1. INTRODUCTION
The six genera Cheiloclinium, Peritassa, Salacia, Salacighia, and Thyrosalacia Tontelea make up the Salacioideae family. Salacioideae have bisexual, three stamens, pentamerous flowers with an extrastaminal disc, and fruits which are berries with arillate mucilaginous pulp. [1]. Salacia chinensis (SC) is commonly known as Sapta chakra and S. reticulata (SR) as Ekanayakam in Kannada. It is distributed in Dry Deciduous, Evergreen Forests of the Western Ghats of Kerala, Karnataka, Tamil Nadu, and also found in forests of Andhra Pradesh, Northeast India [2]. Salacia sp. is used in Ayurvedic medicine to treat diabetes. SR, S. macrosperma (SM), S. oblonga (SO) along with other plants such as Gymnema sylvestre, Curcuma longa, Azadirachta indica, Aegle marmelos, Emblica officinalis, and natural sweetener Stevia rebaudiana is used in the preparation of the polyherbal medicines such as Diajith churna [3], Rajanyamalakadi [4], and Madhujeevan Churna [5]. Salacia species has anti-inflammatory, antimutagenic, nephroprotective, hypolipidemic, antimicrobial activity [6] antitumor, antiviral (H1N1, HIV) [7,8], and immunomodulatory activities. The Western Ghats have a wide range of climatic and topographic variability, which supports a broad range of plant types and animals. The endemism of the Western Ghats’ flora and fauna has been widely documented. The moist zones of the Western Ghats have a greater level of endemism than the dry zones [9].
The previous studies of the Salacioideae, Salacia supported monophyly [10]. Salacioideae have an Old-World origin and within New World, Salacioideae have undergone single successful radiation. In the previous study, Salacia samples from Central-South East Asia and other parts of Asia were excluded except for Kokoona which is sampled from Sri Lanka. The morphological characters and sequences from phyB, 26S, 18S, internal transcribed spacers (ITS), matK, trnL-F, atpB, and rbcL have been used to construct the phylogeny of the Celastraceae [11,12] and Salacioideae, Dev, Anoop [13] have used rbcL, matK, trnH-psbA and ITS2 region for species discriminate of Salacia samples Kerala. Genetic diversity of the four Salacia sp. SM, SC, SO, and S. fruticosa (SF) samples from the Karnataka region was analyzed using ISSR, RAPD, and ITS marker [14]. Using ITS2 sequence secondary structure the variation within the Salacia sp. sampled with the Western Ghats was assessed by Anaz, Sasidharan [15]. The current research focuses on the phylogeny and phylogeography of the subfamily Salacioideae.
2. MATERIALS AND METHODS
2.1. Sampling, DNA Extraction
SM, SC, SO, and SF samples were collected from the Western Ghats of Karnataka. The modified Stange, Prehn [16,17] method of DNA extraction was used. In this method, lithium chloride (LiCl2) and high concentration of sodium chloride (NaCl) selectively precipitates DNA eliminating the use of RNAse in the extraction procedure. Furthermore, the polyphenolic complex is removed using Polyvinylpyrrolidone (PVP). DNA extraction buffer contains two solutions A and B. Solution A (100 mM Tris-HCL [pH 8.0], 20 mM EDTA [pH 8.0], 1.4 M NaCl, 4% PVP, 2% [w/v] CTAB, 2% - mercaptoethanol/DTT) and Solution B (4M LiCl2) which are mixed in a 1:1 ratio just before the extraction. 100-200 mg of sample was weighed, thoroughly washed and rinsed with alcohol to remove any adhering dirt.
Using liquid nitrogen, samples are crushed in a pestle and mortar. The crushed samples were transferred to a 1.5 mL microfuge tube containing 700 uL of extraction buffer and shaken well. Sample tubes were incubated for 30 min at 65°C while shaking intermittently in a heating block. After cooling, 700 uL of chloroform and isoamyl alcohol (24:1) were added to the samples. The tubes were thoroughly shaken and were then centrifuged at 12,000 rpm for 10 min at room temperature. The above step is repeated once more. To sample containing tubes, an equal volume of chilled isopropyl alcohol was added and incubated in −20 refrigerators for 1 h or was kept overnight. Tubes were centrifuged at 12,000 rpm for 10 min to obtain the DNA pellet. The supernatant was decanted, and the DNA pellet was washed with 70% alcohol. The pellet was air-dried and then the DNA pellet dissolved in 20–50 ul Tris10-EDTA1 buffer.
ITS, matK, and rbcL region polymerase chain reaction (PCR), sequencing, and Homology Search were conducted as the method described by Bajpe et al. [18]. For PCR amplification ITS1-5.8s-ITS2 region, ITS4, and ITS5 primers, were used by White et al. [19], for the matK region, the primers 3FKIM and 1RKIM developed by Ki-Joong Kim were used, and for the rbcL region, the primers rbcLa-F and rbcLa-R were used Kress and Erickson [20]. ITS, matK, and rbcL sequences of Salacioideae genera Cheiloclinium, Peritassa, Salacia, and Tontelea were retrieved from the NCBI GenBank database. SM, SC, SO, and SF sequences of the Karnataka Western Ghats deposited in the GenBank repository are listed in Table 1.
Table 1: Sample code, and accession number of ITS, matK, rbcL S. chinensis, S. macrosperma, S. fruticosa, S. oblonga samples sequences deposited in NCBI GenBank database.
| S. No. | Name | Sample code | Sampling area | Accession number | ||
|---|---|---|---|---|---|---|
| ITS | matK | rbcL | ||||
| 1 | S. chinensis | SC1 | Udupi District | SC1-KF986225 | SC1-KX573059 | SC1-KX581076 |
| S. macrosperma | SM2 | Shanivarsanthe, Kodagu District. | SM2-KF860877 | SM3-KX573069 | SM3-KX581083 | |
| SM4 | Abbi falls, Madikeri District | SM4-KF881919 | ||||
| SM5 | Madikeri town, Madikeri District | SM5-KF881920 | SM5-KX573070 | SM5-KX581084 | ||
| SM6 | Bisle ghat, Hassan District | SM6-KF881921 | SM6-KX573071 | |||
| SM7 | Kigga, Chikkamagaluru District | SM7-KF881922 | SM7-KX581085 | |||
| SM8 | Somwarpet, Madikeri District | SM8-KF881923 | SM8-KX573074 | |||
| SM10 | Kigga, Chikkamagaluru District | SM10-KF894421 | ||||
| SM11 | Makuta, Madikeri District | SM11-KF894422 | SM12-KX573067 | SM12-KX581082 | ||
| 3 | S. fruticosa | SF1 | Makuta, Madikeri District | SF1-KF894424 | SF2-KX573064 | SF1-KX581081 |
| 4 | S. oblonga | SO1 | Kigga, Chikkamagaluru District | SO1-KF881913 | SO1-KX573074 | SO1-KX581088 |
ITS: Internal transcribed spacers
2.2. Phylogenetic Analyses
MEGA 5 was used to build the sequences. Using the Mega 5 software’s Model function, the model with the lowest Bayesian information criterion (BIC) was generated [21]. Bayesian inference analysis (BI) [22] and maximum likelihood (ML) [23] approaches were used to perform phylogenetic studies. A Markov chain Monte Carlo (MCMC) technique, as implemented in Mr.Bayes, version 3.2.2, was used to perform the Bayesian analysis [24]. With a sampling frequency of 1000 iterations, four parallel MCMC runs were generated for a total of 10,000,000 iterations. The first 25% of samples were eliminated due to burn-in. The graphical interface in RAxML version 1.3 was utilized to do ML tree searches and bootstrapping (BS) of the data with 1000 repetitions [25].
Using the program Sequence Matrix, ITS, matK, and rbcL sequences were concatenated (Vaidya et al. 2011). MEGA 5 was used to determine genetic divergences for the three markers using the P-distance approach. The samples that exhibited n/c in the distance analysis for concatenated sequences were eliminated. To determine whether the ITS, matK and rbcL sequences could be combined into one dataset, a partition homogeneity test (PHT) with a thousand repetitions of ITS, matk, and rbcl sequences were examined to see if the partitions were homogenous in PAUP* v4.0b1. FigTree v. 1.3 was used to view and modify the trees. The BI analysis consensus tree and the best tree of ML analysis with BS replicates were combined using TreeGraph 2 [26]. The combined tree for user-specified tree topology was analyzed using the Maximum Parsimony (MP) method to calculate tree length (TL), consistency index (CI), retention index (RI), homoplasy index (HI), and combined index (RCI) in MEGA X.
2.3. Estimation of Age
BEAST 2.1.3 was used to calculate divergence times (Drummond et al., 2012). The BEAST analysis was conducted using the GTR + G + I model with the best-fit clock model and tree prior. The most recent common ancestor (MRCA) of Salacia species was calculated using a concatenated data barcode dataset. A taxonomic set has been created. The divergence was calculated using an uncorrelated relaxed lognormal clock model. The mean previous distribution UCLD was set to normal with an initial value of 50 million years (Ma) and a standard deviation of 5 Ma prior to the analysis. The MCMC chains were run for 50 million generations, with 5000 generations being sampled. Tracer 1.6 was used to check for convergence and sufficient effective sample size (ESS) values (>200) (Rambaut et al., 2014). TreeAnnotator 2.1.2 software (available in the BEAST v.2.1.3 package; http://beast.bio.ed.ac.uk/treeannotator) was then used to create maximum clade credibility tree with median branch lengths and 95% greatest posterior density intervals on nodes, with the first 25% of trees removed as burn-in. The maximum-credibility tree and divergence dates were displayed using FigTree.
2.4. Phylogeographic Inference
Utilizing RASP v.2.0b (Reconstruct Ancestral State in Phylogenies) and Bayesian binary MCMC (BBM) analysis for the geographic inferences. The F81 model of evolution was used, and various rates of change across ancestral states were permitted to minimize any limitations on ancestral state reconstruction. For all other MCMC parameters, default values were chosen. In all analyses, the MCMC chains were run for 5x106 generations. Six regions were assigned according to the sampling and distribution ranges. A-central and South America (CSA), B= Western Ghats of India, C = Central and South-East Asia (SEA), D = Madagascar/East Africa, E=West Africa, and F=Australia.
3. RESULTS
3.1. ITS Phylogeny
To build the ITS sequences phylogenetic tree a total of 134 sequences were used. Multiple sequence alignment (MSA) produced a consensus sequence of 1029 bp, with 435 bp, 539 bp, and 452 bp conserved, variable, and parsimony informative sites, respectively. Tamura-Nei model with gamma distribution and invariable sites (T92 + G + I) had the lowest BIC values of 19637.527 and tree log-likelihood of −22756.75. The tree topology of the combined tree had a log-likelihood of −8140.22. Likewise, tree topology was analyzed using the Maximum Parsimony method had TL, CI, RI, and RCI to be 1462, 0.58 (0.55), 0.85 (0.85), and 0.49 (0.47) for all sites and parsimony-informative sites (in parentheses).
The Salacioideae subfamily was separated into two major clades in combined BI and ML trees. The phylogram was clear and free of polytomies. Clade I-old world clade (OWC) comprises Salacia samples from the Western Ghats, Africa, Madagascar, Australia, and Southeast Asia. Six groups made up the first subclade [Figure 1]. Within the OWC, subclades 1 (Western Ghats), 2 (S. Chinensis), 3 (Madagascar), and 4 (Africa) were observed. Along with SM, S. agathamaliana (SA) from Western Ghats, the cladogram contained S. beddomei (SB), SO, and S. oblonga var kakayamana (SOVK), as well as S. vellaniana (SV), S. malabarica (SMa), S. brunoniana (SBr), and S. wayanadica (SW) from Karnataka and Kerala.
 | Figure 1: Combined cladogenetic tree of maximum likelihood bootstrap consensus (above branches boot-strap values >50%) and Bayesian inference (below branches posterior probability values >0.5) of the ITS sequences of Salacioideae. [Click here to view] |
SO samples from the Western Ghats of Karnataka were nested within the SM samples from Karnataka rather than being grouped with samples from Kerala in Group, 1. In Group 1, SM samples from Karnataka were merged with those from Kerala. There was also a polytomic group in SM and SB samples from Karnataka and Kerala. SM and SV from Kerala’s Western Ghats, both of which showed strong posterior probability (PP) support values at the node. There was also a polytomic group of SM samples from Karnataka and Kerala that included SB. There were two species of SM and SV from the Western Ghats of Kerala in Group 2, both of which had high PP support values at the node. SO samples from Karnataka’s Western Ghats were nested within the SM. Kerala’s SO was placed in Group 3 with node support values of 100% (BS) and 0.59 (PP). This group included S. cf. cochinchinensis, Salacia sp. (MR-2013), and SBr. Group 4 included SA, SV, SBr, and SW, all of which exhibited excellent support values. SA and SV were nested inside group 4, while SMa and SW formed a separate group with high node and tip support values. SF samples from Karnataka and Kerala formed a robust monophyletic group in Group 5.
Subclade 2 consisted of SR, SC, S. erythrocarpa, S. polysperma, and S. disepala. This clade included all S. chinensis from Karnataka, Kerala, and Australia. S. erythrocarpa, S. disepala, and SR were members of the clade. Salacia samples from Madagascar were discovered in Subclade 3, a clade with a strong node support value. Subclade 4 included East African samples.
Clade II - New world clade (NWC) includes samples from Central and South America, as well as a disjunct sample from the Ivory Coast, S. nitida. The phylogenetic distribution matched that of the Salacioideae analysis of Coughenour, Simmons [10]. From ITS analysis, it was clear that the principal nodes have a high PP value (>0.9) and a BS of >90%. However, an average and weak support value of 50% was recorded for the internal node, as well as a strong PP support value of >0.5.
3.2. matK Phylogeny
To build the matk sequences phylogenetic tree, a total of 80 sequences were used. MSA produced a consensus sequence of 1383 bp, with 801 bp, 565 bp, and 514 bp conserved, variable, and parsimony informative sites, respectively. T92+G provided the substitution model with the lowest BIC values of 10488.262 and tree log-likelihood of −22756.75. The tree topology of the combined tree had a log-likelihood of −8140.22. Likewise, tree topology was analyzed using the Maximum Parsimony method had TL, CI, RI, and RCI to be 656, 0.93 (0.92), 0.95 (0.95), and 0.89 (0.88) for all sites and parsimony-informative sites (in parentheses).
In the matK sequencing phylogeny, there were no noteworthy clades including NWC Salacia and OWC Salacia species. Samples from several geographical zones were grouped ambiguously. The BS and PP support levels for nodes were low [Figure 2]. The samples from the Western Ghats, as well as those from Africa and Australia, were found to be discordantly positioned throughout the phylogeny. The Western Ghats samples of SM, SO, SC, SA, and SB formed polytomies and were disjunct and distributed within the tree. Only SF formed a monophyletic group with strong nodal BS and PP support values. Madagascar (S. madagascariensis, Salacia sp., S. gerrardii S. owabiensis), Africa (Salacia species and S. loloensis-SL), and central American samples (S. elliptica, S. impressifolia, S. crassifolia, S. alwynii, Cheiloclinium cognatum, S. grandifolia, S. krigsneri, S. petenensis, and S. cordata) were grouped to form a paraphyletic clade with weak and good BS and PP support values. S. lehmbachii and S. polysperma were sister taxa Peritassa laevigata and Salacia samples from Madagascar (S. bussei, S. rehmannii, and S. nitida). The samples from China and South-East Asia (SEA) were nested with those from the Western Ghats, with no different clades visible in the cladogram.
 | Figure 2: Combined cladogenetic tree of maximum likelihood bootstrap consensus (above branches boot-strap values >50%) and Bayesian inference (below branches posterior probability values >0.5) of the matK sequences of Salacioideae. [Click here to view] |
3.3. rbcL Phylogeny
To build rbcL sequences phylogenetic tree, a total of 73 sequences were used. MSA produced a consensus sequence of 733 bp, with 639 bp, 93 bp, and 30 bp conserved, variable, and parsimony informative sites, respectively. Kimura 2-parameter model with gamma distribution (K2+G) provided the best fit substitution model, with the lowest BIC values of 4968.493 and log-likelihood of - 1708.209. Likewise, tree topology was analyzed using the Maximum Parsimony method had TL, CI, RI, and RCI to be 115, 0.83 (0.62), 0.81 (0.81), and 0.68 (0.50) for all sites and parsimony-informative sites (in parentheses).
In the rbcL phylogeny, species resolution was low with very weak BS, and PP support was seen within the cladogram The samples from Karnataka and Kerala were significantly discordantly dispersed in Clade I. SM, SA, SB, SO, SV, and SMa were assigned a low BS, but a high PP. Karnataka’s Western Ghats samples Salacia species from Kerala and Africa were sister taxa to SC, SM, SO, and SF. Salacia species from Africa formed two different clades in Clades II and III, with S. macrophylla and Salacia species from Thailand and China, nested within one of the clades [Figure 3]. Clades IV and V contained samples from Kerala and were different clades of SC and SF. Tontelea micrantha, Peritassa flaviflora, and Tontelea ovalifolia formed a distinct clade (Clade VI) with good support values.
 | Figure 3: Combined cladogenetic tree of maximum likelihood bootstrap consensus (above branches boot-strap values >50%) and Bayesian inference (below branches posterior probability values >0.5) of the rbcL sequences of Salacioideae. [Click here to view] |
3.4. ITS, matK, rbcL Concatenated Sequences and BI, ML Combined Tree Phylogeny
Before constructing a phylogenetic tree, genetic divergences for the concatenated ITS + matK + rbcL were examined in Mega 5 software using the P-distance technique. In the distance analysis for concatenated sequences, samples with n/c were excluded from the dataset. Partition homogeneity test (PTH) results were p = 0.03 indicating that the DNA sequences have a consistent phylogenetic tree topology and no conflicts in the genes’ sequence dataset for the bootstrap trees. To build concatenated sequences phylogenetic tree, a total of 106 sequences were used. The total length of the concatenated sequences was 3166, with 1931 bp, 1187 bp, and 988 bp being conserved, variable, and parsimony informative sites, respectively. The best fit substitution model was T92 + G having the lowest BIC values of 34956.694 and log-likelihood of −16176.919.
The cladogram was separated into two major clades in the combined analysis, namely the OWC and the NWC, with no or very poor support at the branch nodes. Three subclades of the OWC were identified: A, B, and C [Figure 4]. Subclade A nested within the clade comprised samples from Karnataka and Kerala, as well as samples from China and South-East Asia. G1, G2, G3, and G4 are the sub-clade A groupings. G1 included samples from Karnataka’s Western Ghats, SO, and SM. SM, SB, S. gambleana, and SV from Kerala constituted a paraphyletic group. Salacia species nested inside G2 included SO, SOVK, S. brunoniana, SMa, SW, SA from Kerala and S. cf cochinchinensis from China. G3 contains samples of SF as well as SMa. Samples from Africa, SL, and Salacia sp. (PM5235) were found in the clade G4, which had high node branch support values. Both of these samples were taken in Cameroon and other geographically adjacent regions. S. polysperma (China), Salacia sp. (JH-2017-24-4063) from Borneo, and SC-JH06 (Australia) formed a paraphyletic clade in clade G5, and these species were sister taxa to Western Ghats species.
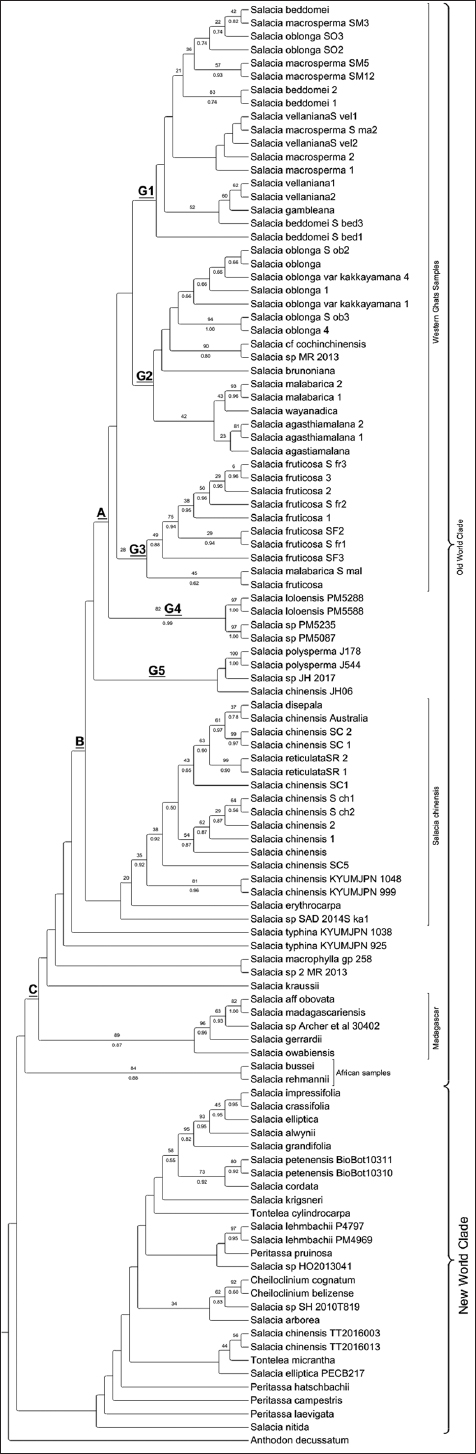 | Figure 4: Concatenated, combined cladogenetic tree of maximum likelihood bootstrap consensus (above branches boot-strap values >50%) and Bayesian inference (below branches posterior probability values >0.5) of the ITS, matK and rbcL sequences of Salacioideae. [Click here to view] |
SC from Karnataka, Kerala, and Australia was found in Subclade B, as were SR, S. disepala, and S. erythrocarpa. Salacia sp. (SAD2014S.ka1), S. macrophylla, Salacia sp. (MR2013), and S. kraussii create polytomies at the clade’s base. S. aff obovata, S. madagascariensis, Salacia sp. (Archer et al. 30402), S. gerrardii, S. owabiensis, and east Africa (Zambia and Mozambique) species S. rehmannii and S. bussei have high support values in sub-clade C. The NWC formed into a paraphyletic group that included all of the species from Central and South America. Within this primary clade, however, samples from West Africa Cameroon-S. lehmbachii, Ivory Coast-S. nitida, and SEA SC (TT2016003 and TT2016013), Thailand, Salacia sp. (HO2013041), Salacia sp. (SH-2010), and S. arborea from Indonesia were nested.
3.5. Divergence Time Analysis -Age Estimation Salacia Species
BEAST analysis had an ESS of more than 500 for all parameters. According to age estimates, the node age of old-world Salacia species and new-world Salacia species was 50.35 M.Y. All Salacia species have stem ages of 32.58 Ma. The OWC and NWC crown ages were 9.82 Ma and 17.4 Ma, respectively. A paraphyletic cluster of Salacia species from Madagascar and East Africa has a shared tip node age of 3.13 Ma within OWC. The SC (Kerala, SEA, and Australia) and SR share the same tip node age of 3.57 Ma. SC from Karnataka and S. erythrocarpa, on the other hand, shared the same tip node age of 4.29, demonstrating a divergent rate of SC divergence throughout the Western Ghats. SF had common tip node d age of 2.11 Ma indicating a similar rate of divergence within species. The NWC Salacia species has the youngest tip node age of 0.28 million years and the oldest tip node age of 11.3 million years. With a tip node age of 1.69 Ma, two China species, S. cf cochinchinensis and Salacia sp. (MR2013) were nested among new world species at the base of the clade. This basal distribution of S. cf cochinchinensis and Salacia sp. (MR2013) was not seen in the ITS, matK, rbcL, or concatenated sequences [Figure 5].
 | Figure 5: Chronogram for the sub-family Salacioideae based on ITS sequence obtained with BEAST analysis. The numbers at nodes refer to the mean age of the nodes blue bars represent 95% confidence intervals for the estimated mean dates. [Click here to view] |
3.6. Geographic Inference
Based on BEAST combined DNA barcode phylogeny and the BBM technique, geographic analysis reveals 50 dispersals and 25 vicariance episodes. According to node 216, the root node of all Salacia species evaluated in the current geographic study had an ancestral area of C-81.84: A-11.46: AC-2.36 with a probability of 0.4623, indicating that Salacia species might have originated in Asia.
The observation that node 208, which contains the majority of the NWC Salacia species represented, had the ancestral state of C-56.74: A-19.98: F-9.57: E-7.60 but a very low probability of 0.0931 in a route of C->->FE->F|E with three Dispersal, a Vicariance, and an extinction event, strengthened this observation. S. bussei (Zambia), S. rehmannii (Mozambique) had ancestral state C-40.47: E-37.31: D-9.77 with two Dispersal, a Vicariance event in the route C->EC->E|C but a low probability of 0.1631 in OWC, Madagascar, and East Africa.
Most Salacia sp. of Western Ghats of Kerala and Karnataka have the origin note 174, which has ancestral state B 66.05 C 21.44 BC 6.36 with a route of B->B^B->B|B and Probability of 0.3116. The clade containing most of SC from the Western Ghats and Australia, along with SR, S. typhina, and S. macrophylla has Indian, and Asia origin, and 16.13% of unknown origin, with a probability of 0.4196 [Figure 6].
 | Figure 6: The output of the BBM analysis from the RASP program showing ancestral geographic reconstructions at each node of the phylogenetic tree of Salacioideae. The proportion of colours in circles at each node represents possible ancestral ranges, letter close to the color are ancestral state and letters in the centers of the circles show the node number. Biogeographical regions: A: Central and South America, B: Western Ghats of India, C: Central and South/East Asia (SEA), D: Madagascar/East Africa, E: West Africa, and F: Australia. [Click here to view] |
Some of the disjunct geographic distributions were seen in east Africa Salacia species SL, S. kraussii west Africa Salacia species – S. lehmbachii, and Salacia species-PM5288, 5588 which were nested along with Western Ghats Salacia species. In the Salacia species from Asia disjunct distribution of Salacia species SH_2010T819, Salacia sp.- HO2013041, SC-JH06 was observed which was nested within the NWC. Details are listed in Table 2.
Table 2: Details of ancestral state, route, probability of some of the Salacia species.
| Node | Species | Ancestral state | Route | Probability | Event |
|---|---|---|---|---|---|
| Node-214 | S. polysperma J178&J544 | C=81.84, A=11.46 | C->C^C->C|C | 0.4623 | N/A |
| NODE 207 | S. disepala | F=41.3, C=34.75, A=15.09 | F->FC->F|C | 0.2332 | D: 2, V: 1, E: 0 |
| NODE 206 | S. chinensis JH06 | C=56.46, A=33.14 | C->CA->C|A | 0.5582 | D: 2, V: 1, E: 0 |
| NODE 183 | Peritassa laevigata, | E 49.21 A 41.15 | E->AE->A|E | 0.4921 | D: 2, V: 1, E: 0 |
| NODE 186 | S. lehmbachii P4797, | E=92.46, AE=5.87 | E->E^E->E|E | 0.9246 | N/A |
| NODE 199 | Tontelea micrantha | A=97.13 | A->AC->A|C | 0.9713 | D: 2, V: 1, E: 0 |
| NODE 202 | Cheiloclinium belizense, Salacia sp SH2010T819, | A=96.98 | A->AC->A|C | 0.9698 | D: 2, V: 1, E: 0 |
| NODE 190 | Salacia sp HO2013041 | A=96.77 | A->CA->C|A | 0.9655 | D: 2, V: 1, E: 0 |
| NODE 151 | S. loloensis PM5288 | E=89.47, BE=7.80 | E->E^E->E|E | 0.9957 | |
| NODE 127 | S. erythrocarpa, | B=47.57, F=21.12, un=31.31 | B->FB->F|B | 0.2402 | D: 2, V: 1, E: 0 |
| NODE 120 | S. chinensis Australia | C=96.88, CF=2.49 | C->FC->F|C | 0.9669 | D: 2, V: 1, E: 0 |
4. DISCUSSION
Following the previous study, the monophyly of Salacioideae has been convincingly established (Coughenour et al. 2010). Salacia species from Central and SEA, the Western Ghats, and East Africa were included in this study to present a more complete overview of species diversity. Even though the Salacioideae have been determined to be monophyletic, the disjunct distribution of Salacia sp. throughout new and OWCs indicates that the genera have undergone substantial divergence. ITS sequences generated a well-resolved, high-support value phylogenetic tree than matK and rbcL sequences among the three DNA barcodes evaluated. Furthermore, ITS sequences have 59.59% efficiency in comparison matK and rbcL sequences have a substantially lower potential for species discrimination [27]. Furthermore, the combination of the ITS + matK and ITS + rbcL has stronger species discrimination capacity, although the ITS sequence alone was able to identify between Salacia sp. when the TaxonDNA tool was used. Anaz, Sasidharan [15] confirm the current finding by analyzing delimited ITS2 sequences utilizing a neighbor-joining tree and ITS2 secondary structure.
The distribution of species in the Western Ghats demonstrates the relic character of the once-continuous distribution of flora and fauna [9]. Various authors have proposed three theories to explain the distribution and dissemination of flora and wildlife in the Western Ghats: convergence, dispersal, and vicariance [28]. The vicariance theories explain the disjunct distribution of SEA samples and monophyly among Salacia samples from the Western Ghats. The geographical distributions of SF and SC have not been changed, as samples from diverse locations have formed a monophyletic group. We hypothesise that sympatric speciation resulted in the evolution of the SM, SO, SB, SMa, SA, SOVK, and SV of Karnataka and Kerala. Based on the evolutionary study and physical characteristics, it is clear that SF is endemic to the Western Ghats. According to Karanth [29], spurious disjuncts are a result of incorrect identification and are not actual sister taxa. Because ambiguous samples were deleted from the sequence matrix, incorrect identification can be ruled out. The samples from Madagascar and Africa are indisputably well-resolved in the OWC. S. rehmanii is the sister species of the NWC and African/Madagascar samples. Salacia sp. from East Africa is related to Madagascar, SC, and Western Ghats samples. S. cf. cochinchinensis and Salacia sp. (China) form polytomies nested inside SEA and Western Ghats samples, implying that disjunction arose owing to convergence.
The estimated age of the Salaciodiaea is based on a geographic examination of the Celastraceae samples from Madagascar (Bacon, Simmons [30] and the crown age of Celastral as inferred by Magallón and Castillo [31] which is 71.6 Ma. Yoder and Nowak [32] estimated Celastraceae Madagascar lineages to be 69.6–1.7 Ma. Madagascar Salacia sp. has stem node age and crown node age to be 4.7 (7.1-2.8) Ma and 3.7 (5.7-1.9) Ma, respectively. However, in the current study, we estimated the crown age of the Madagascar clade to be 4.63 Ma which is higher than the latter studies. S. rehmanii and S. nitida, which were previously nested inside the NWC Salacia sp. were nested alongside the Madagascar Salacia sp. and had tip ages of 1.81 and 1.12 Ma, respectively. Salacia sp. (HO2013041, JH-2017, and SH2010T819) and S. lehmbachii, together with S. arborea, were still nested with Salacia samples from NWC, as previously found in the combined barcode sequence analysis. Furthermore, the Salacioideae divergence period coincides to the estimated divergence time of Asian-North American disjunct lineages, which is between 3 and 25 Ma (Wen et al. 2010).
Salaciodiaea’s ancestral state shows that Salacia sp. spread is due to long-distance dispersion episodes followed by vicariance between Asia and America. The presence of a significant ancestral state of SEA in the NWC suggests a long-distance dispersion event of Salacia sp., followed by allopathic species and radiation among Salacia sp. in Central and South America. Long-distance dispersion might result in the distribution and dissemination of samples S. arborea, Salacia sp. HO2013041, JH-2017, SH2010T819, and S. lehmbachii from West Africa. The disjunct distribution of SEA Salacia sp. among NWC species might be owing to geographic disjunctions between Eastern Asia and North America [33], and S. lehmbachii and S. nitida nesting among NWC species could be due to a vicariance event.
5. CONCLUSION
Salacia species can be misidentified with the genera of sub-family Hippocretoideae. The current study is an attempt to understand the endemism of Salacia sp. of Western Ghats using DNA barcodes. Salacia sp. from the distinct geographic area were discordantly distributed within the phylogenetic tree indicating a high level of sequence similarity. The convergence of the Western Ghats indicated homoplastic speciation of Salacia species. SOVB and SO could be Plesiomorphic species and SC and SR are the same species. Age estimation is a crucial aspect to understand the endemism of a particular geographic region. Salacia species serve as Paleoendemic species of Western Ghats which in turn give clues to the dynamics of the habitat of Western Ghats and its fragile ecosystems. Furthermore, the geographic analysis reveals the origin of Salacia species in Asia. Western Ghats and the forest of SEA are relict for several important flora. Therefore, it is essential to conserve the habitat of the Western Ghats which is one of the significant biodiversity hot spots of the world.
6. AUTHORS’ CONTRIBUTIONS
SNB and KKM carried out the molecular studies, sequence alignment, and analysis and drafted the manuscript. ASB, MG, RR and KRM helped in reviewing and corrections of Manuscript. KRK conceived the study, participated in its design, coordination, and supervised the current research work.
7. FUNDING
There is no funding to report.
8. CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
9. ETHICAL APPROVALS
This article does not contain any studies conducted on human or animal subjects.
10. DATA AVAILABILITY
The original contributions presented in the study are included in the article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Simmons MP, Hedin JP. Relationships and morphological character change among genera of Celastraceae sensu lato (including Hippocrateaceae). Ann Missouri Bot Gard 1999;86:723-57. [CrossRef]
2. Paarakh PM, Patil LJ, Thanga SA. Genus Salacia:Acomprehensive review. J Nat Remed 2008;8:116-31.
3. Rajalakshmy MR, Maniyan AK, Sruthi CV, Sindhu A. Standardisation of a polyherbal anti-diabetic Ayurvedic medicine Diajith. Int J Ayur Allied Sci 2014;3:186-94.
4. Blesson J, Narayanan S, Murali S. In vitro studies on the anti-oxidant property of rajanyamalakadi:An ayurvedic patent medicine. World J Pharm Pharm Sci 2015;4:1431-42.
5. Salunkhe V, Wachasundar N. Clinical evaluation of antidiabetic polyherbal Ayurvedic formulation-Madhujeevan Churna using Stevia rebaudiana. Indian J Pharm Pract 2009;2:58-65.
6. Chawla A, Singh S, Sharma AK. Salacia oblonga wall:A review on its pharmacognostic, phytochemical and pharmacological aspects. Int J Pharm Bio Med Sci 2013;4:1215-28.
7. John KM, Enkhtaivan G, Ayyanar M, Jin K, Yeon JB, Kim DH. Screening of ethnic medicinal plants of South India against influenza (H1N1) and their antioxidant activity. Saudi J Biol Sci 2015;22:191-7. [CrossRef]
8. Narayan C, Rai RV, Tewtrakul S. A screening strategy for selection of anti-HIV-1 integrase and anti-HIV-1 protease inhibitors from extracts of Indian medicinal plants. Int J Phytomed 2011;3:312.
9. Prasad V, Farooqui A, Tripathi S, Garg R, Thakur B. Evidence of late Palaeocene-early Eocene equatorial rain forest refugia in Southern Western Ghats, India. J Biosci 2009;34:777-97. [CrossRef]
10. Coughenour JM, Simmons MP, Lombardi JA, Cappa JJ. Phylogeny of Celastraceae subfamily Salacioideae and tribe Lophopetaleae inferred from morphological characters and nuclear and plastid genes. Syst Bot 2010;35:358-67. [CrossRef]
11. Simmons MP, Clevinger CC, Savolainen V, Archer RH, Mathews S, Doyle JJ. Phylogeny of the Celastraceae inferred from phytochrome B gene sequence and morphology. Am J Bot 2001;88:313-25. [CrossRef]
12. Simmons MP, Savolainen V, Clevinger CC, Archer RH, Davis JI. Phylogeny of the Celastraceae inferred from 26S nuclear ribosomal DNA, phytochrome B, rbcL, atpB, and morphology. Mol Phylogenet Evol 2001;19:353-66. [CrossRef]
13. Dev SA, Anoop B, Anoja K, Udayan P, Muralidharan E. Species discrimination through DNA barcoding in the genus Salacia of the Western Ghats in India. Nordic J Bot 2015;33:722-8. [CrossRef]
14. Bajpe SN, Bharathi TR, Marulasiddaswamy KM, Kumara KK, Prakash HS, Kini RK. Efficiency of RAPD, ISSR and ITS markers in detecting genetic variability among Salacia species sampled from the Western Ghats of Karnataka. Mol Biol Rep 2018;45:931-41. [CrossRef]
15. Anaz M, Sasidharan N, Remakanthan A, Dilsha M. ITS 2 and RNA secondary structure-based analysis reveals a clear picture on phylogeny of South Indian Salacia spp. Comput Biol Chem 2021;91:107438-47. [CrossRef]
16. Stange C, Prehn D, Arce-Johnson P. Isolation of Pinus radiata genomic DNA suitable for RAPD analysis. Plant Mol Biol Rep 1998;16:366. [CrossRef]
17. Ginwal H, Mittal N. An efficient genomic DNA isolation protocol for RAPD and SSR analysis in Acorus calamus L. Indian J Biotechnol 2010;9:213-6.
18. Bajpe SN, Marulasiddaswamy K, Ramu R, Badiger AS, Rudrappa MK, Kini RK. Assessing DNA barcodes species discriminating ability and phylogenetic relation within Embelia species. J Appl Biol Biotechnol 2022;10:2-7. [CrossRef]
19. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In:PCR Protocols:A Guide to Methods and Applications. Vol. 18. United States:Academic Press Inc.;1990. 315-22. [CrossRef]
20. Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants:The coding rbcL gene complements the non-coding trnH-psbAspacer region. PLoS One 2007;2:e508. [CrossRef]
21. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5:Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731-9. [CrossRef]
22. Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 2001;294:2310-4. [CrossRef]
23. Thorne JL, Kishino H, Felsenstein J. An evolutionary model for maximum likelihood alignment of DNA sequences. J Mol Evol 1991;33:114-24. [CrossRef]
24. Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2:Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012;61:539-42. [CrossRef]
25. Silvestro D, Michalak I. raxmlGUI:A graphical front-end for RAxML. Organ Divers Evol 2012;12:335-7. [CrossRef]
26. Stöver BC, Müller KF. TreeGraph 2:Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 2010;11:7. [CrossRef]
27. Badiger AS, Rudrappa MK, Bajpe SN. Assessing genetic diversity and DNA barcodes efficiency of Salacia and related species from Western Ghats, Karnataka. J Appl Biol Biotechnol 2021;9:6-4.
28. Biswas S, Pawar SS. Phylogenetic tests of distribution patterns in South Asia:Towards an integrative approach. J Biosci 2006;31:95-113. [CrossRef]
29. Karanth KP. Evolution of disjunct distributions among wet-zone species of the Indian subcontinent:Testing various hypotheses using a phylogenetic approach. Curr Sci 2003;85:1276-83.
30. Bacon CD, Simmons MP, Archer RH, Zhao LC, Andriantiana J. Biogeography of the Malagasy Celastraceae:Multiple independent origins followed by widespread dispersal of genera from Madagascar. Mol Phylogenet Evol 2016;94:365-82. [CrossRef]
31. Magallón S, Castillo A. Angiosperm diversification through time. Am J Bot 2009;96:349-65. [CrossRef]
32. Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in Madagascar?Only time will tell. Ann Rev Ecol Evol Syst 2006;37:405-31. [CrossRef]
33. Wen J. Evolution of Eastern Asian and Eastern North American disjunct distributions in flowering plants. Ann Rev Ecol Syst 1999;30:421-55. [CrossRef]