1. INTRODUCTION
The emergence of new infectious microorganisms and the global crisis in antibiotic resistance has necessitated the need for the identification of novel antimicrobials and therapies [1]. In that perspective, mycocins that are secreted from killer yeasts are considered a potential alternative to antibiotics, as they present minimal toxicity and resistance [2]. Mycocins are extracellular glycoproteins produced by certain yeasts and fungi and act as potential biocontrol agents against diverse pathogenic organisms. The fact that they are not harmful to humans has caught the attention of academicians and industrialists in recent decades [3]. Mycocins were first identified in Saccharomyces cerevisiae strains by Makower and Bevan [4] and the organisms producing mycocins or killer toxins are also known as killer yeasts. Apart from S. cerevisiae, the killer phenomenon is identified in Cryptococcus, Candida, Debaryomyces, Kluyveromyces, Hanseniaspora, Hansenula, Pichia, Metschnikowia, Ustilago, Torulopsis, Zygosaccharomyces, and Williopsis implying that the killer phenomenon is indeed extensive among yeasts [5]. The attributes like the broad spectrum of antagonistic properties and no resistance development have made mycocin and mycocin-producing organisms benefit with potential applications in many sectors. They are used as starter cultures and flavor enhancers in wineries [6,7], food preservatives [8], probiotics [9,10], and post-harvest biocontrol agents [11]. The killer yeasts have been isolated from diverse habitats like fruits, cereals, fermented vegetables, leaves, flower petals, soil, forest, and marine habitats [12-16].
Psidium gujava is a familiar fruit in tropical and subtropical regions. It is commonly known as guava. India was the leading producer of guava (17,650,000 Metric Tons) in the year 2018–2019 [17]. The fruit is consumed in raw form, juice, jellies, and jams. The plant and its parts have a long history of usage in the treatment of diarrhea, dysentery, hypertension, caries, and pain relief treatments. The Guava fruit used to isolate killer yeasts can be further explored for their antagonistic and biocontrol qualities.
2. MATERIALS AND METHODS
2.1. Media, Chemicals, and Strains
The killer sensitive (MTCC 473) and the killer positive (MTCC 475) strains of S. cerevisiae were procured from Microbial type culture collection (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh, India. The media components such as yeast extract, malt extract, peptone, yeast nitrogen base, and dextrose were purchased from HiMedia Laboratories Pvt. Limited, India. The carbohydrate discs were obtained from Microexpress, India. Primers were purchased from Integrated DNA Technologies, Singapore. DNA ladder was purchased from Takara, India. All other chemicals were purchased from HiMedia Laboratories Pvt. Limited, India unless specifically mentioned.
2.2. Isolation of Yeasts from P. gujava
The P. gujava commonly known as Guava was brought from the local market at Davangere, Karnataka, India. The fruits were cut into small pieces and 25 g were resuspended individually in sterile solutions of 2%, 5%, and 9% sodium chloride (250 mL). The flasks were incubated at room temperature (25–28°C) for 10 days. Later, the resuspension was serially diluted (1:10000 and 1:100000 times) and plated on Yeast extract-Peptone-Dextrose (YPD) agar media in duplicates. The plates were incubated at 25°C for 24–48 h. The obtained unique colonies were saved for further analysis.
2.3. Determination of Killer Activity in the Isolated Yeasts
The obtained yeast colonies were screened for the production of mycocins by killer-zone assay [12]. The killer sensitive S. cerevisiae (MTCC 473), the killer positive S. cerevisiae (MTCC 475), and the test strains were grown in YPD broth for 24 h at 25°C, 180 rpm. Sterile YPD plates with 0.03% methylene blue were prepared and swab inoculated with MTCC 473 cells at a concentration of 0.05 in the optical density scale at 600 nm wavelength. The test strains at a concentration of 0.05 O.D. at 600 nm were inoculated as a line on the swabbed plates. MTCC 475 was used as a positive control. Only MTCC 473 swabbed plate and un-inoculated plate were, respectively, used as negative and media controls. Thoroughly dried plates were incubated at 25°C for 48–72 h. The test was performed in duplicates.
2.4. Morphological and Microscopic Observations of the Isolated Yeasts
The test isolates were grown in YPD broth and agar at 25°C for 24–48 h. The colony characteristics of the isolates on YPD agar were noted. The culture broth was used to record the microscopic characteristics of the test isolates viewed through the bright field microscope after staining with lactophenol cotton blue.
2.5. Germ Tube Test
The germ tube test for all the isolates was performed by the standard method as in the guidelines of fungal identification given by the Indian Council of Medical Research [18]. A drop of the serum suspension from each sample was viewed through the microscope. The presence of germ tubes in more than 30% of the cells seen on the microscopic field was considered to be a positive result in the test.
2.6. Ascospore and Hyphae Formation
The test isolates were streaked on 5% malt extract agar (MEA) and incubated at 25°C for 2–5 days. After incubation, the cultures were stained with lactophenol cotton blue and observed through a microscope for the presence of ascospores [19]. The formation of hyphae was also examined in test isolates using the Dalmau plate assay method [20].
2.7. Biochemical Tests for the Isolates
2.7.1. Urease test
A loopful of freshly grown test isolate (24 h old culture on solid medium) was inoculated into Stuart’s urea broth in duplicates. The inoculated broth was incubated at 37°C for 4 h. The cultures were observed for color change.
2.7.2. Nitrogen assimilation test
The test isolates were grown in YPD broth at 25°C, 180 rpm for 24 h. The yeast carbon base media was mixed individually with ammonium sulfate, potassium nitrate, and sodium nitrite along with 2% agar, sterilized, and poured into appropriately labeled Petri plates in duplicates. The freshly grown test cultures were starved in 0.9% saline for 3 h and 0.05 O.D. of each test culture was spotted on the labeled Petri plates. The plates were incubated at 25°C for 24 h and observed for growth.
2.7.3. Sugar fermentation test
The freshly grown liquid cultures of test isolates were inoculated into sterile peptone water containing sugar discs and Durham’s tube in duplicates. Phenol red (0.01%) was used in the media to observe the color change. Each of the isolates was tested with dextrose, galactose, maltose, lactose, trehalose, cellobiose, and raffinose. The inoculated cultures were incubated at 25°C for 5–7 days. The color change and gas formation were noted.
2.8. Molecular Identification of the Isolates
The genomic DNA of the test isolates was extracted and purified according to CSH protocols [21]. Purified DNA from each of the test isolates was amplified using NS1 (5’-GTAGTCATATGCTTGTCTC-3’) and NS4 (5’-CTTCCGTCAATTCCTTTAAG-3’) primers that are specific to nuclear ribosomal small subunit (SSU) 18s rDNA. The amplified products were run on 1% agarose gel and visualized under a ultraviolet transilluminator. Further, the polymerase chain reaction (PCR) products were gel extracted and purified using Nucleospin gel and PCR clean-up kits. The amplified products were Sanger sequenced using NS1 and NS4 primers. The sequences obtained for NS1 and NS4 primers were assembled using the SnapGene tool. The aligned sequences of each isolate were identified by comparison to nucleotide databases using the basic local alignment search tool (BLAST) from National Centre for biotechnology information (NCBI) with the default parameters. Further, the sequences of all the isolates G1–G6 were submitted to NCBI GenBank.
3. RESULTS AND DISCUSSION
Killer yeasts are natural antimicrobial agents that are utilized for winemaking, food and feed protection, silage treatment, diarrheal treatment, and so on [22]. Many of the so-described killer yeasts are found to be halotolerant and the killer activity is increased with the increase in salt concentration [23]. In recent years, halotolerant yeasts have gained importance because of their potential applications in antimicrobials, food processing, biofuel production, hydrolase production, bioremediation, and other industrial processes [24,25]. They are mostly isolated from marine environments, fermented foods, wastewater, and industrial settings. Therefore, in addition to the killer activity, halotolerant yeasts can be exploited for other prospective applications mentioned above. The present study is the first report on the isolation of halotolerant and killer yeasts from P. gujava, a tropical fruit used in making jams, juices, and jellies. The isolated yeasts were screened for killer activity, characterized by morphological, microscopic, and biochemical tests. It was further identified at the species level by using appropriate molecular tools. The details are as follows.
3.1. Isolation of Yeasts from P. gujava
The sterile saline at 2%, 5%, and 9% was used to isolate killer yeasts from Guava fruits. The mycelial fungus showed its growth in 2% saline suspension and therefore it was not considered for further studies. The other solutions were serially diluted and plated on a YPD agar medium. On average, 14X107 CFU/mL and 16.6X106 CFU/mL colony counts were observed, respectively, in 5% and 9% saline solutions, which mostly belonged to the division Ascomycota. A total of 22 colonies from both solutions were saved for further screening of killer yeasts.
3.2. Killer Activity Determination
The mycocin produced by killer yeast shows a potential antagonistic effect on sensitive yeast [3]. Hence, this property was exploited to screen the obtained isolates. The killer activity of the isolated yeast was determined using methylene blue dye. The isolates possessing killer activity had a blue halo zone around them. Methylene blue, a cationic dye stains negatively charged parts of the cells such as the nucleus and cytoplasm. In the live cells, methylene blue is reduced and therefore they do not take up this stain, whereas, the dead cells cannot reduce methylene blue and therefore stains blue [26,27]. Out of the twenty-two isolates, six of them showed blue halo zones around them [Figure 1]. These six isolates were considered to possess killer activity and were labeled G1, G2, G3, G4, G5, and G6. G1 and G2 were the isolates from 5% sodium chloride suspension. G3, G4, G5, and G6 were the isolates from 9% sodium chloride suspension. All six isolates were taken forward for further characterization.
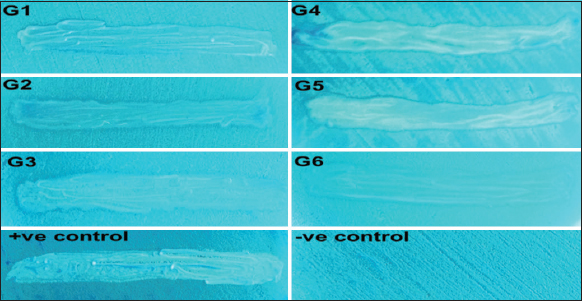 | Figure 1: Killer-zone assay. G1?G6: The test isolates showing killer activity.: The blue halo zone around the test isolates is indicative of the killer activity in test isolates. [Click here to view] |
3.3. Morphologic and Microscopic Observations of the Isolated Killer Yeast
The colony characteristics of the killer yeast isolates on YPD agar were well defined and noted as given in Table 1. All six isolates were yeast-like with raised elevation, mostly spherical. G3, G4, G5, and G6 showed two types of colony morphology at the different time courses of growth [Table 1]. They were observed to be smooth, glistening with entire margins at their younger stage whereas at a later stage of growth the margins became scalloped. This phenomenon is commonly found in S. cerevisiae [28].
Table 1: Summary of the colony characteristics of the killer yeast isolates grown on YPD agar medium, observed after 48 h of incubation at 25°C.
| S. No. | Isolate no. | Pictures of the colony on agar surface | Colony morphology | |||
|---|---|---|---|---|---|---|
| Appearance | Elevation | Margin | Surface | |||
| 01. | G1 |  | Circular | Raised | Entire | Smooth, glistening |
| 02. | G2 |  | Circular | Raised | Entire | Smooth, glistening |
| 03. | G3 |  | Spherical | Raised | Scalloped | Butyrous, wrinkled |
| 04. | G4 |  | Spherical | Raised | Entire or scalloped | Smooth, glistening or butyrous, wrinkled |
| 05. | G5 |  | Spherical | Raised | Entire or scalloped | Smooth, glistening or butyrous, wrinkled |
| 06. | G6 |  | Spherical | Raised | Entire or scalloped | Smooth, glistening or butyrous, wrinkled |
The 24 h old cultures of test isolates grown in YPD broth were observed through the bright field microscope with 400 X magnification [Figure 2]. G1 and G2 appeared spherical and sometimes in the group, whereas G3–G6 isolates were elongated and mostly visualized in aggregates. The microscopic characters confirmed that they were a pure culture of yeast. The isolates G3–G6 were mostly observed in aggregates. The ability to form aggregates plays a determining role in the sexual reproduction, colonization, and pathogenesis of microorganisms [29]. Therefore, the observed aggregation capacity suggests that these probiotic yeasts will be capable of colonizing in the gastrointestinal area of mammals. Further experiments are required to explore their bio-film formation capabilities. This observation categorized the isolates into two morphologically similar groups - the first one with G1 and G2, the second one with G3, G4, G5, and G6.
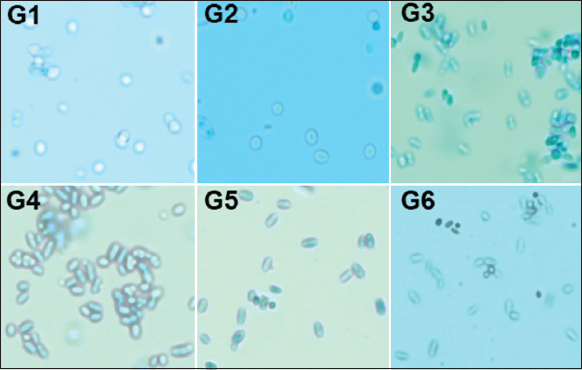 | Figure 2: Microscopic observations. G1?G6 are the microscopic fields showing respective isolates at 400× magnification. [Click here to view] |
3.4. Germ Tube Test
A germ tube test was performed with human serum as described in the materials and methods section. Fortunately, none of the test isolates showed the presence of germ tubes even after 4 h of incubation at 37°C [Figure 3]. This indicated none of them were Candida albicans or Candida dubliniensis. The germ tube test is considered the presumptive test in the identification of C. albicans. It is the most affordable and easy-to-do diagnostic assay for the discrimination of C. albicans and C. dubliniensis from others [30,31]. However, utmost care was taken while handling the isolates during the experiments.
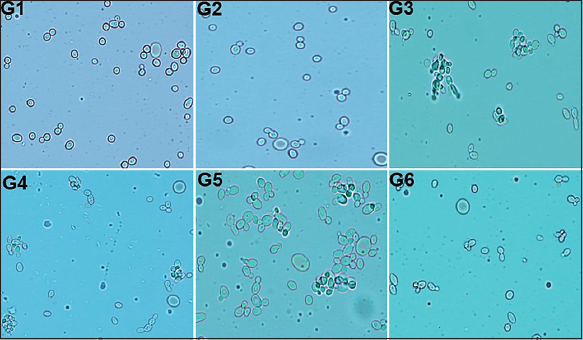 | Figure 3: Results of germ tube test. G1?G6 shows microscopic observations of the respective test isolates after their incubation with human serum at 37°C for 4 h. [Click here to view] |
3.5. Ascospores and Hypha Formation in the Isolated Killer Yeast
Spores are generally formed to survive extreme conditions of heat, nutrition deficiency, or any other physiological and biological stress. Ascospore formation is a defining feature of the phylum Ascomycota. They are the gametic stage of the life cycle produced by meiotic cell division [32,33]. The microscopic observation of isolates grown on MEA plates disclosed that the G3, G4, G5, and G6 isolates possess two to four evanescent ascospores [Figure 4]. However, the isolates G1 and G2 showed only budding but not ascospore formation.
 | Figure 4: Ascospore formation. G1?G6: Lactophenol cotton blue stained cells picked from MEA plates. Arrow marks indicate the presence of ascospores. [Click here to view] |
The change in the environmental conditions can trigger the morphological characteristics of fungi from unicellular to multicellular (hyphal) growth [34,35]. The Dalmau plate technique provided an anaerobic environment locally inside the Petri plate. This helped us to visualize the dimorphic changes in the isolated yeast. All the yeast isolates G1–G6 expressed pseudohypha which was observed microscopically [Figure 5]. G1 and G2 were also seen to produce blastospores.
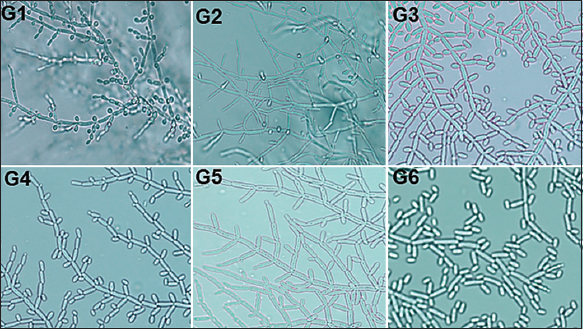 | Figure 5: Results of Dalmau plate technique. Microscopic view of the isolates G1 to G6 plated using Dalmau plate technique. [Click here to view] |
The formation of pseudohyphae with blastospores and the absence of ascospore formation in G1 and G2 indicated that they do not belong to Saccharomyces genera. The presence of evanescent ascospores and pseudohyphae formation in G3, G4, G5, and G6 indicated that they do not belong to either Candida or Saccharomyces genera [36,37].
3.6. Biochemical Tests
The traditional way of organism identification is by the conduction of biochemical tests. The sole carbon utilization test, nitrogen assimilation test, and metabolic product conversion test are some of the conventional experiments to identify and classify microorganisms [38-40]. In our study, we examined and characterized our isolates by urease test, sugar fermentation tests, and nitrogen utilization tests. The presence of urease in microbial pathogens facilitates the breakdown of urea into ammonia molecules, thereby causing toxicity in human cells [41]. Therefore, the urease test aids in the presumptive identification of toxic yeast such as Cryptococcus neoformans, Coccidioides posadasii, Rhodotorula, Trichosporon, and Candida krusei [41-43]. The biochemical test for the detection of urease enzyme in the isolates depicted that urease is not present in any of the isolates [Table 2 and Supplementary Figure 1]. Therefore, all the isolates seemed to be avirulent [44].
Table 2: Results of the biochemical tests conducted for the isolated yeasts G1–G6.
| Name of the isolate | Urease test | Nitrogen assimilation | Carbohydrate fermentation test | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ammonium sulfate | Potassium nitrate | Sodium nitrite | Glucose | Galactose | Maltose | Lactose | Trehalose | Cellobiose | Raffinose | |||||||||
| A | F | A | F | A | F | A | F | A | F | A | F | A | F | |||||
| G1 | ? | + | w | ? | + | ? | + | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| G2 | ? | + | w | ? | + | ? | + | + | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| G3 | ? | + | ? | ? | + | w | + | w | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| G4 | ? | + | ? | ? | + | ? | + | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| G5 | ? | + | ? | ? | + | + | + | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| G6 | ? | + | w | ? | + | + | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
+positive, ? negative, w: Weakly positive, A: Assimilation, F: Fermentation.
Further, all the isolates were able to utilize a nitrogen source in the form of ammonium sulfate, but only G1, G2, and G6 were able to grow poorly on potassium nitrate as the sole nitrogen source. No isolate was able to use nitrogen in the form of nitrite [Table 2 and Supplementary Figure 2].
All the isolates were tested for the assimilation and fermentation of glucose, galactose, maltose, lactose, trehalose, cellobiose, and raffinose. Only the isolates G5 and G6 were able to assimilate and ferment glucose. The other isolates were able to assimilate but unable to ferment glucose. Although the isolates utilized galactose, they were not able to ferment these sugars except for G2. Galactose is an epimer of glucose which is usually not utilized by yeast, in presence of glucose because of catabolite repression. In our study, we observed that the isolates G1 and G2, which are found to be C. orthopsilosis strains, utilize galactose as the sole carbon source. G2 was able to ferment galactose poorly [Supplementary Figure 3b]. The Pichia fermentans isolates G3, G4 and G5 were able to assimilate galactose, which is not a typical characteristic of these species [45], but the G6 isolate was negative to galactose assimilation. The other sugars namely maltose, lactose, trehalose, cellobiose, and raffinose could not be utilized by the isolates G1, G2, G3, G4, G5, and G6 [Table 2 and Supplementary Figure 3].
3.7. Molecular Identification of the Killer Yeast Isolates
The advancements in the availability of molecular tools have aided in the quick identification of microorganisms at the species level [46,47]. 18s rRNA region is conserved in eukaryotes and therefore, we used NS1 and NS4 primers specific to this SSU on the ribosome for molecular identification of the yeast isolates [46,48]. The genomic DNA extracted from the yeast isolates was purified and run on 0.8% agarose gel to verify its purity and integrity [Figure 6a]. The PCR with the extracted genomic DNA was visualized on the agarose gel [Figure 6b]. All the amplified products were found to have a band size around 1200 bp. They were purified and further sequenced by Sanger sequencing. The obtained sequences were analyzed using BLAST from NCBI as described in materials and methods.
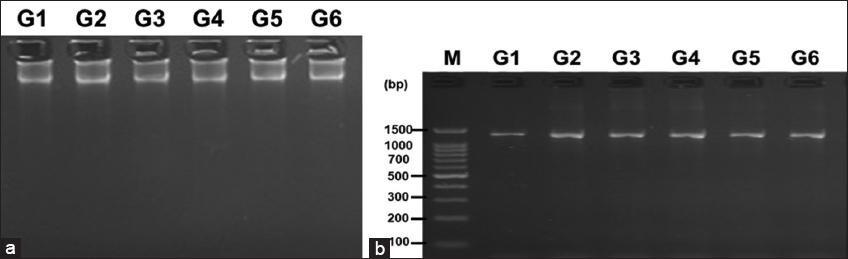 | Figure 6: Gel pictures of (a) DNA extraction from the yeast isolates. Lanes G1 – G6 are DNA obtained from the respective yeast isolates. (b) PCR amplified products, Lane M – 100 bp DNA ladder, Lanes G1 – G6 are the PCR products obtained from the respective yeast isolates. [Click here to view] |
The summary of the BLAST results obtained for the sequences from respective isolates is given in Table 3. Further, each of the partial 18s rDNA sequences was submitted to NCBI Genbank with accession numbers given in Table 3. BLAST analysis revealed G1 and G2 to be C. orthopsilosis. G3, G4, G5, and G6 isolates were found to be P. fermentans. This data were also supported by phylogenetic tree analysis, wherein P. fermentans and C. orthopsilosis isolate formed two different clades altogether [Supplementary Figure 4]. Though the BLAST analysis of the G3, G4, G5, and G6 isolates showed more than 99% similarity to P. fermentans, Pichia kluyveri, Pichia nakasei, and Pichia eremophilia sequences, the phylogenetic tree analysis confirmed it is more closely related to the P. fermentans sequence with accession number DQ489318.1. Overall, the characteristics exhibited by each isolate during morphological, microscopic, biochemical, and molecular examinations were in accordance and matched with attributes of the type strain. Further, the isolates are highly halotolerant and probable mycocin-producers. Therefore, they can be exploited for their antimicrobial and industrial applications.
Table 3: Summary of BLAST results.
| S. No | Isolate ID | Scientific name | Accession no. of the similar species | E-value from BLAST | Percent identity | Genbank accession no. allotted to the isolate |
|---|---|---|---|---|---|---|
| 1. | G1 | Candida orthopsilosis | MN268777.1 | 0.0 | 99.16 | ON422235.1 |
| 2. | G2 | Candida orthopsilosis | MN268777.1 | 0.0 | 99.72 | ON422248.1 |
| 3. | G3 | Pichia fermentans | DQ489318.1 | 0.0 | 99.81 | ON422268.1 |
| 4. | G4 | Pichia fermentans | DQ489318.1 | 0.0 | 99.44 | ON422270.1 |
| 5. | G5 | Pichia fermentans | DQ489318.1 | 0.0 | 99.44 | ON422278.1 |
| 6. | G6 | Pichia fermentans | DQ489318.1 | 0.0 | 99.53 | ON422279.1 |
4. CONCLUSIONS
Killer yeasts are prioritized in recent years because of their biocontrol capabilities. They are looked upon as an alternative to antibiotic therapy because of their “no resistance development” phenomenon. The isolation of killer yeast from P. gujava, a tropical fruit has a fortifying effect on the fruit itself and the delicacies prepared using them. The isolated yeasts from the salt suspensions of P. gujava were characterized by morphologic, microscopic, biochemical, and molecular methods. The tests reveal that yeast isolates majorly consist of P. fermentans species, which are not clinically pathogenic. Therefore, further studies on mycocin extraction from P. fermentans species and evaluation of its antagonistic activities will promote their commercial usage as a potential biocontrol agent.
5. ACKNOWLEDGMENTS
The authors acknowledge the facility given at the Institution of Excellence (IOE), Vijnana Bhavana, Manasagangothri, University of Mysore, Mysuru. Srividya D. is thankful to the Karnataka Science and Technology Promotion Society (KSTePs) for the award of the DST-Ph.D fellowship.
6. AUTHOR CONTRIBUTIONS
Seema J. Patel conceptualized and designed the experiments. Srividya D. executed the experiments and wrote the first draft manuscript. Srividya D. and Anil H. Shyam Mohan analyzed the data and all the authors interpreted the data, commented on previous versions of the manuscript, and read and approved the final manuscript.
7. FUNDING
Srividya D. has received Karnataka DST-Ph.D. fellowship award numbered DST/KSTePS/Ph.D. Fellowship/LIF-11 from the Department of Science and Technology, Government of Karnataka supporting the financial need of the study.
8. CONFLICT OF INTEREST
The authors declare that they have no competing interests.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. National Research Council (US) Committee on New Directions in the Study of Antimicrobial Therapeutics:New Classes of Antimicrobials;National Research Council (US) Committee on New Directions in the Study of Antimicrobial Therapeutics:Immunomodulation. Treating Infectious Diseases in a Microbial World:Report of Two Workshops on Novel Antimicrobial Therapeutics. Washington, DC:National Academies Press (US);2006.
2. Muccilli S, Restuccia C. Bioprotective role of yeasts. Microorganisms 2015;3:588-611. [CrossRef]
3. Nascimento BL, Delabeneta MF, Rosseto LR, Junges DS, Paris AP, Persel C, et al. Yeast mycocins:A great potential for application in health. FEMS Yeast Res 2020;20:foaa16. [CrossRef]
4. Makower M, Bevan EA. The inheritance of the killer character in yeast (Saccharomyces cerevisiae). In:Geerts SJ, editor. Proceedings of the 11th International Conference Generation. XI 1. 1963. 202-3.
5. Bajaj BK, Singh S. Biology of killer yeast and technological implications. In:Yeast Diversity Human Welfare. Singapore:Springer;2017. 163-90. [CrossRef]
6. Zagorc T, Maráz A, Cadez N, Jemec KP, Péter G, Resnik M, et al. Indigenous wine killer yeasts and their application as a starter culture in wine fermentation. Food Microbiol 2001;18:441-51. [CrossRef]
7. De Ullivarri MF, Mendoza LM, Raya RR. Killer activity of Saccharomyces cerevisiae strains:Partial characterization and strategies to improve the biocontrol efficacy in winemaking. Antonie Van Leeuwenhoek 2014;106:865-78. [CrossRef]
8. Lowes KF, Shearman CA, Payne J, Mackenzie D, Archer DB, Merry RJ, et al. Prevention of yeast spoilage in feed and food by the yeast mycocin HMK. Appl Environ Microbiol 2000;66:1066-76. [CrossRef]
9. Lara-Hidalgo CE, Dorantes-Álvarez L, Hernández-Sánchez H, Santoyo-Tepole F, Martínez-Torres A, Villa-Tanaca L, et al. Isolation of yeasts from Guajillo pepper (Capsicum annuum L.) fermentation and study of some probiotic characteristics. Probiotics Antimicrob Proteins 2019;11:748-64. [CrossRef]
10. Hatoum R, Labrie S, Fliss I. Antimicrobial and probiotic properties of yeasts:From fundamental to novel applications. Front Microbiol 2012;3:421. [CrossRef]
11. Perez MF, Contreras L, Garnica NM, Ferna´ndez-Zenoff MV, Fari´as ME, Sepulveda M, et al. Native killer yeasts as biocontrol agents of postharvest fungal diseases in lemons. PLoS One 2016;11:e0165590. [CrossRef]
12. Waema S, Maneesri J, Masniyom P. Isolation and identification of killer yeast from fermented vegetables. Asian J Food Agro Ind 2009;2:126-34.
13. De Lima JR, Gonçalves LR, Brandão LR, Rosa CA, Viana FM. Isolation, identification, and activity in vitro of killer yeasts against Colletotrichum gloeosporioides isolated from tropical fruits. J Basic Microbiol 2013;53:590-9. [CrossRef]
14. Wójcik M, Kordowska-Wiater M. The occurrence of killer activity in yeasts isolated from natural habitats. Acta Biochim Pol 2015;62:821-4. [CrossRef]
15. Labbani FZ, Turchetti B, Bennamoun L, Dakhmouche S, Roberti R, Corazzi L, et al. A novel killer protein from Pichia kluyveri isolated from an Algerian soil:Purification and characterization of its in vitro activity against food and beverage spoilage yeasts. Antonie Van Leeuwenhoek 2015;107:961-70. [CrossRef]
16. Medina-Córdova N, López-Aguilar R, Ascencio F, Castellanos T, Campa-Córdova AI, Angulo C, et al. Biocontrol activity of the marine yeast Debaryomyces hansenii against phytopathogenic fungi and its ability to inhibit mycotoxins production in maize grain (Zea mays L.). Biol Control 2016;97:70-9. [CrossRef]
17. Ahmad R, Hussain B, Ahmad T. Fresh and dry fruit production in Himalayan Kashmir, Sub-Himalayan Jammu and Trans-Himalayan Ladakh, India. Heliyon 2021;7:e05835. [CrossRef]
18. Indian Council of Medical Research New Delhi, India. Standard Operating Procedures for Fungal Identification and Detection of Antifungal Resistance, Antimicrobial Resistance Surveillance and Research Network. New Delhi:Division of Publication and Information on behalf of the Secretary;2019. 23-4.
19. Suh S, Zhang N, Nguyen N, Gross S, Blackwell M. Lab Manual for Yeast Study. United States:Mycology Lab, Louisiana State University;2008. 1-38.
20. Beena MS. Characterization and virulence of Candida isolated from children with dental caries and its susceptibility to various antimicrobial agents. In:Dental Caries. London:IntechOpen;2020. [CrossRef]
21. Heintz N, Gong S. Small-scale preparations of yeast DNA. Cold Spring Harb Protoc 2020;2020:10. [CrossRef]
22. Mannazzu I, Domizio P, Carboni G, Zara S, Zara G, Comitini F, et al. Yeast killer toxins:From ecological significance to application. Crit Rev Biotechnol 2019;39:603-17. [CrossRef]
23. Aguiar C, Lucas C. Yeasts killer/sensitivity phenotypes and halotolerance. Food Technol Biotechnol 2000;38:39-46.
24. Margesin R, Schinner F. Potential of halotolerant and halophilic microorganisms for biotechnology. Extremophiles 2001;5:73-83. [CrossRef]
25. Musa H, Kasim FH, Gunny AA, Gopinath SC. Salt-adapted moulds and yeasts:Potentials in industrial and environmental biotechnology. Process Biochem 2018;69:33-44. [CrossRef]
26. Bapat P, Nandy SK, Wangikar P, Venkatesh KV. Quantification of metabolically active biomass using methylene blue dye reduction test (MBRT):Measurement of CFU in about 200 s. J Microbiol Methods 2006;65:107-6. [CrossRef]
27. Kwolek-Mirek M, Zadrag-Tecza R. Comparison of methods used for assessing the viability and vitality of yeast cells. FEMS Yeast Res 2014;14:1068-79. [CrossRef]
28. Goldschmidt A, Kunert-Graf J, Scott AC, Tan Z, Dudley AM, Kutz JN. Quantifying yeast colony morphologies with feature engineering from time-lapse photography. Sci Data 2022;9:216. [CrossRef]
29. Vallejo JA, Sánchez-Pérez A, Martínez JP, Villa TG. Cell aggregations in yeasts and their applications. Appl Microbiol Biotechnol 2013;97:2305-18. [CrossRef]
30. Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis:Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev 2012;36:288-305. [CrossRef]
31. Hilmioglu S, Ilkit M, Badak Z. Comparison of 12 liquid media for germ tube production of Candida albicans and C. tropicalis. Mycoses 2007;50:282-5. [CrossRef]
32. Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 2005;69:565-84. [CrossRef]
33. Kurtzman C, Fell J, Boekhout T. The Yeasts, a Taxonomic Study. 5th ed. Amsterdam:Elsevier Publishers;2011.
34. Doiphode N, Joshi C, Ghormade V, Deshpande MV. Biotechnological applications of dimorphic yeasts. In:Satyanarayana T, Kunze G, editors. Yeast Biotechnology:Diversity and Applications. Berlin:Springer, Dordrecht;2009. 635-50. [CrossRef]
35. Boyce KJ, Andrianopoulos A. Fungal dimorphism:The switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol Rev 2015;39:797-811. [CrossRef]
36. Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP. Candida. In:The Yeasts. London:Elsevier;2011. 987-1278. [CrossRef]
37. Vaughan-Martini A, Martini A. Saccharomyces Meyen ex reess (1870). In:The Yeasts. Amsterdam:Elsevier;2011. 733-46. [CrossRef]
38. Chanchaichaovivat A, Ruenwongsa P, Panijpan B. Screening and identification of yeast strains from fruits and vegetables:Potential for biological control of postharvest chilli anthracnose (Colletotrichum capsici). Biol Control 2007;42:326-5. [CrossRef]
39. Pitt TL, Barer MR. Classification, identification and typing of microorganisms. Med Microbiol 2012:24-38. [CrossRef]
40. Filippousi R, Antoniou D, Tryfinopoulou P, Nisiotou AA, Nychas GJ, Koutinas AA, et al. Isolation, identification and screening of yeasts towards their ability to assimilate biodiesel?derived crude glycerol:Microbial production of polyols, endopolysaccharides and lipid. J Appl Microbiol 2019;127:1080-100. [CrossRef]
41. Rutherford JC. The emerging role of urease as a general microbial virulence factor. PLoS Pathog 2014;10:e1004062. [CrossRef]
42. McTaggart L, Richardson SE, Seah C, Hoang L, Fothergill A, Zhang SX. Rapid identification of Cryptococcus neoformans var. grubii, C. neoformans var. neoformans, and C. gattii by use of rapid biochemical tests, differential media and DNA sequencing. J Clin Microbiol 2011;49:2522-7. [CrossRef]
43. Marcos JY, Pincus DH. Fungal diagnostics:Review of commercially available methods. Methods Mol Biol 2013;968:25-54. [CrossRef]
44. Mora D, Arioli S. Microbial urease in health and disease. PLoS Pathog 2014;10:e1004472. [CrossRef]
45. Kurtzman CP. In:Hansen PE. The Yeasts. Amsterdam:Elsevier;2011. 685-707. [CrossRef]
46. Raja HA, Miller AN, Pearce CJ, Oberlies NH. Fungal identification using molecular tools:A primer for the natural products research community. J Nat Prod 2017;80:756-70. [CrossRef]
47. Ollinger N, Lasinger V, Probst C, Pitsch J, Sulyok M, Krska R, et al. DNA barcoding for the identification of mold species in bakery plants and products. Food Chem 2020;318:126501. [CrossRef]
48. Singh P, Raghukumar C, Meena RM, Verma P, Shouche Y. Fungal diversity in deep-sea sediments revealed by culture-dependent and culture-independent approaches. Fungal Ecol 2012;5:543-53. [CrossRef]
SUPPLEMENTARY DATA
1. Urease test
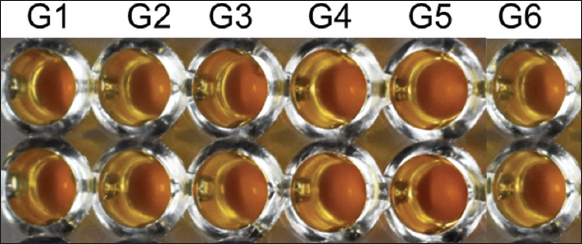 | Supplementary Figure 1: Results obtained from urease test. The wells are labelled G1–G6 indicating the respective yeast isolate. [Click here to view] |
2. Nitrogen assimilation test
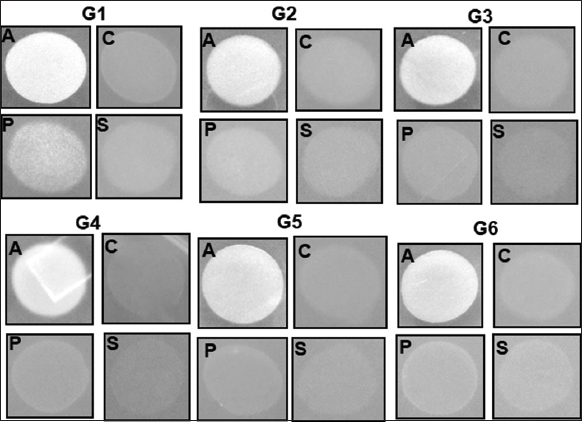 | Supplementary Figure 2: The growth of G1–G6 isolates on minimal media with the respective nitrogen sources. A: Ammonium sulfate, P: Pottasium nitrate, S: Sodium nitrite, C: Control (without nitrogen source). [Click here to view] |
3. Sugar assimilation and fermentation in liquid media
 | Supplementary Figure 3: (a-j) Results of sugar assimilation and fermentation examined in liquid media. [Click here to view] |
4. Phylogenetic tree
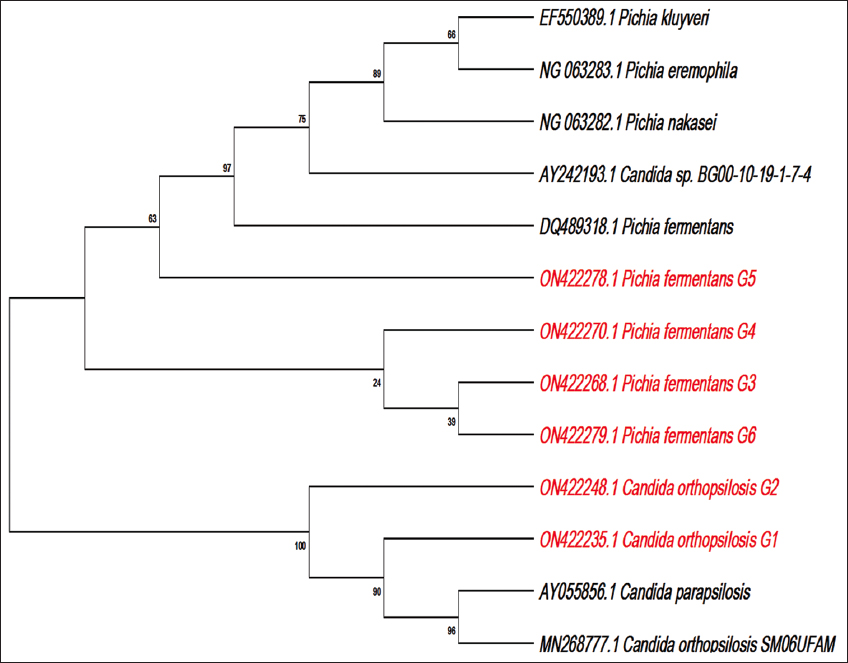 | Supplementary Figure 4: Phylogenetic tree analysis of the isolated killer yeasts. [Click here to view] |
The 18s rDNA sequences of all the six killer isolates along with Pichia fermentans (DQ489318.1), Candida sp (AY242193.1), Pichia kluyveri (EF550389.1), Pichia nakasei (NG_063282.1_), Pichia eremophilia (NG_063283.1), Candida orthopsilosis (MN268777.1), and Candida parapsilosis (AY055856.1) sequences were used to construct phylogenetic tree with maximum likelihood method. Initially the sequences were aligned using Clustal W wit default parameters and then the phylogenetic tree was constructed by applying the maximum likelihood method with a confidence level of 1000 bootstrap replicates. The phylogenetic tree was constructed using MEGA 11 (11.0.11) software.