterococcus faecalis on Muller Hinton Agar media. [Click here to view] |
3.3. Evaluation of MICs in E. faecalis
The strains that showed resistance to gentamycin and vancomycin were screened using the 96-well plate method at different dilutions. The MIC of gentamycin for E. faecalis showed 10 isolates inhibited at the concentration ≥32 μg/ml (25%), 07 isolates for ≥64 μg/ml (17.5%), and 23 isolates inhibited at ≥128 μg/ml (57.5%), respectively, as mentioned in Table 3. For vancomycin, out of 40 isolates screened, no isolate showed inhibition activity at ≥32 μg/ml, ≥64 μg/ml, and ≥128 μg/ml as shown in Table 5. Similarly, the MIC of vancomycin for E. faecalis showed inhibition at ≥1024 μg/ml for 17 strains (42.5%), ≥512 μg/ml for 14 strains (35%), and ≥256 μg/ml for ≥9 strains (22.5%).
Table 3: MIC determination of gentamycin and vancomycin resistance E. faecalis isolated form urine samples using micro dilution method.
| Total number of isolates | MIC level of gentamycin | |||||
|---|---|---|---|---|---|---|
| 32 (μg/ml) | 64 (μg/ml) | 128 (μg/ml) | 256 (μg/ml) | 512 (μg/ml) | 1024 (μg/ml) | |
| E. faecalis AJEF 01–40 | 10 | 07 | 23 | NIL | NIL | NIL |
| Total number of isolates | MIC level of vancomycin | |||||
| 32 (μg/ml) | 64 (μg/ml) | 128 (μg/ml) | 256 (μg/ml) | 512 (μg/ml) | 1024 (μg/ml) | |
| E. faecalis AJEF 01–40 | NIL | NIL | NIL | 09 | 14 | 17 |
NIL indicates the absence of MIC for gentamycin and vancomycin antibiotics of isolates. MIC: Minimal inhibitory concentration, E. faecalis: Enterococcus faecalis
3.4. Quorum Sensing Assay
Quorum sensing assay was performed in a 96-well microtiter plate, a total of 10 E. faecalis isolates were used based on the resistance to both antibiotic gentamycin and vancomycin used. Among these 10 isolates, two isolates normalized relative intensity units were 0.45–0.50, four isolates 0.65–1.0, and four isolates ranges from 1.0 to 1.3 as mentioned in Figures 3 and 4, as compared with control.
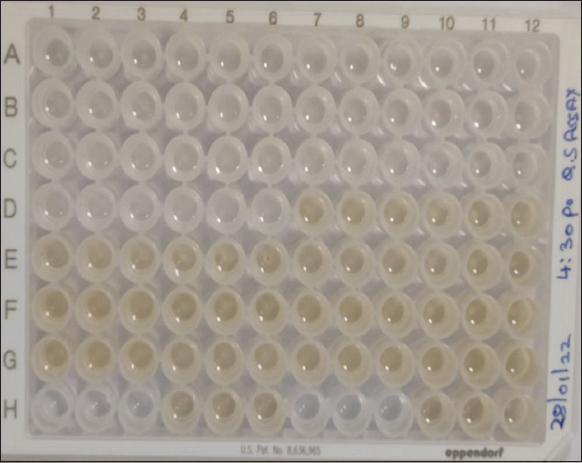 | Figure 3: Quorum sensing assay of Enterococcus faecalis isolates in a 96-well microplate. [Click here to view] |
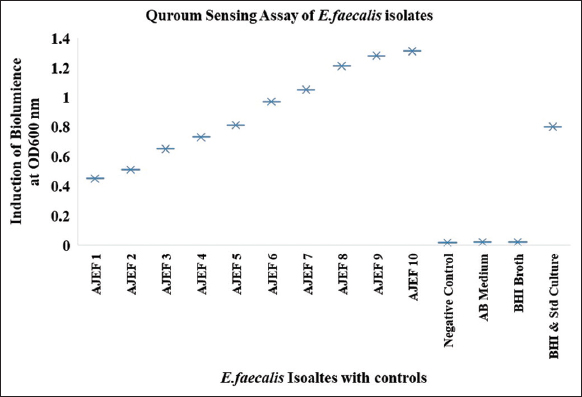 | Figure 4: Induction of bioluminescence (relative light unit) in Enterococcus faecalis isolates from clinical samples for the detection of the AI-2 autoinducer. [Click here to view] |
3.5. Genomic DNA, PCR Detection, and Sequence Analysis
A total of 40 isolates were selected on the basis of drug resistance to gentamycin and vancomycin antibiotics for the extraction of genomic DNA. The extracted genomic DNA separation was represented in Figure 5. Five isolates out of 40 were resistant to both antibiotics utilized in the PCR amplification of 16s rRNA. Our PCR results revealed that of the ten isolates tested positive for test universal primers. After PCR and sequencing, the quality of the produced products was evaluated using agarose gel electrophoresis Figure 6, bacterial species were then determined by blasting known 16s rRNA sequence represented with phylogenetic a tree showed that the seven isolates were identified as E. faecalis represented in Figure 7 and three were S. gallolyticus, respectively, and sequences were submitted to the GenBank with accession no OP164322, OP164427.
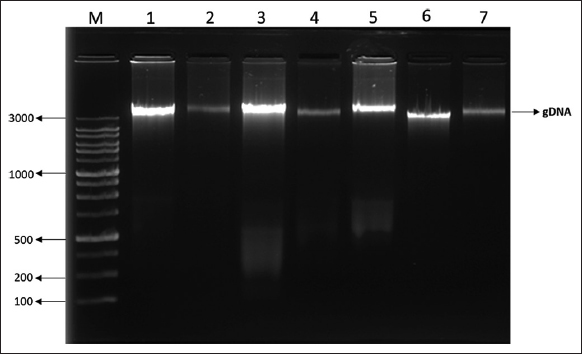 | Figure 5: Separation of genomic DNA from Enterococcus faecalis isolates on 1% of Agarose gel electrophoresis. [Click here to view] |
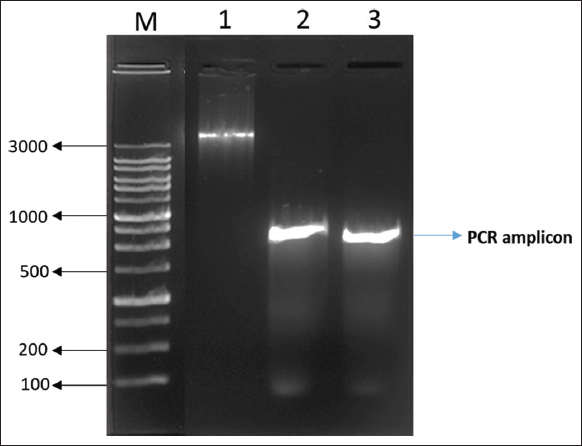 | Figure 6: Polymerase chain reaction amplification of 16s rRNA gene from Enterococcus faecalis isolates. [Click here to view] |
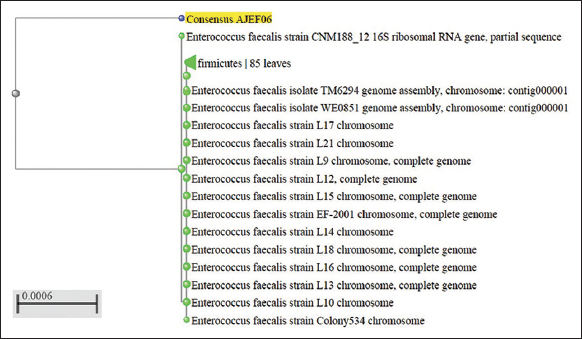 | Figure 7: Phylogenetic tree of the consensus sequence AJEF06. [Click here to view] |
4. DISCUSSION
UTIs are infectious diseases that hamper humans. E. faecalis are the second leading cause of UTIs, which accounts for 10% of all nosocomial UTIs. These species have a major pathogen in India developing multidrug resistance [22]. The predominant isolates were obtained from the urine sample with 86.2% of enterococcus reported by Yadav and Agarwal [23]. Another study carried out by Sangwan et al. [27], reported that E. faecalis are the major occurring species with an occurrence of 40.4% reported from urine samples and followed by E. faecium species. In our study, E. faecalis isolates showed with an incidence of 47.74% (74/155) isolated from urine samples of Kalaburagi city, Karnataka-India. The variation in outcomes could be different characteristics used in participants and the use of conventional methods for the identification of Enterococci [24].
E faecalis isolates are resistant to various antibiotics and vancomycin is the antibiotic of last resort for treating these isolates [25]. In addition, it posed a serious risk to the patients receiving treatment. Clinical samples of Enterococcus species revealed resistance to ampicillin of 80%, gentamycin of 60%, doxycycline of 73.3%, and vancomycin of 6.7% in Ethiopia [26]. According to the study from the northern region of India [27], no isolates of Enterococcus species were found to be resistant to the antibiotics vancomycin and high levels of gentamycin and streptomycin were both effective against Enterococcus species. According to Yadav and Agarwal [23], vancomycin resistance was present in nine isolates of E. faecium and only four isolates of E. faecalis, with doxycycline resistance being the greatest at (69.6%), tetracycline resistance coming in at (66.2%), ciprofloxacin resistance at (60.6%), and ampicillin resistance at (59.3%) in clinical isolates of enterococci. In our investigation, isolates of E. faecalis demonstrated resistance to the antibiotics ceftriaxone (27.02%), imipenem (13.50%), chloramphenicol (13.51%), gentamycin (54.05%), and vancomycin (54.05%).
Vancomycin MIC tested with E-strip has shown two isolates of E. faecalis presenting MIC of 32μg/ml and 48 μg/ml among the 75 isolates of Enterococci species [28]. Out of the 75 isolates, 15 (30%) were resistant to high-level gentamicin of E. faecalis. In a study report by Iseri et al. [29], the MIC range for VRE was 0.064 and 0.094 μg/ml, and for HLAR (high level of aminoglycoside resistance) enterococci was from 0.023 to 0.125 μg/ml. In our study, high-level-resistance gentamycin was 128 μg/ml which showed by the 23 isolates, 07 ranged from 64 μg/ml, and ten isolates showed a concentration of 32 μg/ml. High-level vancomycin resistance has been shown by the 17 isolates with concentration of 1024 μg/ml, 14 isolates with concentration of 512 μg/ml, and nine isolates with a concentration of 256 μg/ml which was confirmed by the broth dilution method with a control strain E. faecalis NCIM 5025 used.
Quorum sensing systems control virulence determinants in E. faecalis which causes nosocomial infections. E. faecalis quorum sensing systems which consist of several factors regulated by the cytolysin operon, encode the cytolysin toxin [31]. A quorum-sensing assay for four isolates of E. faecalis has normalized relative light unit levels ranging between 0.89 and 1.59 as reported by Da Silva Fernandes et al. [20], confirming the detection of bioluminescence and the presence of gene luxS. In our study, the bioluminescence detection has been shown from 10 isolates of E. faecalis which were isolated from the clinical sample of urine ranging from 0.4 to 1.4.
Rapid identification of bacterial pathogens is crucial for appropriate and adequate antibiotic treatment, which significantly improves patient outcomes. 16S ribosomal RNA (rRNA) gene amplicon sequencing has proven to be a powerful strategy for diagnosing bacterial infections [32]. Sequencing of 16S rDNA has been widely used to reconstruct phylogenetic relationships of microorganisms [32]. E. faecalis E-12 was identified as E. faecalis with deposited nucleotide sequence in the GenBank with accession number KT3667333]. In our study, seven isolates were positive for E. faecalis and three were identified as Streptococcus gallolyticus and two isolates sequence were submitted to the NCBI GenBank with accession no OP164322, OP164427. The 16s rRNA gene amplification is a very useful technique for discriminating the main groups of Enterococc and also to overcome false positive results with the biochemical test.
5. CONCLUSION
The present study indicates the high rate of prevalence of gentamycin and vancomycin resistance E. faecalis isolated from urine samples. Quorum sensing assay determines the presence of virulence factors for the development of resistance to antibiotics. The PCR technique harbor has shown the appropriate method for the identification of E. faecalis isolates at the species level. The emergency of the vancomycin resistance E. faecalis should be monitored, avoiding the spreading of these isolates, and critical primitive measures should be taken in using the antibiotics.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
The present study is not being funded by any funding agencies or organizations.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
The present study was approved by the Institutional Ethics Committee of Gulbarga University Kalaburagi, Karnataka- India (Ref. No: GUK/IEC/2023-24/179 dated 09.02.2023).
10. DATA AVAILABILITY
All the data obtained in the study are represented as tables or figures.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Ahmed MO, Baptiste KE. Vancomycin-resistant enterococci:A review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microbial Drug Resist 2018;24:590-606. [CrossRef]
2. Ali L, Goraya MU, Arafat Y, Ajmal M, Chen JL, Yu D. Molecular mechanism of quorum-sensing in Enterococcus faecalis:Its role in virulence and therapeutic approaches. Int J Mol Sci 2017;18:960. [CrossRef]
3. Arpitha PM, Rani NR, Sumana K. Prevalence of vancomycin-resistant enterococci from urinary tract infected patients. Int J Pharm Pharm Sci 2022;15:1-7. [CrossRef]
4. Barghouthi SA. A universal method for the identification of bacteria based on general PCR primers. Indian J Microbiol 2011;51:430-44. [CrossRef]
5. Bin-Asif H, Ali SA. The genus Enterococcus and its associated virulent factors. In:Blumenberg M, Shaaban M, Abdelaziz E, editors. Microorganisms. London:IntechOpen;2019. 109-30. [CrossRef]
6. Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. Enterococci in the environment. Microbiol Mol Biol Rev 2012;76:685-706. [CrossRef]
7. Calzada FC, Aguilera-Correa JJ, Gonzalez JC, Moreno JE, Biosca DR, Perez-Tanoira R. Urinary tract infection and antimicrobial susceptibility of bacterial isolates in Saint Joseph Kitgum hospital, Kitgum, Uganda. Antibiotics (Basel) 2022;11:504. [CrossRef]
8. Clinical and Laboratory Standards Institute. M100 Performance Standards for Antimicrobial Susceptibility Testing. Wayne, Pennsylvania:Clinical and Laboratory Standards Institute;2021.
9. Da Silva Fernandes M, Esper LM, Kabuki DY, Kuaye AY. Quorum sensing in Enterococcus faecium, Enterococcus faecalis and Bacillus cereus strains isolated from ricotta processing. Cien Rural 2018;48:e20161111. [CrossRef]
10. Dadashi M, Sharifian P, Bostanshirin N, Hajikhani B, Bostanghadiri N, Khosravi-Dehaghi N, et al. The global prevalence of daptomycin, tigecycline, and linezolid-resistant Enterococcus faecalis and Enterococcus faecium strains from human clinical samples:A systematic review and meta-analysis. Front Med (Lausanne) 2021;8:720647. [CrossRef]
11. Daniel DS, Lee SM, Gan HM, Dykes GA, Rahman S. Genetic diversity of Enterococcus faecalis isolated from environmental, animal and clinical sources in Malaysia. J Infect Public Health 2017;10:617-23. [CrossRef]
12. De Oliveira DM, Forde BM, Kidd TJ, Harris PN, Schembri MA, Beatson SA, et al. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev 2020;33:1-49. [CrossRef]
13. Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol 1989;27:731-4. [CrossRef]
14. Farooqui HH, Selvaraj S, Mehta A, Heymann DL. Community level antibiotic utilization in India and its comparison vis-a-vis European countries:Evidence from pharmaceutical sales data. PLoS One 2018;13:e0204805. [CrossRef]
15. Ferede ZT, Tullu KD, Derese SG, Yeshanew AG. Prevalence and antimicrobial susceptibility pattern of Enterococcus species isolated from different clinical samples at Black Lion specialized teaching hospital, Addis Ababa, Ethiopia. BMC Res Notes 2018;11:793. [CrossRef]
16. Goh HM, Yong MH, Chong KK, Kline KA. Model systems for the study of Enterococcal colonization and infection. Virulence 2017;8:1525-62. [CrossRef]
17. Hassan MM, Belal ES. Antibiotic resistance and virulence genes in Enterococcus strains isolated from different hospitals in Saudi Arabia. Biotechnol Biotechnol Equip 2016;30:726-32. [CrossRef]
18. Hassan MM, Ismail AI. Isolation and molecular characterization of some pathogenic mobile phone bacteria. Int J Biochem Biotechnol 2014;3:516-22.
19. Iseri L, Sahin E, DolapçI, Yürüken Z. Minimum inhibitory concentration values and problematic disk break points of tigecycline against vancomycin and/or high level aminoglycoside-resistant enterococci. Alex J Med 2016;52:125-9. [CrossRef]
20. Jani K, Srivastava V, Sharma P, Vir R, Sharma A. Easy access to antibiotics;spread of antimicrobial resistance and implementation of one health approach in India. J Epidemiol Glob Health 2021;11:444-52. [CrossRef]
21. Jinglin Y, Mingxi H, Nan C, Jiarui L, Xinzhe L, Ang D, Huizhu W, Pengcheng D, Chengbo R, Duo Y, Chen C. Role of the fsr Quorum-Sensing System in Enterococcus faecalis Bloodstream infection. Appl. Environ. Microbiol 2022;88;1-10. [CrossRef]
22. Johnston LM, Jaykus LA. Antimicrobial resistance of Enterococcus species isolated from produce. Appl Environ Microbiol 2004;70:3133-7. [CrossRef]
23. Kai S, Matsuo Y, Nakagawa S, Kryukov K, Matsukawa S, Tanaka H, et al. Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinION™ nanopore sequencer. FEBS Open Bio 2019;9:548-57. [CrossRef]
24. Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial antibiotic resistance:The most critical pathogens. Pathogens 2021;10:1310. [CrossRef]
25. Martín-Rodríguez AJ, Fernández JJ. A bioassay protocol for quorum sensing studies using Vibrio campbellii. Bio Protoc 2016;6:1-7. [CrossRef]
26. Noroozi N, Momtaz H, Tajbakhsh E. Molecular characterization and antimicrobial resistance of Enterococcus faecalis isolated from seafood samples. Vet Med Sci 2022;8:1104-12. [CrossRef]
27. Said MS, Tirthani E, Lesho E. Enterococcus Infections. Treasure Island, FL:Stat Pearls Publishing;2022.
28. Sangwan J, Lohan K, Mane P, Kumar M. Emerging crisis of multidrug-resistant enterococci from a rural tertiary care hospital of North India. Indian J Health Sci Biomed Res KLEU 2021;14:260-4. [CrossRef]
29. Tao JJ, Xiang JJ, Jiang M, Kuang SF, Peng XX, Li H. A microtitre plate dilution method for minimum killing concentration is developed to evaluate metabolites-enabled killing of bacteria by β-lactam antibiotics. Front Mol Biosci 2022;9:878651. [CrossRef]
30. Varghese V, Menon AR, Nair PK. Speciation and susceptibility pattern of Enterococcal species with special reference to high-level gentamicin and vancomycin. J Clin Diagn Res 2020;14:DC08-12. [CrossRef]
31. Vu J, Carvalho J. Enterococcus:Review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front Biol 2011;6:357-66. [CrossRef]
32. Yadav RK, Agarwal L. Enterococcal infections in a tertiary care hospital, North India. Ann Afr Med 2022;21:193-7. [CrossRef]
33. Zhen X, Lundborg CS, Sun X, Hu X, Dong H. Economic burden of antibiotic resistance in ESKAPE organisms:A systematic review. Antimicrob Resist Infect Control 2019;8:137. [CrossRef]