1. INTRODUCTION
An alkaloid name noscapine belonging to the benzylisoquinoline class is obtained naturally from the plant (Papaver somniferum) opium poppy. It is usually effective in treating coughs of various etiologies, including acute and chronic bronchitis, asthma, and other respiratory conditions [1]. Beyond its cough-suppressing properties, noscapine has demonstrated antitumor, anti-inflammatory, and immunomodulatory effects [2]. Experimentally, it is demonstrated that noscapine binds to microtubules (MTs) in a 1:1 stoichiometric ratio, resulting in a modification of tubulin compliance, leading to arrest in cell cycle at the mitosis phase, and targeting apoptosis in cancer cells. Significantly, noscapine did not induce any over-polymerization, depolymerization, or any alterations in the overall interphase in MT organization, in contrast to vinca alkaloids and taxanes. By affecting the kinetic parameters of dynamic instability of MTs, noscapine inhibits mitosis at the prometaphase stage, leading to the arrest of both dividing cancer cells and normal cells in mitosis [3-5]. However, unlike normal cells, cancer cells due to their compromised cell cycle checkpoints, and cells do not maintain this halted mitosis state for extended periods and instead experience apoptosis, a type of programmed cellular demise. Besides, noscapine provides several advantages for cancer treatment compared to other MT binding drugs, such as taxanes and vinca alkaloids [2,4]. These include the ability to arrest drug-resistant cancer cells in mitosis and induce apoptosis and having a low susceptibility to drug pumps that cause drug resistance. Noscapine was shown to inhibit tumor progression in various types of cancer in animal models without detectable toxicity to normal cells [6-8]. It can be administered orally without the risk of anaphylactic reactions or infections. Despite these advantages, noscapine has limited cytotoxicity to cancer cells, with IC50 values remaining in the high micromolar range [1,3,6,7,9].
Numerous synthetic derivatives of noscapine, called noscapinoids, have been developed since its discovery as an antitumor agent to magnify its anticancer activity. These derivatives were improved therapeutic indices and pharmacological profiles. One of the promising derivatives of noscapine includes coupling a urea pharmacophore [1,3,4]. The anticancer activity of this derivative has been validated using a Hela cell line. The results showed that it inhibits the proliferation of Hela cell line. It arrested the cells at G2/M phase, followed by the induction of apoptosis [7,10]. Moreover, the exact mechanism of action of urea-coupled noscapine derivative has not been explored. In this study, we approached the novel series of urea noscapine (7 h) chemical synthesis and validated their anticancer activity in Hela cervical cancer cell line to find the differentially expressed genes (DEGs) and pathways in the treated cells compared to the untreated cells based on transcriptome sequencing.
2. MATERIALS AND METHODS
2.1. Chemical Synthesis of Urea Noscapine Conjugates
The production of noscapine derivatives poses considerable challenges owing to the delicate C-C bond linking two heterocyclic phthalide and isoquinoline components within the molecular structure. Nevertheless, we have effectively fine-tuned the reaction conditions to safeguard the stability of this bond. To initiate the synthesis, we utilized natural α-noscapine (4) as the starting material and achieved a high yield (90%) in the synthesis of 9-bromonoscapine (5) by modifying the reaction conditions described in the literature [9]. Subsequently, we converted 9-bromonoscapine to 9-aminonoscapine (6) in a 62% yield using CuI, Sodiumazide, and L-Proline in DMSO at 135°C for 3 h [Reaction Scheme 1]. It is important to note that the reaction scheme employed in our study differs from the previously reported facile synthesis of 9-aminonoscapine [10], and our reaction conditions have no impact on the sensitive C-C bond. Next, we introduced approximately measured alkyl/aryl isocyanates (7a-h) at a quantity of 1.2 millimoles to a solution containing 9-aminonoscapine, 6 (1.0 millimole), in DCM (10 mL), and stirred the mixture for 12 h at room temperature. Following confirmation of reaction completion through TLC analysis, we subjected the contents to washing with a brine solution [10]. The organic layer was gathered, filtered through a Na2SO4 bed, and subsequently evaporated under reduced pressure. The resulting crude residue underwent chromatography over a triethylamine silica bed using pet. Ether/ethyl acetate (3:2) as eluents, yielding solid products, including the pure compound 7 h. The final products were characterized using IR, 1H, and 13C NMR spectroscopy, and mass spectrometry techniques, with yields ranging from 65% to 90% [Reaction Scheme 2].
 | Reaction Scheme 1: The chemical synthesis of 9-bromonoscapine (3) and 9-aminonoscapines (6) using the drug noscapine as starting material. [Click here to view] |
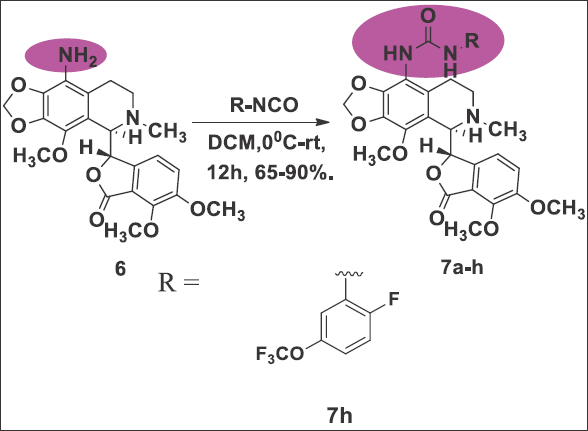 | Reaction Scheme 2: The chemical synthesis of urea-noscapine 7 h by coupling of alkyl/aryl isocyanates with 9-aminonoscapine. [Click here to view] |
2.1.1. 1-((R)-5-((S)-4,5-dimethoxy-3-oxo-1,3-dihydroisobenzofuran-1-yl)-4-methoxy-6-methyl-5,6,7,8-tetrahydro-[1,3]dioxolo[4,5-g]isoquinolin-9-yl)-3-(2,4,6-trichlorophenyl)urea (7h)
Nature: White solid. M.P: 125-127°C. IR (KBr): 3368, 2925, 2805, 1757, 1713, 1659, 1496, 1441, 1270, 1200, 1034, 789, 747 cm-1. 1H NMR (400 MHz, CDCl3): δ 7.31 (s, 2H, Ar-H), 6.99 (d, J = 8.3 Hz, 1H, Ar-H), 6.47 (d, J = 8.3 Hz, 1H, Ar-H), 5.93 (s, 2H, O-CH2-O), 5.49 (d, J = 4.1 Hz, 1H, Ar-CH, (C3-phthalide)), 4.33 (d, J = 4.1 Hz, 1H, Ar-CH, (C5’-isoquinoline)), 4.05 (s, 3H, -OCH3), 3.92 (s, 3H, -OCH3), 3.79 (s, 3H, -OCH3), 2.84-2.75 (m, 1H, -CHH-N-CH3 (C7’-isoquinoline)), 2.70-2.58 (m, 1H, -CHH-N-CH3 (C7’-isoquinoline)), 2.51 (s, 3H, N-CH3), 2.48-2.39 (m, 1H, Ar-CHH (C8’-isoquinoline)), 2.12-2.02 (m, 1H, Ar-CHH (C8’-isoquinoline)). 13C NMR (100 MHz,CDCl3): δ 165.3, 160.2, 151.1, 150.0, 144.6, 142.5, 138.6, 136.6, 132.9, 132.3, 131.2, 129.4, 128.2, 126.0, 117.2, 116.6, 114.2, 110.5, 99.3, 79.3, 59.5, 58.7, 57.3, 54.4, 47.0, 43.7, 20.6. MS (ESI-MS) m/z: 652[M+H]+, HRMS (ESI): Calcd for C29H27Cl3N3O8 [M+H]+: 650.08582, found: 650.08880.
2.2. Cell Culture and Establishment of the Hela Cell Line
Hela cell line was obtained from the National Centre for Cell Science in Pune, Maharashtra, India. The cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Pan Biotech) supplemented with 10% fetal bovine serum (FBS) and 0.1% antibiotics (penicillin-streptomycin). Cell maintenance was conducted at 37°C in a humidified atmosphere with 5% CO2. The nutrient solutions were replaced every alternate day. A 100 mM stock solution of the newly synthesized urea-noscapine derivative was prepared using dimethyl sulfoxide (DMSO, SRL) and stored at 4°C for subsequent use.
2.3. Cell Viability Assay of Urea-Noscapine Derivative using Hela Cell Line
The cell viability was estimated through the 3-(4,5-dimethylthiazol-2-yl)-2,5-ditrazolium bromide (MTT) assay. Following cellular adherence in 96-well plates, cells were exposed to the urea-noscapine derivative and sustained in complete medium for 48 h, with an extension to 72 h. To assess cell viability, 10 μFollowing trypsinization, the cellLs of MTT solution (5 mg/ml) was introduced to the cells and incubated for 4 h at 37°C, facilitating the generation of formazan crystals. Subsequently, the formazan crystals were dissolved in dimethyl sulfoxide (DMSO). Optical density (OD) was gauged at 570 nm using a microplate reader (BIO-RAD, Hercules, CA, USA). The half-maximal inhibitory concentration (IC50), indicative of the concentration at which cancer cell proliferation is restrained by 50%, was figured out using the online Quest GraphTM IC50 Calculator tool provided by AAT Bioquest, Inc. in Sunnyvale, CA, USA.
2.4. Cell Cycle Analysis by Flow Cytometry
Hela cells were treated with derivative of urea-noscapine for 24 h at the IC50 concentration (8.1 µM). Following trypsinization, the cells were subjected to two washes using the cold phosphate-buffered saline (PBS) and subsequently secure overnight at 20°C in 70% cold ethanol to facilitate staining, cells were exposed to a staining solution comprising PI/Triton X-100 (0.1% Triton X-100, 2 mg/mL propidium iodide, and 0.2 mg/mL DNase-free RNase) for a duration of 30 min. Subsequent cell cycle analysis was conducted employing flow cytometry (Beckman-Coulter, Fullerton, CA, USA).
2.5. Apoptosis Assay by Flow Cytometry
Hela cells were exposed to a derivative of urea-noscapine 7h derivative at the IC50 concentration (8.1 µM) for a 24-h duration. Following the manufacturer’s guidelines, cells underwent trypsinization, were double-washed using with cold PBS, and underwent staining done using Alexa Fluor® 488, Annexin V, and propidium iodide (PI) (BD Pharmingen, San Diego, CA, USA). Apoptotic cells were subsequently discerned utilizing a flow cytometer. Each sample contained 10,000 events, and the data were examined using CXP analysis software (Beckman-Coulter, Fullerton, CA, USA).
2.6. Preparation of Cell Lines for mRNA Sequencing
The Hela cell line was cultured using Dulbecco’s modified Eagle medium (DMEM, Pan Biotech) supplemented with keeping 10% FBS and 0.1% antibiotics (penicillin-streptomycin) at 37°C in a 5% CO2 humidified environment condition. Media replacement occurred every other day. A 100 mM stock solution of the urea-noscapine derivative 7h, prepared using dimethyl sulfoxide (DMSO), was utilized. The compound’s IC50 concentration, established at 8.1 µM, was employed for a 72-h treatment of the Hela cell line. Post-treatment, cells were incubated in 2 ml of RNAlater at 2-8 °C freezer for overnight, allowing for RNAlater permeation into the tissue. The triplicate replica of biological samples was transferred to Clevergene Private Ltd., Bengaluru, for mRNA sequencing.
2.7. Enhancement of mRNA and Preparation of the Library
Approximately 250 ng total mRNA was isolated using NEBNext Poly (A) mRNA Magnetic Isolation Module (Catalog: E7490, New England Biolabs) following the provided guidelines. The enriched mRNA was subsequently employed for this library preparation with the NEBNext Ultra II RNA Library Prep Kit for Illumina (Catalog: E7775S, New England Biolabs). In summary, the enriched mRNAs were primed with NEBNext Random Primers and subjected to chemical fragmentation at 94°C for 10 min in a magnesium-based buffer, resulting in ~200 nucleotide inserts. The fragmented mRNAs underwent reverse transcription to generate cDNA, and the resulting first-strand cDNA reactions were converted into double-stranded DNA (dsDNA). Purification of dsDNA fragments was achieved using 1.8X AMPure XP beads (Catalog: A63881, Beckman Coulter). End repair was performed, converting overhangs from fragmentation into blunt ends through an end repair mix. The 3’ to 5’ exonuclease activity of the end repair mix removed 3’ overhangs, while polymerase activity filled in 5’ overhangs. Adenylation involved adding a single ‘A’ nucleotide to the 3’ ends of blunt-ended fragments. Ligation of loop adapters to adenylated fragments was followed by cleavage with the uracil-specific excision reagent (USER) enzyme. Size selection aimed for a library size of 400-600 bp using AMPure beads (Catalog: A63881, Beckman Coulter) according to the manufacturer’s instructions. In addition, cDNA underwent 12 cycles of PCR amplification using NEBNext Ultra II Q5 master mix and “NEBNext Multiplex Oligos for Illumina” for sequencing multiplexing. Purification of amplified products was accomplished with the 0.9X AMPure XP beads (Catalog: A63881, Beckman Coulter), and the final DNA library was eluted inm, p’d of 0.1X TE buffer (source: https://international.neb.com/products/e7335-nebnext-multiplex-oligos-for-illumina-index-primers-set-1#Product%20Information).
2.8. Library Quantification
Concentration of the library was assessed using the Qubit 3 Fluorometer (Catalog: Q33216, Life Technologies) along with the Qubit dsDNA HS (High Sensitivity) assay kit (Catalog: Q32854, Thermo Fisher Scientific). The QubitTM 1X dsDNA HS assay kit includes a highly sensitive DNA reagent, buffers, and two DNA standards. The DNA HS assay reagent employs a detection dye recognized for its heightened sensitivity in precisely quantifying DNA/libraries in solution. It facilitates linear fluorescence detection in the range of 10 pg/μl to 100 ng/μl of DNA. The dye and buffers were diluted at a 1:200 ratio, and 1 μl of the library was combined with the dye mix. The mixture underwent incubation at room temperature for 2 min, and fluorescence readings were acquired using the Qubit 3 Fluorometer (Catalog: Q33216, Life Technologies). Before sample measurement, the instrument underwent calibration using the two standards provided in the kit (source: https://international.neb.com/products/e7770-nebnext-ultra-ii-rna-library-prep-kit-for-illumina#Product%20Information).
2.9. Library Validation
Quality assessment of the library was carried out utilizing the Agilent D5000 ScreenTape System (Catalog: 5067-5588, Agilent) and Agilent D1000 ScreenTape System (Catalog: 5067-5582, Agilent) on a 4150 TapeStation System (Catalog: G2992AA, Agilent), specifically designed for analyzing DNA molecules within the range of 100 to 5000 bp. Approximately 1 μl of the purified library was mixed with 10 μl of D5000 sample buffer (Catalog: 5067-5589) and subjected to vortexing using an IKA vortexer at 2000 rpm for 1 min. Following vortexing, the sample was centrifuged to collect it at the bottom of the strip. Subsequently, the strip was loaded onto the Agilent 4150 TapeStation system (source: https://international.neb.com/products/e7770-nebnext-ultra-ii-rna-library-prep-kit-for-illumina#Product%20Information).
2.10. Gene Expression and Transcriptome Analysis
2.10.1. Quality control and preprocessing of raw data involve measures to guarantee the integrity and reliability of the data.
All transcriptome activity, including coding and non-coding genes, can be characterized using transcriptome analysis experiments. The work processing of RNA-seq data encompasses multiple crucial steps, including raw read sequences analysis, adapter trimming, read alignment, gene quantification, and meticulous quality assessment at each stage of the analysis. The tool FastQC was used to examine the quality control of the raw readings (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/Babraham Bioinformatics, UK). The adapter sequence and low-quality bases were removed from the reads using Cutadapt (v2.6) to eliminate alignment biases [11].
2.10.2. Read alignment of raw data
To mitigate the potential loss of unannotated regions of the genome, the raw read sequences underwent alignment against human genome using as reference in the tool HISAT2 aligner (v2.1.0) [12]. The indexing algorithms used by the aligner vary in terms of readings’ map ability and required amounts of memory. With the help of the featureCount (v2.0.0) package, the expression count reads were produced [13]. The tool featureCount was used to calculate the gene expression of the MCF7 cells treated with urea-noscapine 7h derivative and untreated cells.
2.10.3. Expression quantification of the clean data
Transcript abundance quantification was carried out utilizing StringTie (v2.1.1), which utilizes a network flow algorithm grounded in optimization theory. StringTie improves the reconstruction of the transcriptome from RNA-seq reads by building a splice graph derived from the alignment, iteratively discerning highly covered splice paths, and employing maximum flow calculations to estimate gene abundance [14].
2.10.4. Analysis of DGE
Clean data were utilized to remap the assembled transcriptome, and the mapping results were used to calculate each gene in the read. To assess the differential expression between treated and control samples, the EdgeR package (http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html) developed by Robinson and McCarthy (2010) [15] was employed. This package utilizes a Poisson dispersion model to account for biological and technical variations and employs a Bayesian empirical model to optimize the level of over-dispersion across transcripts. The data are normalized to counts per million using the trimmed mean of M values (TMM) normalization (CPM). The p-value was modified using the q-value. Log2 (fold change) > 1 (for upregulated genes) and −1 (for significantly changed expression) were the cut-off values (for downregulated genes).
2.11. Gene Ontology Profiling
DEGs were functionally annotated using various databases, including Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and Clusters of Orthologous Groups (COG). These databases provided valuable insights into the biological pathways, molecular functions, and evolutionary relationships associated with the identified DEGs. Employing a significance level of p 0.05, the DEGs underwent functional enrichment analysis through DAVID web-accessible tool (http://david.abcc.ncifcrf.gov). G: Profiler (https://biit.cs.ut.ee/gprofiler/gost) was utilized to classify Gene Ontology (GO) and construct a GO tree, presenting distinct GO ids and P values for both up and downregulated DEGs after obtaining Gene Ontology annotation for each unigene. The DEGs were further categorized into functional pathways through KEGG analysis (https://www.genome.jp/kegg/). KEGG Orthology (KO) and KEGG pathway annotation were carried out using the KEGG automatic annotation service.
2.12. Network Analysis through Protein–Protein Interaction (PPI)
The query gene sequences underwent comparison with subject sequences through BLAST X, utilizing multiple databases for protein–protein functional interactions with an E-value set as cut-off at 10-5 (0.00001) [16]. By counting physical binding, genetic connections, and functional relationships, an analysis of “protein–protein” interaction networks was conducted to explore downstream associations among proteins. The STRING web Brower (https://string-db.org) was employed to investigate the relationship between specifically identified DEGs responsible for the anticancer action of noscapinoids and other genes functionally associated with them.
2.13. Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Gene Expression Study
Following the RNA-seq data analysis, quantitative real-time PCR (RT-PCR) was employed to validate the results. Two micrograms of total RNA will be used for cDNA synthesis, utilizing the PrimeScript™ RT reagent Kit with the gDNA Eraser Kit, following the manufacturer’s guidelines (TaKaRa Bio, China), for reverse transcription of 2 mg mRNA aliquots. Subsequently, SYBR Green-based qPCR analysis will be performed on an ABI ViiA 7 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The housekeeping gene GAPDH will be employed as the endogenous control, and all reactions will be conducted in triplicate. Relative gene expression will be determined using the comparative cycle threshold (2−DDCT) method.
3. RESULTS
3.1. Urea Derivative of Noscapine Inhibited Proliferation of Cancer Cells without Affecting the Normal Cells
Noscapine and the one of its derivative urea-noscapine (0–100 µM) were analyzed for their anti-proliferative activity using Hela cell line based on MTT assay. The drug urea-noscapine exhibited potent cytotoxic activity compared to noscapine. The IC50 value for the urea-noscapine is 4.8 µM and was found to be significantly less compared to noscapine (IC50 value is 43.9 µM) for Hela cells. Surprisingly noscapine and urea-noscapine inhibited proliferation of normal healthy cells, 293T to a value of <5% at a concentration of 100 µM, whereas it was found to be above 5% with a concentration above 100 µM, pointing the normal healthy cells are not affected by these compounds [Figure 1].
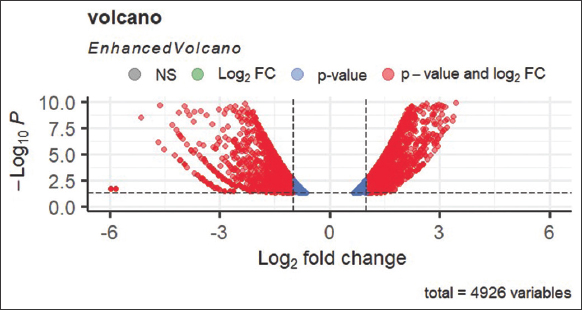 | Figure 1: The volcano plot visually represents the distribution of differentially expressed genes. Red dots indicate upregulated genes, blue dots represent non-differentially expressed genes, and downregulated genes are depicted by red dots on the negative side. This plot helps identify significant gene expression changes and allows researchers to focus on relevant genes for further analysis (P-value). It shows that the downregulated distribution of genes is more than upregulated gene distribution, which is showing in Figure 1. [Click here to view] |
3.2. Urea Derivative of Noscapine-induced Cell Death in Hela Cells
We conducted to investigate the induction of cell death in Hela cells upon treatment with urea-noscapine. The extent of apoptosis was assessed using Annexin/FITC fluorescent dyes and quantified by flow cytometry (FACS). Figure 2a presents the compiled data on the percentage of early apoptotic and late apoptotic cells in Hela cells treated with the1 IC50 concentration of urea-noscapine (4.8 µM) after 72 h. The control group, consisting of untreated cells, exhibited minimal levels of early apoptotic cells (2%) and late apoptotic cells (3%). These values were considered as the background cell death resulting from routine cellular stress during culture. In contrast, the treatment of urea-noscapine resulted in a significantly higher percentage of early apoptotic cells (14%) and late apoptotic cells (70%) compared to control group. “IC50” refers to the half-maximal inhibitory concentration and “mM” represents micromolar concentration [Figure 2].
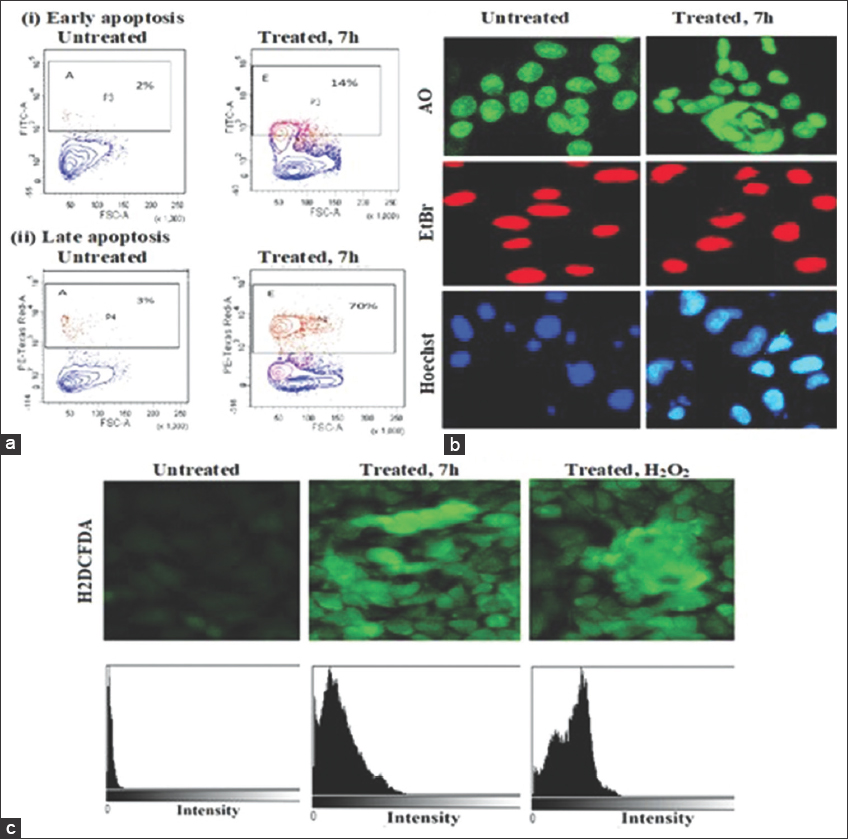 | Figure 2: Induction of apoptosis by the urea derivative of noscapine (urea-noscapine). Flow cytometry analysis of cells showing increased percentage of early and late apoptosis with the treatment of urea-noscapine 7 h derivative for 72 h compared to untreated cells, (a) flow cytometry of apoptosis cell, (b) morphological changes in cell line, (c) analysis of reactive oxygen species activity in cell line. [Click here to view] |
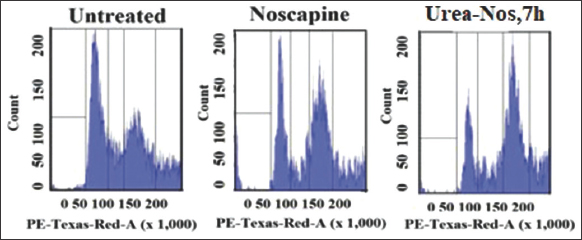
3.3. Urea Derivative of Noscapine Change in the Cell Cycle Profile and Cause Mitotic Arrest at G2/M Phase
We studied the outcome of urea-noscapine at its IC50 concentration (4.8 µM) on the cell cycle distribution changes in Hela cells using flow cytometry (FACS) analysis. Following a 72-h treatment with urea-noscapine, notable alterations in the cell cycle profile were observed. The FACS analysis demonstrated a substantial aggregation of cells in the G2/M phase compared to the untreated control cells. In addition to the G2/M block, a distinct peak corresponding to hypodiploid DNA content (sub-G1) was observed to increase after 72 h of drug treatment. This progressive generation of cells exhibiting hypo diploid DNA content signifies fragmented DNA, indicative of apoptotic or dying cells.
In summary, the observed alterations in cell cycle profile of Hela cells treated with urea-noscapine at its IC50 concentration suggest that the drug is causing G2/M phase arrest and inducing apoptosis. The accumulation of cells in the G2/M phase may be a cellular response to DNA damage, and the presence of a sub-G1 peak strongly suggests that cells are undergoing programmed cell death [8,9]. These findings provide valuable insights into the mechanism of action of urea-noscapine and its potential as an anti-cancer agent or in the context of other therapeutic applications that are also enable the researcher’s community for further analysis. Further studies and experiments may be needed to elucidate the precise molecular pathways involved in these observed effects.
3.4. Transcriptome Sequencing Data Analyses
Toward quality assurance and accurate trimming, a combined 29.4 GB of raw data from untreated and treated Hela cells with urea-noscapine was used and presented in Table 2.
Table 1: IC50 values of noscapine and its urea derivative (urea-Nos) using Hela cell line.
| IC50 (mM) | |
|---|---|
| Compounds | MCF-7 |
| Noscapine | 43.9±4.6*** |
| Urea-Nos | 4.8±0.5*** |
Significance value
*** p < 0.001
Table 2: Statistical summary of quality control of transcript sequencing.
| Filename | Hela-T_R1_1.fastq/Hela-T_R2_1.fastq | Hela-T_R1_2.fastq/Hela-T_R2_2.fastq | Hela-T_R1_3.fastq/Hela-T_R2_3.fastq | Hela-UT_R1_1.fastq/Hela-UT_R2_1.fastq | Hela-UT_R1_2.fastq/Hela-UT_R2_2.fastq | Hela-UT_R1_3.fastq/Hela-UT_R2_3.fastq |
|---|---|---|---|---|---|---|
| File type | Conventional base calls | Conventional base calls | Conventional base calls | Conventional base calls | Conventional base calls | Conventional base calls |
| Encoding | Sanger/Illumina 1.9 | Sanger/Illumina 1.9 | Sanger/Illumina 1.9 | Sanger/Illumina 1.9 | Sanger/Illumina 1.9 | Sanger/Illumina 1.9 |
| Total Sequences | 18162184 | 17169177 | 18662987 | 19416660 | 20446950 | 19456669 |
| Sequences flagged as poor quality | 0 | 0 | 0 | 0 | 0 | 0 |
| Sequence length | 151 | 151 | 151 | 151 | 151 | 151 |
| % GC | 51 | 51 | 51 | 53 | 53 | 53 |
Hela-T_R1_1.fastq/Hela-T_R2_1.fastq, Hela-T_R1_2.fastq/Hela-T_R2_2.fastq, Hela-T_R1_3.fastq/Hela-T_R2_3.fastq is representing the Urea-Nos/Hela (Treated) and Hela-UT_R1_1.fastq/Hela-UT_R2_1.fastq, Hela-UT_R1_2.fastq/Hela-UT_R2_2.fastq, Hela-UT_R1_3.fastq/Hela-UT_R2_3.fastq is representing the Hela (Control)
The alignment of reads was conducted using the HISAT2 software against the GRCh38 reference genome sequence. For the Hela (Control) and urea-noscapine/Hela (Treated) cells, the mapping percentages for human reads were 96.32%, 95.30%, 96.25% and 96.69%, 96.03%, 95.91%, respectively. Detailed statistical data on the mapping results can be found in Table 3.
Table 3: Statistical summary of the mapping results.
| SAMPLES | Hela_1 (Control) | Hela_2 (Control) | Hela_3 (Control) | Urea-Nos/Hela_1 (Treated) | Urea-Nos/Hela_2 (Treated) | Urea-Nos/Hela_3 (Treated) |
|---|---|---|---|---|---|---|
| Reads paired | 19416660 (100.00%) | 20446950 (100.00%) | 19456669 (100.00%) | 18162184 (100.00%) | 17169177 (100.00%) | 18662987 (100.00%) |
| Aligned | 18702126 | 19485943 | 18727043 | 17561015 | 17441145 | 17899670 |
| Overall alignment rate | 96.32% | 95.30% | 96.25% | 96.69% | 96.03% | 95.91% |
Transcript abundances were assessed through the utilization of the StringTie package. Read estimations were performed using the featureCount package, and the identification of DEGs was carried out using the EdgeR R tool (version 4.3). The p value was adjusted using the q value, and the threshold for determining significantly altered expression was established at q value 0.05 with a log2 (fold change) >1 for upregulated genes and −1 for downregulated genes. The tabulated results illustrating the calculation of the number of DEGs can be found in Table 4.
Table 4: Statistical summary of the DEGs.
| DEGs | P value | Fold change | Calculates DEGs |
|---|---|---|---|
| Total significant genes | <0.05 | 14442 | |
| Upregulated genes | <0.05 | ≥1 | 3824 |
| Downregulated genes | <0.05 | ≥−1 | 10618 |
DEGs: Differentially expressed genes
3.4.1. Volcano plot
Figure 1 displays a volcano plot illustrating the contrast in gene expression analysis levels between two groups: Control and treatment samples, along with the statistical significance analysis of these differences. A comprehensive analysis involving 4926 variables was conducted to calculate the presence of both upregulated and downregulated genes.
3.4.2. Venn diagram
Figure 3 illustrates the comparison of upregulated and downregulated genes, as well as unique and shared genes, in urea-noscapine-treated samples in relation to the control. Set A denotes the count of upregulated genes, while Set B represents the count of downregulated genes. Among 3824 variables, 1895 unigenes were identified in upregulated genes, and for downregulated genes, 8689 unigenes were identified out of 10618 variables. In addition, a total of 1929 genes were identified as common to both the upregulated and downregulated categories.
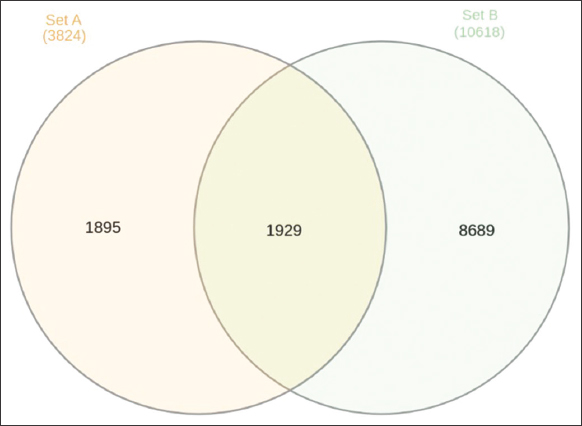 | Figure 3: Venn diagram showing the number of upregulated and downregulated genes, unique genes, and common genes in the urea-noscapine-treated samples compared to control. [Click here to view] |
3.4.3. Gene ontologies
The DEGs were annotated in “g: profiler” (https://biit.cs.ut.ee/gprofiler/) and Enrichr (https://maayanlab.cloud/Enrichr/) database, respectively. A total of 1895 upregulated and 8689 downregulated gene annotations were done in the David database. These genes were distributed in five categories: Biological process, Cellular component, Molecular function, KEGG pathways, and MiRNA. The DEGs were found to be distributed in the Biological processes containing highest portion (975 “upregulated” genes and 2127 “downregulated” genes). The second-highest count of DEGs, comprising 169 “upregulated” and 433 “downregulated,” was observed in the Cellular Component category, specifically within the subcategory of cell or cellular part. Within the Molecular Function category, there were 162 upregulated and 419 downregulated DEGs. Furthermore, 83 upregulated and 222 downregulated DEGs were distributed among KEGG pathways. In the MiRNA category, 661 upregulated and 1844 downregulated DEGs were identified, presenting potential markers. In addition, 502 upregulated genes and 618 downregulated genes were calculated within the Human Protein Atlas, as depicted in Figure 4.
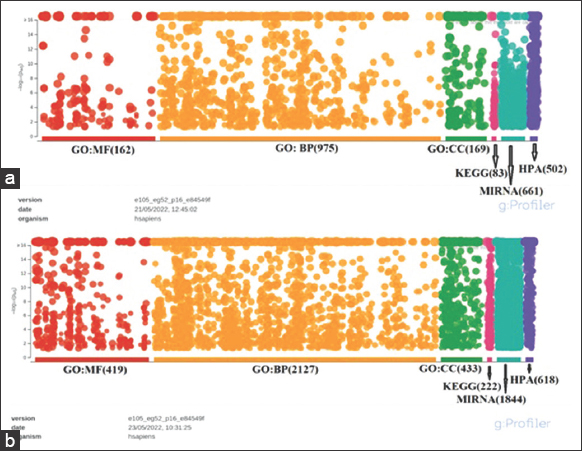 | Figure 4: (a and b) Cluster of orthologous (COGs) genes function classification of the annotated database of essential genes (DEGs). A total of 1895 differentially expressed genes were found to be upregulated, while 8689 DEGs were downregulated. These DEGs were classified into five functional categories based on the COG group classification system. [Click here to view] |
The COG categories are displayed on the horizontal axis of a graph, with gene numbers and proportions plotted on the vertical axis. This analysis provides insights into the functional distribution of annotated DEGs according to COG categories, allowing for a comprehensive understanding of the different biological processes and functions associated with the upregulated and downregulated genes [Figure 4].
We find that the functional ontology of the upregulated genes is using “g: Profiler” (https://biit.cs.ut.ee/gprofiler/gost) tool. DEGs are distributed in different functions of Biological Process, Molecular Function, and Cellular component [Figure 3]. The 20 functional categories and the pathways information are represented in Table 5. All the 1895 DEGs are distributed into four functional categories as shown in Figure 5 and Table 5. The 3987 DEGs belonging to Biological process, 764 DEGs in Molecular Function, 332 DEGs in Cellular Component, and 291 DEGs in KEGG pathways (Supplementary_Biological_Process_Up, Supplementary_Cellular_Component_Up, Supplementary_Molecular_Function_Up, Supplementary_KEGG_Up) are included in the supplementary materials. We found that the largest number of DEGs (8689) are involved in downregulation [Figure 6 and Table 6], which are functioning in different GO (Gene ontology) functions like Biological process (5755 number of DEGs), Molecular function (1175 number of DEGs), Cellular Component (490 number of DEGs), and KEGG pathways (316 number of DEGs) which are included in the Supplementary material (Supplementary_Biological_Process_down, Supplememtary_Molecular_Function_down, Supplementary_Cellular_Component_down, Supplementary_KEGG_down).
Table 5: The top KEGG pathways data of upregulated DEGs according to p-value. The total genes involve and KEGG Id.
| Term (KEGG Pathway) | Genes | P-value |
|---|---|---|
| Legionellosis (hsa05134) | 16 | 5.30652E-05 |
| Synthesis and degradation of ketone bodies | 5 | 0.001273379 |
| TNF signaling pathway (hsa04668) | 19 | 0.008803736 |
| Fluid shear stress and atherosclerosis (hsa05418) | 22 | 0.011448563 |
| NOD-like receptor signaling pathway (hsa04621) | 26 | 0.021061566 |
| Peroxisome (hsa04146) | 14 | 0.02132007 |
| Basal cell carcinoma (hsa05217) | 11 | 0.033282963 |
| NF-kappa B signaling pathway (hsa04064) | 16 | 0.035500797 |
| Arginine and proline metabolism (hsa00330) | 9 | 0.043307776 |
| Aminoacyl-tRNA biosynthesis (hsa00970) | 11 | 0.044866304 |
| Pertussis (hsa05133) | 12 | 0.053227619 |
| Neutrophil extracellular trap formation (hsa04613) | 25 | 0.054969604 |
| IL-17 signaling pathway (hsa04657) | 14 | 0.059096363 |
| Shigellosis (hsa05131) | 31 | 0.061880773 |
| RIG-I-like receptor signaling pathway (hsa04622) | 11 | 0.064214688 |
| Selenocompound metabolism (hsa00450) | 4 | 0.070376254 |
| Axon guidance (hsa04360) | 23 | 0.094284467 |
| Non-alcoholic fatty liver disease (hsa04932) | 20 | 0.096221565 |
| Nicotinate and nicotinamide metabolism (hsa00760) | 6 | 0.108514167 |
| Butanoate metabolism (hsa00650) | 5 | 0.119990454 |
KEGG: Kyoto Encyclopedia of Genes and Genomes
Table 6: The top KEGG pathways data of downregulated DEGs according to p-value. The total genes involve and KEGG Id.
| Term | Overlap | P-value |
|---|---|---|
| Mitophagy (hsa04137) | 42 | 0.001766 |
| SNARE interactions in vesicular transport (hsa04130) | 21 | 0.015461 |
| Proteasome (hsa03050) | 27 | 0.026676 |
| Base excision repair (hsa03410) | 20 | 0.03533 |
| Fanconi anemia pathway (hsa03460) | 30 | 0.049022 |
| Endocytosis (hsa04144) | 121 | 0.079713 |
| Citrate cycle (TCA cycle) (hsa00020) | 17 | 0.101164 |
| Ribosome (hsa03010) | 77 | 0.10305 |
| Homologous recombination (hsa03440) | 22 | 0.122724 |
| RNA polymerase (hsa03020) | 17 | 0.135998 |
| Spliceosome (hsa03040) | 72 | 0.147603 |
| Ubiquitin-mediated proteolysis (hsa04120) | 67 | 0.165638 |
| mRNA surveillance pathway (hsa03015) | 47 | 0.210994 |
| Circadian rhythm (hsa04710) | 16 | 0.229769 |
| Vitamin B6 metabolism (hsa00750) | 04 | 0.23013 |
| DNA replication (hsa03030) | 18 | 0.264507 |
| Chronic myeloid leukemia (hsa05220) | 36 | 0.281504 |
| Lysine degradation (hsa00310) | 30 | 0.29268 |
| Non-small cell lung cancer (hsa05223) | 34 | 0.29737 |
KEGG: Kyoto Encyclopedia of Genes and Genomes
Table 7: Protein–protein interaction network analysis table, showing the interaction, co-expression, and combined score of upregulated genes from n = 3824 DEGs. The table shows the data of top 20 expressed gene from the PPI network.
| Node 1 | Node 2 | Co-expression | Combined score |
|---|---|---|---|
| BAG2 | HSPA4 | 0.103 | 0.602 |
| HSPA4 | BAG2 | 0.103 | 0.602 |
| EMC2 | MMGT1 | 0.166 | 0.972 |
| MMGT1 | EMC2 | 0.166 | 0.972 |
| CASP8 | CFLAR | 0.098 | 0.981 |
| CFLAR | CASP8 | 0.098 | 0.981 |
| CASP8 | TRAF2 | 0.062 | 0.87 |
| TRAF2 | CASP8 | 0.062 | 0.87 |
| TNFAIP3 | TRAF1 | 0.2 | 0.695 |
| TRAF1 | TNFAIP3 | 0.2 | 0.695 |
| TNFAIP3 | TRAF2 | 0.062 | 0.798 |
| TRAF2 | TNFAIP3 | 0.062 | 0.798 |
| CASP8 | TRAF1 | 0.064 | 0.678 |
| TRAF1 | CASP8 | 0.064 | 0.678 |
| HSPA4 | HSPD1 | 0.43 | 0.374 |
| HSPD1 | HSPA4 | 0.43 | 0.374 |
| CFLAR | TRAF1 | 0.106 | 0.682 |
| TRAF1 | CFLAR | 0.106 | 0.682 |
| CFLAR | TRAF2 | 0.062 | 0.486 |
| TRAF2 | CFLAR | 0.062 | 0.486 |
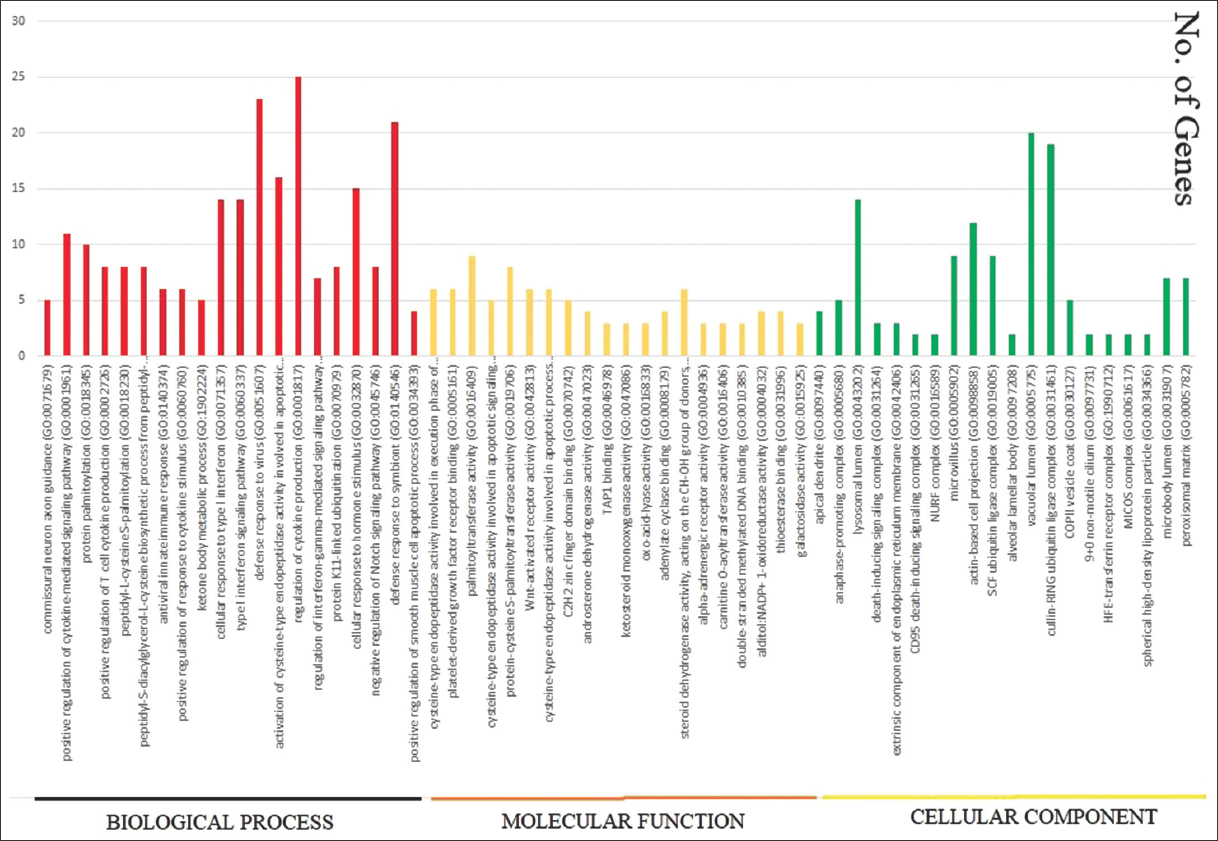 | Figure 5: The representation of upregulated gene-ontology functions such as Biological Process, Molecular Function, and Cellular Component. The lower side shows name of the functions and the horizontal axis shows number of genes regulated by each function in Figure 5. [Click here to view] |
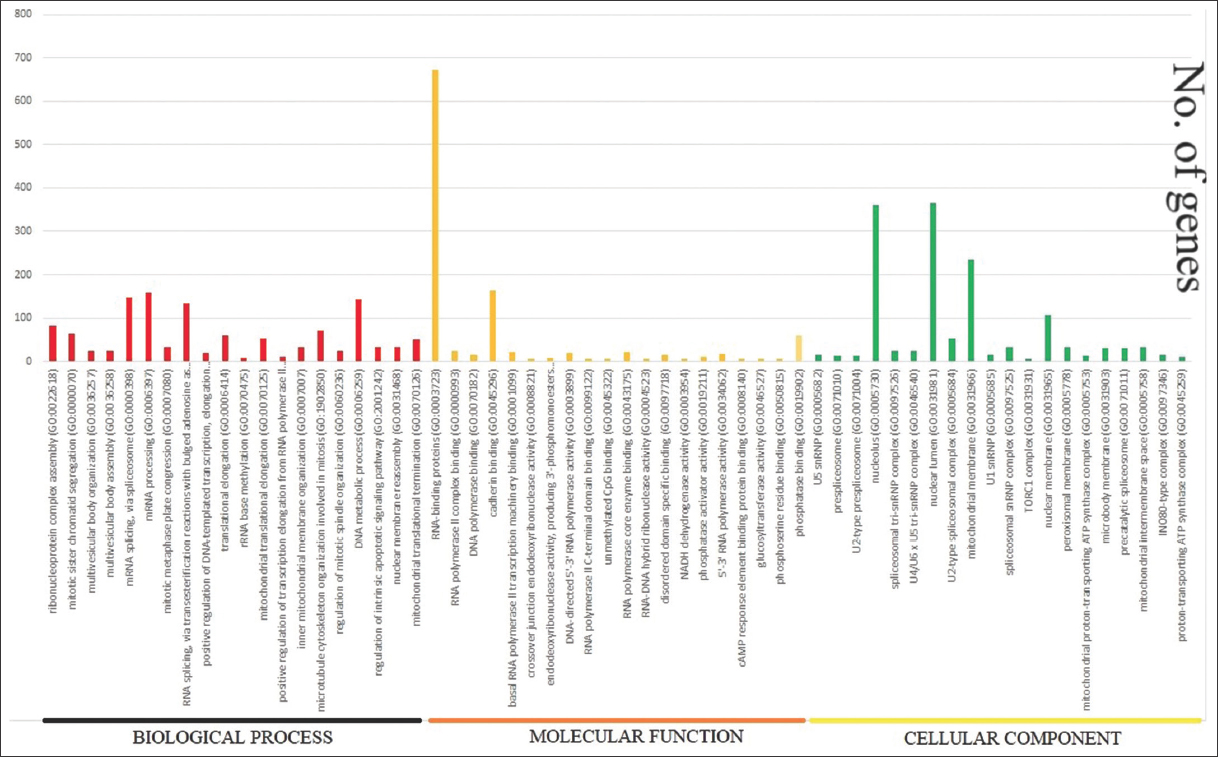 | Figure 6: Downregulated gene-ontology function such as Biological Process, Molecular Function, and Cellular Component. The lower side shows the name of the functions and the horizontal axis shows the number of genes regulated by each function [Figure 6]. [Click here to view] |
Caspases constitute a family of endoproteases that play a crucial role in cellular regulatory networks, influencing inflammation and cell death processes. Caspase-mediated processing can result in substrate inactivation and the generation of active signaling molecules, contributing to well-regulated processes like apoptosis and inflammation [17]. Our study revealed active involvement of Caspase family genes, including CASP14, CASP12, CASP8, CASP10, CASP1, CFLAR, along with NGFR, TRAF2, HSPE1, HSPD1, DAP, NLRP12, KHDC1, PPARG, PDCD2, NLRP1, and BOK, in the activation of cysteine-type endopeptidase activity associated with the apoptotic process in biological pathways. In addition, upregulation was observed in genes such as PDCD4, PPARG, and SOD2, which are involved in the positive regulation of vascular-associated smooth muscle cell apoptotic processes. In the molecular function category, upregulation was identified in genes CASP12, CASP8, CASP10, CASP1, and CFLAR, specifically associated with the cysteine-type endopeptidase activity of the apoptotic signaling pathway. Tumor necrosis factor (TNF) is a versatile cytokine that performs essential functions in various cellular processes, including cell viability, proliferation, differentiation, and apoptosis [18]. In our analysis, we found that the genes IL15; TNFAIP3; CXCL1; TRAF2; VEGFD; TRAF1; CFLAR; CXCL3; CXCL2; MAPK13; IL6; CASP8; CREB1; CASP10; IL1B; IRF1; MAP3K7; IL18R1; and MAP2K6 involved in TNF signaling pathway are over expressed. Further, the genes of the caspases family such as CASP3, CASP7, CASP8, and CASP10 are also over-expressed. It has been reported that cancer cell death is induced by RIG-I-like receptor signaling pathway followed by the release of cytochrome c from mitochondria. Following caspase-9 activation, the intrinsic apoptotic pathway is carried out [19]. In our analysis, we found that the genes CXCL8, CASP8, CASP10, DDX58, IFNE, IRF7, TRAF2, ISG15, MAP3K7, ATG12, and MAPK13 involved in RIG-I-like receptor signaling pathway leading to apoptosis and cell growth arrest are overexpressed.
Our analysis identified the IL-17 signaling pathway, which includes genes such as CXCL8, TRAF3IP2, TNFAIP3, and CXCL1, as well as CASP genes that respond to apoptosis (hsa04657_Supplementary). In addition, we found that Shigellosis can activate Shigella FimA to crosstalk between extrinsic and intrinsic apoptotic pathways (hsa05131_Supplementary). These findings highlight the upregulated function and interaction of Caspases in apoptosis.
Mitophagy is an essential function in regulating the homeostasis of cells and tissues by limiting the build-up of defective mitochondria, which can increase reactive oxygen species (ROS) generation and cause cell damage. Recent research has shown that mitophagy has a role in controlling tumor initiation and growth [20]. We found that the genes (BECN1; USP15; CALCOCO2; SRC; AMBRA1; HIF1A; BCL2L13; NRAS; RHOT2; TBK1; UBB; UBC; E2F1; MFN2; USP30; RPS27A; HRAS; TBC1D15) involved in mitophagy functional pathway are under-expressed with the treatment of urea-noscapines. Many endocytic proteins that control tumor metastasis, including migration and invasion, are deregulated in cancer [21]. In this study, we found 121 functional genes involved in the pathway of endocytosis [hsa04144_Supplememtary and Table 5]. Chronic myeloid leukemia pathways are a crucial functional pathway in cancer development, where P53 tumor suppressor gene inhibits or suppresses cell survival and proliferation process in the pathway (hsa05220_Supplementary). Our analysis of Chronic Myeloid Leukemia and Non-Small Cell Lung Cancer revealed that the tumor suppressor genes P53, RARB, and FHIT were suppressed, while the genes RAS, EGFR, and STAT were upregulated. These genes are involved in regulating tumor progression and uncontrolled cell proliferation in these cancers. Specifically, the suppression of tumor suppressor genes and the activation of oncogenes such as RAS, EGFR, and STAT play a critical role in the development and progression of these cancers [22]. These outcomes offer insights into the molecular mechanisms underlying the pathogenesis of these cancers and highlight potential targets for novel therapies [23]. These functions are downregulated after the treatment of Urea noscapine in Hela cell line.
3.4.4. Study of PPI network analysis
To gain a deeper understanding of the biological implications of the identified DEGs at the protein level, we constructed a PPI network specifically focusing on the expressed proteins encoded by the upregulated DEGs. The network analysis unveiled significant interactions among these proteins, comprising 56 nodes and 77 edges. The protein enrichment p-value for both up and downregulated DEGs was calculated as 3.63e-11. For upregulated DEGs, we calculated the average node degree and average local clustering coefficient were determined to be 2.75 and 0.426, respectively.
The PPI network analysis of upregulated genes indicated heightened co-expression between CCNA2 and KIF18A genes. Notably, genes such as BAG2, BCL2L11, HSPA4, CASP8, and CASP1 exhibited substantial co-expression with other selected genes. Of particular interest, BAG2 and HSPA4 demonstrated the highest level of co-expression. Further, the genes CASP8 and CFLAR, as well as CASP8 and CASP1, displayed homology scores of 0.656 and 0.578, respectively, as illustrated in Figure 7.
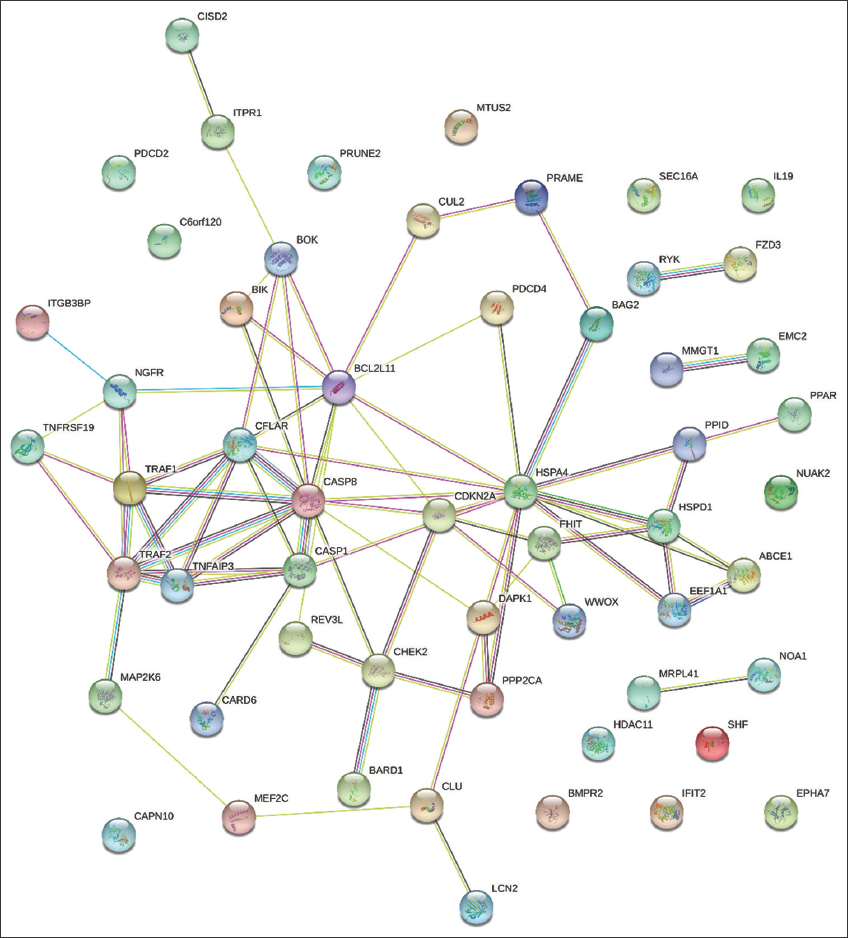 | Figure 7: An analysis of the protein–protein interaction network was conducted, focusing on genes associated with apoptosis, programmed cell death, tumor growth, and proliferation. The construction of the PPI network involved integrating genes participating in various signaling pathways using the “STRING” tool to investigate the interconnections among these genes. The findings revealed that three shared DEGs–BAG2, BCL2L11, HSPA4, CASP8, and CASP1 demonstrated close co-expression with other genes, as illustrated in Figure 7. [Click here to view] |
The table shows the data of top 20 expressed genes from the PPI network.
3.5. Analysis of Gene Expression through RT-PCR
Gene expression analysis determined by RT-PCR to examine the expression of 15 major DEGs in untreated and treated Hela cells with urea-noscapines in carried to validate the transcriptomic sequencing results. An outline of the transcriptome sequencing and RT-PCR data is presented in Figure 8. Through log2FC and Ct-value, the relative expression was described. Compared to control, the regulated patterns of 14 downregulated DEGs were compared with the RT-PCR expression value (Supplementary RT-PCR).
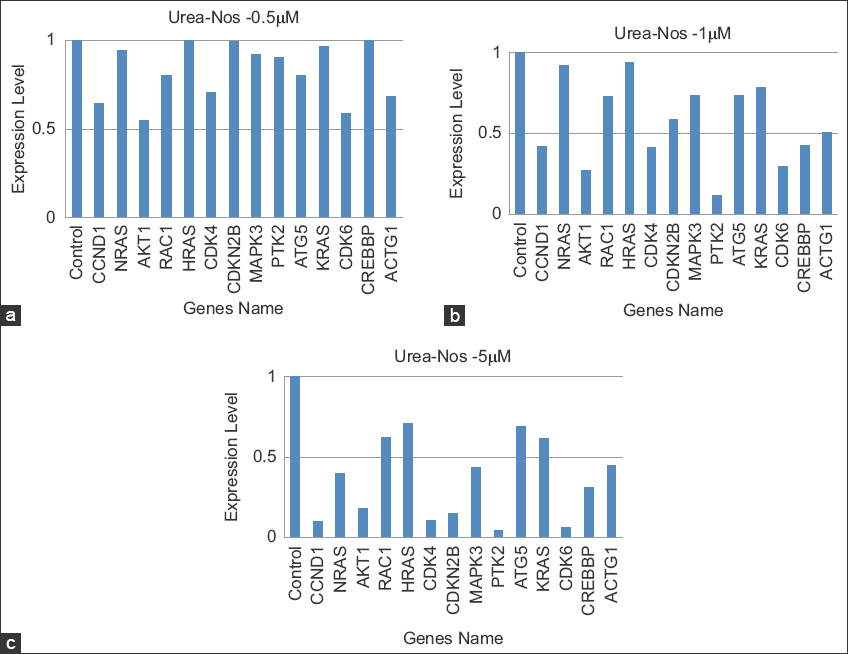 | Figure 8: Comparison of gene expression levels involves assessing the correlation between reverse transcription polymerase chain reaction (RT-PCR) results and transcriptome data to validate their expression levels. (a-c) Figures illustrate the downregulated genes and their expression levels under varying concentrations of urea-noscapine treatment. In the RT-PCR analysis, 14 genes affected by urea-noscapine treatment were specifically selected for examination. The expression levels of each gene were normalized in relation to untreated Hela cells for comprehensive analysis. [Click here to view] |
4. DISCUSSION
Numerous studies have conducted thorough examinations of urea derivatives due to their potential as anticancer agents. Among these compounds, aromatic compounds of urea derivatives like N-aryll-N’-(2-chloroethyl) ureas and benzoylureas have exhibited promising anticancer properties. These compounds have been found to effectively interact with tubulin, a protein crucial for cell division, which further underscores their potential as valuable components in the fight against cancer [21]. Urea derivatives containing heterocyclic structures have been identified as inhibitors of several key proteins involved in various stages of tumor development. These proteins include receptor tyrosine kinases (RTKs), RAF kinases, protein tyrosine kinases (PTKs), and NADH oxidase different biological processes and pathways [24]. These molecules are crucial players in different aspects of tumorigenesis, and the urea derivatives have been found to effectively block their activity, suggesting their potential significance as therapeutic agents in cancer treatment [24,25].
A collection of novel urea-noscapine compounds was created by combining isocyanates with 9-amino-noscapine, as illustrated in Reaction Scheme 1. Synthesizing noscapine derivatives has historically presented challenges because of the fragile carbon-carbon (C-C) bond between two heterocyclic components, namely phthalide and isoquinoline units. However, the reaction conditions employed in this study were specifically designed not to affect this delicate C-C bond linkage [as depicted in Reaction Scheme 2]. Consequently, all eight urea-noscapine conjugates were successfully synthesized with high yields and underwent thorough characterization using techniques such as NMR, IR spectroscopy, and mass spectrometry, providing a comprehensive understanding of their molecular properties. These molecules were already tested in vitro for their antiproliferative activity using different human cancer cell lines: Hela cell line (immortalized human cervical cancer cell line). The wide difference in inhibitory concentration (IC50) values obtained using Hela call line suggests that these compounds inhibit cellular proliferation of cancer cells and were cell-type dependent. Based on the minimum inhibitory concentration (IC50 value), one of the most potent derivatives 7 h having IC50 value 8.1 µm using Hela cell line cell lines was selected [Table 1]. Subsequently, this highly effective derivative was employed for a more in-depth exploration of its mode of action. In general, tumor cells have the propensity to spread and establish themselves in various parts of the body. Therefore, it is valuable to assess the drug molecule’s ability to hinder the colonization capability of these tumor cells. Through the utilization of a colony formation assay, it was determined that the urea-noscapine conjugate effectively suppresses the capacity of Hela cells to form colonies, with an IC50 value of 8.1 µM, as illustrated in Figure 2a. This finding underscores the compound’s potential to impede the critical process of tumor cell colonization. The observed suppression of cancer cell proliferation was attributed to the halt in the progression of the cell cycle, specifically at the G2/M phase, as demonstrated in Figure 9. This finding aligns with prior research, as numerous noscapine derivatives previously developed have been shown to impede cell proliferation by interfering with the progression of the cell cycle at the G2/M phase. This consistency is supported by studies conducted [26-31] further emphasizing the significance of this mechanism in the anticancer activity of noscapine derivatives. Hence, we conducted an examination to understand the initiation of cell death, focusing on the most powerful urea derivative of noscapine, denoted as 7h. This investigation involved the use of three distinct fluorescent dyes, namely AO (Acridine Orange), EtBr (Ethidium Bromide), and Hoechst, which enable differentiation between live and deceased cells. The results from the fluorescent imaging employing these dyes clearly revealed that 7h had a more pronounced and effective role in inducing cell death, as illustrated in Figure 2b. In addition, we employed FACS analysis to quantify the proportions of apoptotic cells. The apoptotic process involves distinct biochemical changes in the lipid composition of the cell membrane. Specifically, phosphatidylserine, which typically resides on the inner leaflet of the cell membrane, relocates to the outer leaflet during apoptosis and can be detected through its binding to Annexin V. Conversely, a cell-impermeable DNA-binding fluorescent dye known as propidium iodide can only penetrate cells in the late stages of apoptosis when membrane integrity is compromised. In our study, when treating Hela cells with the IC50 concentration of 7h, we observed that 14% of cells were in the early apoptotic stage, while a substantial 70% were in the late apoptotic stage. These percentages significantly exceeded those of untreated cells, as depicted in Figure 9. This suggests that the treatment with 7 h induced a notable increase in both early and late apoptotic cells in the Hela cell line. We again conducted an investigation to assess the generation of ROS when cells were treated with 7h, using DCFDA as the probe. What we observed was that the treated cells produced a notably higher quantity of ROS compared to untreated cells, as illustrated in Figure 2c. This increased ROS production was associated with the disruption of cellular MTs and the loss of mitochondrial transmembrane potential, as depicted in Figure 2c. This could serve as a potential mechanism behind the induction of apoptosis. From a therapeutic perspective, it is worth noting that cancer cells typically exhibit elevated ROS levels compared to normal cells [2]. Thus, the elevation of ROS levels through treatment could selectively target and eliminate cancer cells, as indicated in previous research by Pelicano et al. [32]. Furthermore, in vivo experiments demonstrated that the urea derivative of noscapine, specifically 7 h, significantly inhibited the growth of human breast tumors when implanted in nude mice as xenografts of Hela cells.
 | Figure 9: The impact of 7 h at its IC50 concentration (8.1 μM) on the cell cycle distribution of Hela cells was assessed after a 72-h treatment period. The bar graph displays the proportion of cells present in specific phases of the cell cycle. [Click here to view] |
The transcriptome sequences between untreated and treated Hela cells with urea-noscapine were analyzed and compared to find the list of upregulated and downregulated DEGs to explore the mechanism of action of urea-noscapine toward induction of apoptosis. Apoptosis is a process of programmed cell death that is important for the normal functioning of tissues and organs, as well as for the removal of damaged or cancer cells [33]. Noscapine and its derivatives were shown to induce apoptosis in cancer cells of different tissues [3]. However, the exact mechanism of induction of apoptosis is not yet known. This study focused on testing the efficacy of a potent derivative of noscapine coupled with urea pharmacophore 7h against cervical cancer cell lines (Hela). This compound was found to inhibit the proliferation of Hela cell line effectively (IC50 value is 8.1 µM), perturb the cell cycle at G2/M phase and effectively induced apoptosis.
Gene expression studies using transcriptome data analysis are crucial in understanding the underlying mechanisms involved in cancer development and treatment. The primary focus of this study is to identify DEGs that are either downregulated or upregulated in the Hela cells treated with urea-noscapine compared to untreated cells based on transcriptome analysis. Various up or downregulated DEGs were identified, with a majority of these genes involved in cancer development and treatment. Our analysis reveals that several genes involved in the process of apoptosis, inflammation response, cell death, and cysteine-type endopeptidase activity are upregulated. Specifically, genes from the Caspase family (CASP14, CASP12, CASP8, CASP10, CASP1, and CFLAR) play a crucial role in regulating the apoptotic signaling pathway. These findings provide valuable insights into the molecular mechanisms involved in noscapinoid treatment and can aid in the development of novel therapies for breast cancer. Further, CCND1, NRAS, AKT1, RAC1, HRAS, CDK4, CDKN2B, MAPK3, PTK2, ATG5, KRAS, CDK6, CREBBP, and ACTG1 genes are downregulated in different functions and are involved in cell growth, proliferation, initiation, development, and migration.
To validate our findings, we have selected 15 important genes to perform RT-PCR analysis to check their expression regulation in the cell line. Our analysis revealed that the selected genes were downregulated, and their functions were primarily involved in cancer cell growth, proliferation, initiation, development, and migration. This finding indicates that the noscapinoid treatment is effective in suppressing the expression of these genes, which are crucial for cancer cell survival and proliferation. Overall, our study provides valuable insights into the molecular mechanisms of noscapinoid treatment, which can aid in the development of effective cancer therapies.
5. CONCLUSION
Our analysis of Hela cell line (Cervical cancer) RNA-seq data, which was treated with urea derivative of noscapinoids has revealed the relationships among DEGs and shed light on the potential mechanisms of this medicine. These DEGs gene may serve therapeutic targets for cancer resistance. In addition, the downregulated genes identified in our study may be utilized to detect the disease. Furthermore, the upregulated genes and their associated protein complexes could serve as potential antibodies to fight the disease, highlighting their potential utility as targets for novel therapies. By understanding the molecular mechanisms action underlying the effects of noscapinoids, it may be possible to limit disease progression and improve patient outcomes in cervical cancer. Overall, our findings enable the researcher to comprehend the potential applications of noscapinoids in cancer treatment and provide a foundation for further research in this area. While noscapine, the foundational compound, is currently undergoing clinical trials, the urea derivative of noscapine offers a distinct advantage due to its increased potency. Importantly, this heightened efficacy is achieved without jeopardizing the inherent non-toxic nature of noscapine.
6. ACKNOWLEDGMENTS
The authors express gratitude to the Center of Excellence, Natural Products and Therapeutics (COE-NPT), OHEPEE, World Bank, the Department of Biotechnology and Bioinformatics at Sambalpur University for their backing of this research. In addition, appreciation is extended to Centurion University of Technology and Management, Jatni, Bhubaneswar, for their support and provision of infrastructure.
7. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. FUNDING
The funding was provided by Centre of Excellence, Natural Products and Therapeutics (COE-NPT), OHEPEE, World Bank.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
The study protocol was approved by University with the reference SU/BTBI/IAEC/2023/04.
11. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
13. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Diab N, Patel M, O'Byrne P, Satia I. Narrative review of the mechanisms and treatment of cough in asthma, cough variant asthma, and non-asthmatic eosinophilic bronchitis. Lung 2022;200:707-16. [CrossRef]
2. Ye K, Ke Y, Keshava N, Shanks J, Kapp JA, Tekmal RR, et al. Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells. Proc Natl Acad Sci U S A 1998;95:1601-6. [CrossRef]
3. Halim F, Azhar Y, Suwarman S, Hernowo B. p53 mutation as plausible predictor for endocrine resistance therapy in luminal breast cancer. F1000Res 2022;11:330. [CrossRef] [CrossRef]
4. Landen JW, Lang R, McMahon SJ, Rusan NM, Yvon AM, Adams AW, et al. Noscapine alters microtubule dynamics in living cells and inhibits the progression of melanoma. Cancer Res 2002;62:4109-14.
5. Groneberg-Kloft B, Feleszko W, Dinh QT, van Mark A, Brinkmann E, Pleimes D, et al. Analysis and evaluation of environmental tobacco smoke exposure as a risk factor for chronic cough. Cough 2007;3:6. [CrossRef]
6. Pengnam S, Plianwong S, Yingyongnarongkul BE, Patrojanasophon P, Opanasopit P. Delivery of small interfering RNAs by nanovesicles for cancer therapy. Drug Metab Pharmacokinet 2022;42:100425. [CrossRef]
7. Landen CN Jr., Goodman B, Katre AA, Steg AD, Nick AM, Stone RL, et al. Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer. Mol Cancer Ther 2010;9:3186-99. [CrossRef]
8. Jordan MA, Toso RJ, Thrower D, Wilson L. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc Natl Acad Sci U S A 1993;90:9552-6. [CrossRef]
9. Manchukonda NK, Naik PK, Santoshi S, Lopus M, Joseph S, Sridhar B, et al. Rational design, synthesis, and biological evaluation of third generation a-noscapine analogues as potent tubulin binding anti-cancer agents. PLoS One 2013;8:e77970.
10. Pattnaika A, Naikb PK. Transcriptome profiling and identification of differentially expressed genes and pathways with the treatment of N-Alkyl amine-noscapinoids in human breast cancer. Int J Res Anal Rev 2023;10:378-455.
11. Kim D, Langmead B, Salzberg SL. HISAT:A fast spliced aligner with low memory requirements. Nat Methods 2015;12:357-60. [CrossRef]
12. Liao Y, Smyth GK, Shi W. featureCounts:An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923-30. [CrossRef]
13. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290-5. [CrossRef]
14. Robinson MD, McCarthy DJ, Smyth GK. edgeR:A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. [CrossRef]
15. Sobol MS, Hoshino T, Delgado V, Futagami T, Kadooka C, Inagaki F, et al. Genome characterization of two novel deep-sea sediment fungi, Penicillium pacificagyrus sp. Nov. And Penicillium pacificasedimenti sp. Nov., From South Pacific Gyre subseafloor sediments, highlights survivability. BMC Genomics 2023;24:249. [CrossRef]
16. McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol 2013;5:a00⇐. Erratum in:Cold Spring Harb Perspect Biol 2015;7:a026716.
17. Wang X, Lin Y. Tumor necrosis factor and cancer, buddies or foes?Acta Pharmacol Sin 2008;29:1275-88.
18. Martinez I, Oliveros JC, Cuesta I, de la Barrera J, Ausina V, Casals C, et al. Apoptosis, toll-like, RIG-I-like and NOD-like receptors are pathways jointly induced by diverse respiratory bacterial and viral pathogens. Front Microbiol 2017;8:276. [CrossRef]
19. Denisenko TV, Gogvadze V, Zhivotovsky B. Mitophagy in carcinogenesis and cancer treatment. Discov Oncol 2021;12:58. [CrossRef]
20. Khan I, Steeg PS. Endocytosis:A pivotal pathway for regulating metastasis. Br J Cancer 2021;124:66-75. [CrossRef]
21. Mounetou E, Legault J, Lacroix J, Gaudreault RC. Antimitotic antitumor agents:Synthesis, structure-activity relationships, and biological characterization of N-Aryl-N'-(2-chloroethyl) ureas as new selective alkylating agents. J Med Chem 2001;44:694-702. [CrossRef]
22. Viswas RS, Pundir S, Lee H. Design and synthesis of 4-piperazinyl quinoline derived urea/thioureas for anti-breast cancer activity by a hybrid pharmacophore approach. J Enzyme Inhib Med Chem 2019;34:620-30. [CrossRef]
23. Swain SK, Samal S, Mohanty JN, Choudhury J. Nasopharyngeal carcinoma among the pediatric patients in a non-endemic region:Our experience at a tertiary care teaching hospital in Eastern India. Egypt Pediatr Assoc Gaz 2020;68:1-6.
24. Li HQ, Lv PC, Yan T, Zhu HL. Urea derivatives as anticancer agents. Anticancer Agents Med Chem 2009;9:471-80. [CrossRef]
25. El-Naggar M, Almahli H, Ibrahim HS, Eldehna WM, Abdel-Aziz HA. Pyridine-ureas as potential anticancer agents:Synthesis and in vitro biological evaluation. Molecules 2018;23:1459. [CrossRef]
26. Anderson JT, Ting AE, Boozer S, Brunden KR, Crumrine C, Danzig J, et al. identification of novel and improved antimitotic agents derived from noscapine. J Med Chem 2005;48:7096-8. [CrossRef]
27. Sajadian S, Vatankhah M, Majdzadeh M, Kouhsari SM, Ghahremani MH, Ostad SN. Cell cycle arrest and apoptogenic properties of opium alkaloids noscapine and papaverine on breast cancer stem cells. Toxicol Mech Methods 2015;25:388-95. [CrossRef]
28. Shen W, Liang B, Yin J, Li X, Cheng J. Noscapine increases the sensitivity of drug-resistant ovarian cancer cell line SKOV3/DDP to cisplatin by regulating cell cycle and activating apoptotic pathways. Cell Biochem Biophys 2015;72:203-13. [CrossRef]
29. Nagireddy PK, Kumar D, Kommalapati VK, Pedapati RK, Kojja V, Tangutur AD, et al. 9-Ethynyl noscapine induces G2/M arrest and apoptosis by disrupting tubulin polymerization in cervical cancer. Drug Dev Res 2021;83:605-14. [CrossRef]
30. Devine SM, Yong C, Amenuvegbe D, Aurelio L, Muthiah D, Pouton C, et al. Synthesis and pharmacological evaluation of noscapine-inspired 5-substituted tetrahydroisoquinolines as cytotoxic agents. J Med Chem 2018;61:8444-56. [CrossRef]
31. Rahmanian-Devin P, Baradaran Rahimi V, Jaafari MR, Golmohammadzadeh S, Sanei-Far Z, Askari VR. Noscapine, an emerging medication for different diseases:A mechanistic review. Evid Based Complement Alternat Med 2021;2021:8402517. [CrossRef]
32. Nambiar N, Nagireddy PK, Pedapati R, Kantevari S, Lopus M. Tubulin-and ROS-dependent antiproliferative mechanism of a potent analogue of noscapine, N-propargyl noscapine. Life Sci 2020;258:118238. [CrossRef]
33. Nguyen TT, Wei S, Nguyen TH, Jo Y, Zhang Y, Park W, et al. Mitochondria-associated programmed cell death as a therapeutic target for age-related disease. Exp Mol Med 2023;55:1595-619. [CrossRef]