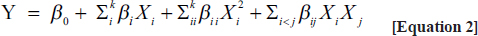1. INTRODUCTION
The practice of incorporating beneficial microorganisms into feeds in the form of single-cell proteins and probiotics represents a promising strategy in aquaculture, as it enhances the growth and immunological resilience of farmed fish. Single-cell protein, or microbial protein, refers to the dried cells of bacteria, fungi, algae, and yeast, which has been regarded as a potential substitute for fish meal up to 25-50% in aquaculture diets. Bacterial cells outpace other microbial proteins in terms of their rich crude protein concentration (50-80 wt.%), large amino acid spectrum, carbohydrate, vitamin content, etc. Although bacterial meal serves as a valuable source of protein, the efficiency of its mass production relies on the final product yield; therefore, using cost-effective methods to maximize cellular growth is a crucial prerequisite for microbial protein production [1-3]. A culture medium exacting in terms of the nutritional requirements of the selected bacteria can exert greater influence on its growth, nutrient content, and biomass yield [4]. The nutritional requirements and composition of a bacterial cell are in fact attributed to the species and strain to which it belongs. Since their growth tactics are nutrient-sensitive, finding the optimal growth medium and conditions for the bacterial strain under study is pivotal for obtaining the desired biomass yield with a high crude protein content.
Bacillus subtilis is a common probiotic bacterium used in aquafeeds. They are frequently incorporated into aquaculture diets as live or heat-killed probiotics, and research indicates that they enhance fish nutrition and exhibit potent immunomodulatory effects. [5-7]. Furthermore, different Bacillus species have been proven to produce single cell proteins with high crude protein content [8-10]. This microbial protein is highly sought after for its essential amino acid profile, which complies with Food and Agriculture Organization and World Health Organizations standards [2,3,11]. B.subtilis PW12 (MTCC 10402) is a non-pathogenic, salt tolerant, probiotic bacterium with anti-microbial activity against aquaculture pathogens. The ability of the bacteria to thrive in normal to higher salinities renders it a valuable water probiotic that can find application across diverse aquaculture sectors spanning freshwater, brackish water, and marine environments [12,13]. The bacterium has been found to have anti-microbial properties, particularly against Vibrio species, and the primary compounds produced are anti-microbial chemicals of aquaculture grade, such as N-substituted phenazinecarboxylate, propyl/phenethyl 2-oxoacetates [14]. Microbial formulations, comprising B. subtilis strains and other bacterial species producing similar antimicrobial compounds, have been developed with elevated nutritional value and antagonistic properties that are intended for incorporation into aquafeeds, aiming to improve fish health and promote growth [15]. Production of high-quality microbial products, however, demands enhancement of microbial biomass production. Since biomass production of B. subtilis PW12 has not yet been the subject of extensive research, the current study deals with pioneering efforts on optimizing the medium to maximize the biomass production of the bacteria in batch culture to overcome the low yield challenge associated with the production of microbial proteins. Moreover, the research fosters the opportunity to explore the potentiality of this microbial biomass in aquafeed as a single-cell protein.
Statistical medium optimization using Response Surface Methodology (RSM) is an excellent strategy to formulate nutritionally balanced as well as cost-effective culture mediums for obtaining desired products from microorganisms before stepping into their large-scale manufacture. Statistical medium optimization surpasses conventional strategies such as the one-factor-at-a-time approach in terms of cost, time efficiency, accuracy in result interpretation, and understanding of the interactive effects of variables. This approach combines statistical and mathematical techniques to build models, analyze the impact of numerous independent variables, and determine the optimal values for each variable. In essence, response surface methodology is a combination of steps comprising experimental design, mathematical modeling, and statistical inference that, when used together, enable the researcher to analyze the response that is influenced by a number of variables [16].
Statistical medium optimization studies have successfully yielded various growth-associated secondary metabolites and facilitated the preparation of bio-control formulations utilizing different strains of B.subtilis. However, the current investigation aims at optimizing a medium specifically for enhanced biomass production of B. subtilis PW12 (MTCC10402) in a batch culture, to unlock its potential in aquafeeds. In the present study, Plackett-Burman design (PBD) was used to select the medium components and culture conditions that significantly affect biomass production. The selected factors were then optimized using RSM with a Central Composite Design (CCD) to achieve a higher biomass yield for the bacterium.
3. RESULTS AND DISCUSSION
3.1. Screening of Significant Nutrient Components Using Plackett-Burman Design
The Plackett-Burman design is a key tool in screening the effects of the variables on the final response [18]. The biomass yield showed variation across the 15 experimental runs (Table 3). ANOVA results for PBD revealed that only 4 out of 11 variables had a significant effect on the response, i.e., biomass yield. Soya peptone, glucose, pH, and inoculum size had a significant influence on biomass production (p < 0.05) [Table 4, Figure 1]. Hence, only these factors were used for further optimization experiments. The polynomial equation for biomass yield is represented by Equation 3.
Table 3 : Design and responses of the Plackett - Burman Design (PBD).
| Run | A | B | C | D | E | F | G | H | J | K | L | Biomass yield (g/L) |
|---|
| 1 | - | + | + | + | - | + | + | - | + | - | - | 9.34 |
| 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.55 |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.06 |
| 4 | + | - | - | - | + | + | + | - | + | + | - | 9.85 |
| 5 | - | + | + | - | + | - | - | - | + | + | + | 7.98 |
| 6 | + | - | + | + | - | + | - | - | - | + | + | 8.36 |
| 7 | - | - | - | + | + | + | - | + | + | - | + | 9.22 |
| 8 | - | - | + | + | + | - | + | + | - | + | - | 9.00 |
| 9 | + | + | - | + | - | - | - | + | + | + | - | 10.45 |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.50 |
| 11 | + | + | + | - | + | + | - | + | - | - | - | 8.57 |
| 12 | - | + | - | - | - | + | + | + | - | + | + | 7.33 |
| 13 | + | + | - | + | + | - | + | - | - | - | + | 9.08 |
| 14 | - | - | - | - | - | - | - | - | - | - | - | 6.74 |
| 15 | + | - | + | - | - | - | + | + | + | - | + | 7.58 |
Table 4: Analysis of variance for the experimental results of the Plackett - Burman Design (PBD).
| Source | DF | Adj SS | Adj MS | F-Value | P-Value |
|---|
| Model* | 12 | 16.6529 | 1.38774 | 19.84 | 0.049 |
| Linear | 11 | 12.8638 | 1.16943 | 16.72 | 0.058 |
| Glucose* | 1 | 1.538 | 1.53797 | 21.99 | 0.043 |
| Peptone | 1 | 0.338 | 0.33802 | 4.83 | 0.159 |
| Yeast extract | 1 | 0.2815 | 0.28152 | 4.03 | 0.183 |
| Soya Peptone* | 1 | 4.5485 | 4.54855 | 65.04 | 0.015 |
| NaCl | 1 | 1.2727 | 1.27271 | 18.2 | 0.051 |
| MgSO..7H.O | 1 | 0.2797 | 0.27969 | 4 | 0.184 |
| KH.PO. | 1 | 0.0608 | 0.06078 | 0.87 | 0.45 |
| Temperature | 1 | 0.0531 | 0.05307 | 0.76 | 0.476 |
| pH* | 1 | 2.3745 | 2.37452 | 33.95 | 0.028 |
| Agitation | 1 | 0.4978 | 0.49776 | 7.12 | 0.116 |
| Inoculum size* | 1 | 1.6192 | 1.61921 | 23.15 | 0.041 |
| Curvature* | 1 | 3.7891 | 3.7891 | 54.18 | 0.018 |
| Error | 2 | 0.1399 | 0.06994 | | |
| Total | 14 | 16.7927 | | | |
| R2 = 0.9917 |
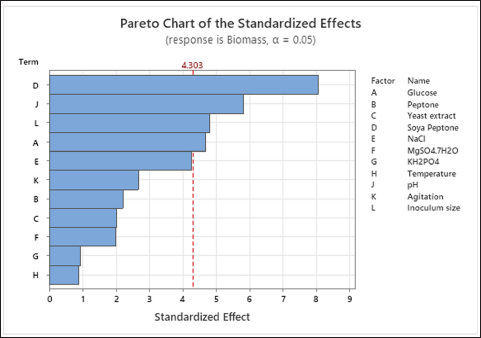 | Figure 1: Pareto chart showing standard effects of 11 variables on biomass production of B. subtilis PW 12.
[Click here to view] |

The linear regression coefficient of determination, R2, was 0.99, which indicated that the predicted model could account for 99.17% of the variability in the experimental data. The model was fit to the data, as the lack of fit was found to be insignificant (p > 0.05). Since the curvature was significant (p < 0.05), a higher order model was necessary to optimize the levels of significant variables.
The Selection of medium components and their concentrations is fundamental to medium optimization, as the availability of substrates influences growth rate, and the limitations of different substrates can significantly affect the nutritional value of microbial biomass [19]. An adequate supply of carbon and nitrogen, together with ideal growing conditions, are the most essential requirements for bacterial growth. The variables investigated in the present study were selected to account for the sources of carbon (glucose), nitrogen (peptone, yeast extract, and soya peptone), inorganic nutrients for the supply of phosphate, sulfate, and minerals (KH2PO4, MgSO4.7H2O, NaCl) and culture conditions (temperature, pH, agitation, and inoculum size). Since no other optimization experiments had been conducted on B. subtilis PW12, the medium components and culture conditions for the study were chosen based on prior research on other strains of B. subtilis. Prior to screening, OFAT studies provided information on the ranges of concentrations for each individual component that should be used in the Plackett-Burman design.
Screening experiments using PBD showed that soya peptone, glucose, pH, and inoculum size had substantial impacts on biomass yield. The nitrogen source has a key role in determining the rate of bacterial growth [20]. Soya peptone had the highest effect on biomass production by B. subtilis PW12. Other nitrogen sources used in the PBD study, such as yeast extract and peptone, had an insignificant effect on biomass production. The effective use of soya peptone or soya protein hydrolysate as nitrogen sources by some strains of B. subtilis has been reported, where high cell density biomass production was a prerequisite for the synthesis of certain secondary metabolites and industrially important enzymes [21-23]. Several prior investigations, in contrast to the current study, have utilized other organic nitrogen sources, such as peptone and yeast extract, to produce biomass using some specific strains of B. subtilis [24-26]. In the present study, the enhanced production of biomass may be attributed to the complex mixture of amino acids, short peptides, carbohydrates, etc. contained in soya peptone. Despite being a non-essential amino acid, glutamate is the one amino acid that B. subtilis prefers [27,28]. According to the study by Leibs et al. [29] the exponential growth phase of B. subtilis showed considerable intake of glutamate, aspartate, serine, and alanine from the growth media. Moreover, soya peptone has a high reserve of amino acids and a total glutamic acid concentration higher than other amino acid contents [30]. Together with other growth factors, the presence of these amino acids might have helped the bacteria produce more biomass. Additionally, it is possible that B. subtilis may efficiently use soya peptone as it can produce the enzyme protein glutaminase (PG), which has the ability to hydrolyze amides of glutamine in proteins and thereby enhance the solubility of plant proteins. During fermentation, B. subtilis may produce extracellular proteases that can self-activate protein glutaminase, which is released in the inactive pro-enzyme form and can facilitate efficient substrate utilization by the bacterium [31].
The effect of glucose was also statistically significant on the biomass production of the bacteria. Previous studies also show that glucose is a preferred carbon source for B. subtilis and other Bacilli [32-34]. The phosphoenolpyruvate: sugar phosphotransferase system (PTS) in B. subtilis aids them to take up and phosphorylate the sugar, and the presence of PTS dependent and independent glucose transporters also facilitates the bacteria in efficiently utilizing glucose as the carbon source [32,35]. Additionally, as glucose is a readily metabolized carbon source, it may accelerate the fermentation process and enhance the production of bacterial biomass. Previous studies have found that glucose was the preferred carbon source for obtaining a high biomass yield in B. subtilis strains for the synthesis of several secondary metabolites and enzymes that are linked to high cell densities using batch or fed batch fermentation methods [21,36,37].
Among the culture conditions, pH and inoculum size significantly affected biomass production. Since pH is the primary control of bacterial metabolism, optimum pH is necessary for their maximum biomass production. Besides pH, inoculum size also showed a significant impact on biomass production. As inoculum concentration determines the lag phase of bacterial growth, the addition of adequate inoculum of bacteria at the appropriate developmental stage is important to achieve increased biomass yield [38].
In the present study, temperature and agitation had the least effect on biomass yield among the culture conditions. Previous studies on B. subtilis strains suggest that their optimal temperature and agitation ranges are 30°C-37°C and 200–250 rpm, respectively [34,39,40]. The temperature and agitation ranges applied in the present PBD study were ideal for B. subtilis PW12 growth. Consequently, the temperature and agitation values in the CCD optimization study were set at 37°C and 200 rpm, respectively.
3.2. Optimization of Biomass Production by RSM and CCD
The Central Composite Design was carried out to optimize the levels of significant variables to maximize the biomass yield. The design matrix and the associated experimental results show that biomass yield varied across the experimental runs (Table 5). The second-order polynomial equation for biomass yield is represented by the following equation [Equation 4].
Table 5 : Design and responses of the central composite design (CCD).
| Run | A | B | C | D | Obtained biomass yield | Predicted biomass yield |
|---|
| 1 | 1 | −1 | −1 | 1 | 9.76 | 9.82 |
| 2 | −1 | 1 | −1 | −1 | 9.54 | 9.90 |
| 3 | −1.414 | 0 | 0 | 0 | 10.17 | 9.70 |
| 4 | 1 | −1 | 1 | 1 | 12.00 | 11.65 |
| 5 | 0 | 0 | −1.414 | 0 | 7.99 | 8.12 |
| 6 | −1 | −1 | −1 | −1 | 5.89 | 5.79 |
| 7 | 1 | −1 | 1 | −1 | 7.05 | 7.32 |
| 8 | 0 | 1.414 | 0 | 0 | 12.93 | 12.79 |
| 9 | 0 | −1.414 | 0 | 0 | 10.89 | 10.93 |
| 10 | 0 | 0 | 0 | 0 | 13.06 | 13.19 |
| 11 | 0 | 0 | 0 | 0 | 13.83 | 13.19 |
| 12 | 0 | 0 | 0 | 0 | 13.43 | 13.19 |
| 13 | 0 | 0 | 0 | 0 | 13.05 | 13.19 |
| 14 | 0 | 0 | 0 | 0 | 13.61 | 13.19 |
| 15 | 0 | 0 | 1.414 | 0 | 7.94 | 7.71 |
| 16 | 0 | 0 | 0 | 0 | 12.53 | 13.19 |
| 17 | −1 | 1 | 1 | 1 | 7.73 | 8.03 |
| 18 | −1 | −1 | 1 | 1 | 7.35 | 7.53 |
| 19 | 1 | 1 | −1 | −1 | 9.15 | 9.00 |
| 20 | −1 | −1 | 1 | −1 | 5.69 | 5.69 |
| 21 | −1 | 1 | −1 | 1 | 9.30 | 9.07 |
| 22 | −1 | 1 | 1 | −1 | 7.52 | 7.50 |
| 23 | 1 | 1 | 1 | 1 | 10.04 | 10.18 |
| 24 | 0 | 0 | 0 | 0 | 12.63 | 13.19 |
| 25 | 1 | 1 | −1 | 1 | 10.65 | 10.65 |
| 26 | 1 | −1 | −1 | −1 | 7.15 | 6.86 |
| 27 | 0 | 0 | 0 | −1.414 | 10.99 | 11.09 |
| 28 | 1 | 1 | 1 | −1 | 7.33 | 7.15 |
| 29 | −1 | −1 | −1 | 1 | 6.07 | 6.26 |
| 30 | 1.414 | 0 | 0 | 0 | 11.58 | 11.96 |
| 31 | 0 | 0 | 0 | 1.414 | 13.75 | 13.56 |

The coefficient of determination, R2, which was 0.98, indicated that 98.71% of the variability in the response could be elucidated by the model (Table 6). ANOVA was conducted to assess the statistical significance of the polynomial equation, and the results showed that the regression was statistically significant (p < 0.05). The significance of the regression coefficient of the model reveals that the linear effect of all variables except pH (p > 0.05) on the maximum biomass production was significant (p < 0.05) (Table 6). However, the squared effects of all the variables were significant (P < 0.05) and except for the soya peptone - pH interaction (P > 0,05), all other interactions were statistically significant (P < 0.05). The lack of fit of the model was insignificant (P > 0.05) indicating that the model fits the data. The significant interactions of variables described by the model are shown in contour plots and three-dimensional response surface plots [Figure 2].
Table 6 : Analysis of variance for the experimental results of the Central Composite Design (CCD).
| Source | DF | Adj SS | Adj MS | F-Value | P-Value |
|---|
| Model | 14 | 203.135 | 14.5096 | 87.26 | 0.000 |
| Linear | 4 | 37.249 | 9.3122 | 56 | 0.000 |
| Soya peptone* | 1 | 12.876 | 12.8757 | 77.43 | 0.000 |
| Glucose* | 1 | 8.711 | 8.7109 | 52.39 | 0.000 |
| pH | 1 | 0.411 | 0.4111 | 2.47 | 0.135 |
| Inoculum size* | 1 | 15.251 | 15.2513 | 91.72 | 0.000 |
| Square | 4 | 146.599 | 36.6498 | 220.41 | 0.000 |
| Soya peptone*Soya peptone* | 1 | 13.124 | 13.1244 | 78.93 | 0.000 |
| Glucose*Glucose* | 1 | 4.159 | 4.1594 | 25.01 | 0.000 |
| pH*pH* | 1 | 65.587 | 65.587 | 394.43 | 0.000 |
| Inoculum size*Inoculum size* | 1 | 1.765 | 1.7647 | 10.61 | 0.005 |
| 2-Way Interaction | 6 | 19.287 | 3.2145 | 19.33 | 0.000 |
| Soya peptone*Glucose* | 1 | 3.882 | 3.8819 | 23.35 | 0.000 |
| Soya peptone*pH | 1 | 0.314 | 0.3139 | 1.89 | 0.188 |
| Soya peptone*Inoculum size* | 1 | 6.191 | 6.1914 | 37.23 | 0.000 |
| Glucose*pH* | 1 | 5.321 | 5.3211 | 32 | 0.000 |
| Glucose*Inoculum size* | 1 | 1.705 | 1.705 | 10.25 | 0.006 |
| pH*Inoculum size* | 1 | 1.873 | 1.8735 | 11.27 | 0.004 |
| Error | 16 | 2.66 | 0.1663 | | |
| Lack-of-Fit | 10 | 1.237 | 0.1237 | 0.52 | 0.827 |
| Pure Error | 6 | 1.424 | 0.2373 | | |
| Total | 30 | 205.795 | | | |
| R2 = 0.9871 |
 | Figure 2: Counter (left) and response surface (right) plots representing significant interaction of variables on biomass yield: (A) Glucose vs. soya peptone, (B) Inoculum size vs. Glucose, (C) Inoculum size vs. soya peptone, (D) pH vs. glucose, and (E) Inoculum size vs. pH.
[Click here to view] |
Carbon and nitrogen sources are absolutely essential for bacterial growth. In the present study, the interactive effects of glucose and soya peptone also significantly contributed to biomass production. The response surface and contour plots show that the optimal concentrations of glucose and soya peptone were around 14 g/L and 27 g/L, respectively [Figure 2A]. In concordance with the current investigation, a single factor optimization study by Naveed et al. [36] revealed that the addition of 1.5% glucose and 2.5 % soya peptone in the fermentation medium of a newly isolated strain, B. subtilis BSN314, caused enhanced biomass production in the bacterium. However, studies by Zhong et al. [21] showed that an optimized medium containing 30.70 g/L of glucose and 2.4 g/L of total nitrogen from soya bean meal hydrolysate was beneficial for high cell density cultivation of B. subtilis ZK8 in fed batch fermentation. Investigation of Yue et al. [41] demonstrated that cell concentration of B. subtilis ZK-H2 could be increased to 7x108 cfu/mL in an optimized medium containing glucose and soya peptone, where the optimal concentration for glucose and total nitrogen concentration from soybean protein were 21 g/L and 4.0 g/L, respectively. Zhong et al. [42] used soya bean meal hydrolysate and glucose in the medium of a wild type strain, Bacillus sp. H-18W, for the production of chiral acetoin, a metabolite associated with fast cell growth. Nguyen [22] used glucose at 5.62 g/L and soya bean peptone at 13 g/L in the optimized medium for higher biomass production by B. subtilis Natto, whereas a study by Ullah et al. [43] showed that soya peptone and glucose at 1% w/v in the medium resulted in improved cell growth of a probiotic strain of B. subtilis and in contrast to the present study, higher concentrations of soya peptone caused a decline in cell growth. Stamenkovic et al. [33] reported that biomass production of B.subtilis NCIM2063 could be enhanced when the growth medium was supplemented with 10 g/L of glucose. Study by Cho et al. [25] demonstrated that supplementation of the initial medium with 10 g/L glucose and 50 g/L peptone in a fed batch culture caused an increase in the cell density of B. subtilis however, exhaustion of glucose after 6 h was also reported. Since B. subtilis can work on soya-based proteins, Wang et al. [44] included soy bean curd as well as soya peptone at concentrations of 12.2% w/v and 5.7% w/v respectively, in the medium for metabolite production and growth of B. subtilis. Study by Yanez et al. [45] reported that even defatted soy flour (40 g/L) could be used in the growth and metabolite production medium of B. subtilis CtpxS2-1 and was found to improve bacterial growth. From the present study, it can be concluded that B.subtilis PW 12 strain could also successfully utilize glucose at less than 15 g/L and soya peptone at less than 30 g/L in the culture medium for attaining maximum growth.
Soya peptone and glucose had significant interactions with inoculum size as well [Figures 2B,]. In the current investigation, it was shown that biomass production rose in tandem with inoculum size. A high inoculum concentration can promote growth up to a certain point, but afterwards, it causes the microbial activity to decline because of the depletion of nutrients in the medium. Conversely, a low inoculum concentration decreases the cell concentration, which may affect the amount of bioproduct produced. At lower cell concentrations, it takes a long time to attain optimum growth [46]. The optimum inoculum size in the current investigation was 11.03% v/v, indicating that the soya peptone and glucose-based optimized medium had sufficient nutrients to maintain a higher inoculum concentration to enhance the growth of the bacterium. Study by Huang et al. [47] on B. subtilis biomass production shows that 8% v/v of inoculum size may be ideal for achieving optimal production.
Though the linear effect of pH was not significant, its interaction with glucose and inoculum size was significant [Figures 2D, E]. Response surface and contour plots show that maximum production of biomass was obtained at pH 7. The optimal pH for B. subtilis strains varies depending on the specific strain and the conditions of the study. In general, the optimal pH for B. subtilis strains appears to range from pH 4.0 to pH 8.0, depending on the specific strain and the activity being measured, and previous studies show that most of the B. subtilis strains prefer pH 7 for their optimal growth [22,26,36]. Acidic pH may result in rigid and ordered membranes, which can affect the membrane dynamics of the bacterium [48].On the other hand, alkaline conditions may cause membrane disorder [49] and hence maintaining optimal pH is crucial for the membrane properties and thereby the growth of B. subtilis. At the optimum pH, glucose at a concentration of 14.07 g/L contributed to the maximum biomass production [Figure 2D]. pH variation during bacterial growth is greatly influenced by the medium composition, particularly the carbon source. Since glucose is a reduced carbon source, more energy may be derived from its oxidation, leading to greater bacterial proliferative potential, and there might be an increased proton flux causing acidification of the medium during the exponential phase of bacterial growth [50]. Therefore, maintenance of optimal pH throughout fermentation is desirable. At the optimal pH, an inoculum size of 11% v/v was significant for enhanced biomass production [Figure 2E].
By using the equation, the optimal concentrations for the variables were predicted. Concentrations of soya peptone (a) glucose, (b) pH (c) and inoculum size (d) obtained from the maximum point of the model were 27.56 g/L for A 14.07 g/L for B, 7.09 for C and 11.03% v/v for D respectively. The model predicted a maximum biomass yield of 14.19 g/L for this point.
In the present investigation, inorganic nutrients, including NaCl, MgSO4 .7H2O, and KH2PO4 had no significant effect on biomass production. While this does not necessarily negate their importance for bacterial growth, the findings of this study suggest that among the media components, soya peptone and glucose notably influence biomass production. Inorganic nutrients are the most common inclusions in bacterial growth mediums as bacteria require anions like phosphates and sulfates, and cations like sodium, potassium, magnesium, iron, and calcium. These compounds are essential for the synthesis of nucleic acids, proteins, and essential cofactors. The quantity of inorganic salts that must be added to the fermentation medium varies depending on the nutritional requirements of the organism, the composition of the culture medium, and the type of desired end product [51]. In the present study, inorganic nutrients were found to have an insignificant impact on bacterial biomass production. Since this study did not center on the production of specific primary or secondary metabolites by the bacteria, there might be room for flexibility in the required quantities of these inorganic salts, thus making their optimal concentration less strict in the bacterial growth medium. The bacteria were able to proliferate at both low and high levels of the inorganic nutrients applied in the PBD. As a result, in the final optimized medium, the concentrations of these inorganic nutrients were set at the lowest level utilized in PBD as 5 g/L, 2 g/L, and 2 g/L for NaCl, MgSO4, 7.H2O, and KH2PO4, respectively (Table 1). Furthermore, soya peptone, the nitrogen source in the medium, usually contains an ash content of less than 15% which indicates the presence of minerals like calcium, magnesium, and potassium. However, soya peptone is reported to have a very low inorganic phosphorous content [52].The essential minerals and vitamins present in the nitrogen sources might have been utilized by the bacteria for growth.
3.3. Validation of the Optimized Culture Medium
To validate the modeling results, triplicate experiments were done using the optimized conditions. The predicted maximum yield was 14.19 g/L, and the average value obtained in the experiments was 14.29 ± 0.23 g/L, which was in agreement with the predicted yield of the current optimization study. The biomass yield obtained in this investigation was similar to that reported in the study of Ma et al. [23] in which a fed batch fermentation experiment with a mutant strain of B. subtilis 168 mut-16# was conducted. Feeding of soya peptone and hydrolyzed starch to the medium increased the dry cell weight of the bacteria, and the highest dry cell weight obtained was 14.3 g/L which was comparable to the findings of the present study. However, a statistical optimization study conducted by Zhong et al. [21] on B. subtilis ZK8 reported 77.5 g/L dry cell weight of the bacterium in fed batch culture and 20.38 g/L in batch culture. Yadav et al. [53] used peptone as the nitrogen source in the growth medium of B. subtilis and a maximum dry cell weight of 7.5 g/L was obtained after optimization. Koim-Puchowska et al. [27] used yeast extract (4 g/L) and soluble starch (40 g/L) as nitrogen and carbon sources, respectively, for the growth of B. subtilis natto BS19 and resulted in a biomass yield of less than 6mg/mL. Studies of Nguyen [22], Stamenkovic et al. [33], Naveed et al. [36], and Ghasemi and Ahmadzadeh [54] reported biomass yields as 3.033 g/L, 6 g/L, 3.81 g/L, and 0.5 g/L, respectively, for different B. subtilis strains.
The biomass yield achieved in this study emphasizes the importance of medium optimization in augmenting microbial biomass production. It also suggests the potential scalability of this medium to an industrial level for the production of single-cell protein from B. subtilis PW12. Considering the former in regard to manufacturing costs, finding alternatives to the main nutrient of the optimized medium i.e., soya peptone, could prove effective in enhancing the production efficiency of the scale-up process. However, cost issues associated with this nitrogen source have been addressed in some prior research, and cost-effective methods of soya peptone synthesis using enzymatic hydrolysis of soya beans have been proposed [55,56]. Substrates based on soya protein, such as defatted soy flour and soy bean curd, have been used in experiments to produce biomass and metabolites by fermentation with B. subtilis [44,45]. Considering the adeptness of B. subtilis PW 12 at utilizing soya peptone, delving deeper into the effects of cost-effective nitrogen sources extracted from soybean meal on bacterial growth could offer valuable insights.
Moreover, the most critical juncture of an entire scale-up process is the transition from shake flask to laboratory bioreactor. It is imperative to understand the hydrodynamic behavior of the bioreactor, including mass and heat transfer, mixing, and aeration, especially in aerobic fermentation, to verify the efficacy of the scale-up process. [34] This information is pertinent to the current study as well, so as to achieve yields in the bioreactor that are equivalent to or surpass those in shake flasks under the same conditions.
The goal of the current study was to optimize the biomass yield as well as the nutritional quality of a beneficial probiotic bacterium, B. subtilis PW 12 that could be used as a single cell protein or protein supplement in aquafeeds. According to Sakarika et al., the nitrogen content of the culture medium may be positively correlated with the nutritional quality, especially the protein content of the microbial biomass [19]. Since the optimized medium is rich in nutrients, the bacteria may synthesize and accumulate more protein due to their ability to proliferate extensively, resulting in a high cell density. Consequently, the biomass can be utilized as microbial protein in aquafeeds. The current investigation validates that B. subtilis grows optimally in a medium based on soya peptone, and glucose, which are the standard ingredients of microbiological media. Since the medium and culture conditions have been optimized to enhance the growth and nutritional quality of the test bacterium, the growth of other strains of B. subtilis, particularly those that can achieve high cell densities and other probiotic strains, can also be evaluated in this optimized medium for increased biomass production. This study has only taken into account the maximal biomass yield; nevertheless, due to the fact that bacterial cells are dynamic, they may produce extracellular and intracellular enzymes, proteins, and metabolites when stimulated by external factors, including medium components or the culture conditions. However, as the study has not examined the production of any particular antimicrobial compound, metabolite, or enzyme by B. subtilis PW12 in the optimized medium, future investigations on these facets may assist in evaluating the transferability of the medium to other similar B. subtilis strains. Given the under-researched status of B. subtilis PW12, thorough exploration is needed for a comprehensive grasp of its metabolic, antibacterial, probiotic, and nutritional properties to probe its potential application in aquaculture and related fields.
REFERENCES
1. Nasseri AT, Rasoul-Amini S, Morowvat MH, Ghasemi Y. Single cell protein: production and process. Am J Food Technol. 2011;6(2):103-16. [CrossRef]
2. Jannathulla R, Sravanthi O, Moomeen S, Gopikrishna G, Dayal JS. Microbial products in terms of isolates, whole-cell biomass, and live organisms as aquafeed ingredients: production, nutritional values, and market potential—a review. Aquac Int. 2021;29(2):623-50. [CrossRef]
3. Anupama, Ravindra P. Value-added food: Single cell protein. Biotechnol Adv. 2000; \18(6):459-79. [CrossRef]
4. Chisti Y, Moo-Young M. Bioreactors. In: Meyers RA, editor. Encyclopedia of Physical Science and Technology. 3rd ed. Vol. 2. San Diego: Academic Press; 2003:247–71. [CrossRef]
5. Shah S, Chesti A, Rather M, Hafeez M, Aijaz A, Yousuf I, et al. Effect of Probiotics (Bacillus subtilis) on the Growth and Survival of Fingerlings of Grass Carp, Ctenopharyngodon idella. Curr J Appl Sci Technol. 2021;10(7):31-7. [CrossRef]
6. Büyükdeveci ME, Cengizler I, Balcazar JL, Demirkale I. Effects of two host-associated probiotics Bacillus mojavensis B191 and Bacillus subtilis MRS11 on growth performance, intestinal morphology, expression of immune-related genes and disease resistance of Nile tilapia (Oreochromis niloticus) against Streptococcus iniae. Dev Comp Immunol. 2023;138:104553. [CrossRef].
7. Liao Z, Liu Y, Wei H, He X, Wang Z, Zhuang Z, et al. Effects of dietary supplementation of Bacillus subtilis DSM 32315 on growth, immune response and acute ammonia stress tolerance of Nile tilapia (Oreochromis niloticus) fed with high or low protein diets. Anim Nutr. 2023;15:375-85. [CrossRef]
8. Al-Awadhi NM, Razzaque MA, Jonker D, Banat IM, Hamdan IY. Nutritional and toxicological evaluation of single-cell protein produced from Bacillus Sp. Kisri-Tm1a in Rats. J Food Qual. 1995;18(6):495-509. [CrossRef]
9. Kurbanoglu EB, Algur OF. Single-cell protein production from ram horn hydrolysate by bacteria. Bioresour Technol. 2002;85(2):125-9. Doi: [CrossRef]
10. Feliatra F, Mardalisa M, Mukti PR, Feliatra VA, Effendi I. Potential of Bacillus cereus SN7 as a single cell protein source. J Phys Conf Ser. 2021;2049(1). [CrossRef]
11. WHO J. Protein and amino acid requirements in human nutrition. World Health Organization technical report series. 2007(935):1.
12. Tank PR, Vadher KH, Patel MP. Isolation of probiotic bacteria from gastrointestinal tract of pacific white shrimp Litopenaeus vannamei and antibacterial activity of probiotic bacteria against Vibrio spp. J Entomol Zool Stud. 2018; 6:974-78.
13. Tank PR, Vadher KH, Patel MP. Effect of salinity on growth performance of probiotics for its potential use in shrimp aquaculture. J Exp Zool India. 2018;21(2):1111-3.
14. Chakraborty K, Vinaya KK, Antony T, Joy M, Sreemol CK. Marine microbes as a source of antimicrobial compounds. In: Winter School on “Recent advances in bioactive compounds from marine organisms and development of high value products for health management, ICAR-Central Marine Fisheries Research Institute, Kochi; 2018;189-198. Available from: http://eprints.cmfri.org.in/id/eprint/13024
15. Nair VA. Bioprospecting of novel antimicrobial metabolites from Bacillus subtilis MBTDCMFRI Ba37 and Pseudomonas aeruginosa MBTDCMFRI Ps04 of tropical estuarine habitats of Cochin, India and its application in fish health management [Dissertation]. ICAR-Central Marine Fisheries Research Institute; 2016.
16. Breig SJ, Luti KJ. Response surface methodology: A review on its applications and challenges in microbial cultures. Mater Today Proc. 2021;42:2277-84. [CrossRef]
17. Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946;33(4):305-25. [CrossRef]
18. Cavazzuti M. Optimization Methods: from Theory to Design, London: Springer; 2013. [CrossRef]
19. Sakarika M, Kerckhof FM, Van Peteghem L, Pereira A, Van Den Bossche T, Bouwmeester R, et al. The nutritional composition and cell size of microbial biomass for food applications are defined by the growth conditions. Microb Cell Factories. 2023;22(1). [CrossRef]
20. Ashino K, Sugano K, Amagasa T, Ying BW. Predicting the decision making chemicals used for bacterial growth. Sci Rep. 2019; 9(1):1. [CrossRef]
21. Zhong J, Zhang X, Ren Y, Yang J, Tan H, Zhou J. Optimization of Bacillus subtilis cell growth effecting jiean-peptide production in fed batch fermentation using central composite design. Electron J Biotechnol. 2014;(3):132-6. [CrossRef]
22. Nguyen Anh T. Optimization of the fermentation medium to receive the highest biomass yield by Bacillus subtilis Natto and the initial test of nattokinase yield. IOSR J Eng. 2014;4(12):35-40. [CrossRef]
23. Ma Y, Shen W, Chen X, Liu L, Zhou Z, Xu F, et al. Significantly enhancing recombinant alkaline amylase production in Bacillus subtilis by integration of a novel mutagenesis-screening strategy with systems-level fermentation optimization. J Biol Eng. 2016;10(1). [CrossRef]
24. Sreekumar G, Krishnan S. Enhanced biomass production study on probiotic Bacillus subtilis SK09 by medium optimization using response surface methodology. Afr J Biotechnol. 2010;9(47):8078-84. [CrossRef]
25. Cho YH, Song JY, Kim KM, Kim MK, Lee IY, Kim SB et al. Production of nattokinase by batch and fed-batch culture of Bacillus subtilis. N Biotechnol. 2010;27(4):341-6. [CrossRef]
26. Koim-Puchowska B, Klosowski G, Drózdz-Afelt JM, Mikulski D, Zielinska A. Influence of the medium composition and the Culture Conditions on Surfactin Biosynthesis by a native Bacillus subtilis natto BS19 strain. Molecules. 2021;26(10):2985. [CrossRef]
27. Fisher SH. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol Microbiol. 1999;32(2):223-32. [CrossRef]
28. Atkinson MR, Fisher SH. Identification of genes and gene products whose expression is activated during nitrogen-limited growth in Bacillus subtilis. J Bacteriol. 1991;173(1):23-7. [CrossRef]
29. Liebs P, Riedel K, Graba JP, Schrapel D, Tischler U. Formation of some extracellular enzymes during the exponential growth of Bacillus subtilis. Folia Microbiol. 1988;33(2):88-95. [CrossRef]
30. Davami F, Eghbalpour F, Nematollahi L, Barkhordari F, Mahboudi F. Effects of peptone supplementation in different culture media on growth, metabolic pathway and productivity of CHO DG44 cells; a new insight into amino acid profiles. Iran Biomed J. 2015;19(4):194. [CrossRef]
31. Yin X, Zhang G, Zhou J, Li J, Du G. Combinatorial engineering for efficient production of protein-glutaminase in Bacillus subtilis. Enzyme Microb Technol. 2021;150:109863. [CrossRef]
32. Stülke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu Rev Microbiol. 2000;54(1):849-80. [CrossRef]
33. Stamenkovic-Stojanovic S, Karabegovic I, Beškoski V, Nikolic N, Lazic M. Bacillus subtilis NCIM2063 batch cultivation: The influence of the substrate concentration and oxygen transfer rate on the biomass yield. Adv Technol. 2020;9(1):44-9. [CrossRef]
34. Stamenkovic-Stojanovic S, Karabegovic I, Danilovic B, Mancic S, Lazic M. High cell density cultivation of Bacillus subtilis NCIM 2063: modeling, optimization and a scale-up procedure. J Serb Chem Soc. 2023;88(11):1103-17. [CrossRef]
35. Schilling O, Frick O, Herzberg C, Ehrenreich A, Heinzle E, Wittmann C, Stulke J. Transcriptional and metabolic responses of Bacillus subtilis to the availability of organic acids: transcription regulation is important but not sufficient to account for metabolic adaptation. Appl Environ Microbiol. 2007;73(2):499-507. [CrossRef]
36. Naveed M, Tianying H, Wang F, Yin X, Chan MW, Ullah A, et al. Isolation of lysozyme producing Bacillus subtilis Strains, identification of the new strain Bacillus subtilis BSN314 with the highest enzyme production capacity and optimization of culture conditions for maximum lysozyme production. Curr Res Biotechnol. 2022;4:290-301. [CrossRef]
37. Kwon EY, Kim KM, Kim MK, Lee IY, Kim BS. Production of nattokinase by high cell density fed-batch culture of Bacillus subtilis. Bioprocess Biosyst Eng. 2011;34(7):789-93. [CrossRef]
38. Prats C, Giró A, Ferrer J, López D, Vives-Rego J. Analysis and IbM simulation of the stages in bacterial lag phase: basis for an updated definition. J Theor Biol. 2008;252(1):56-68. [CrossRef]
39. Mohamed H, Awad MF, Shah AM, Sadaqat B, Nazir Y, Naz T et al. Coculturing of Mucor plumbeus and Bacillus subtilis bacterium as an efficient fermentation strategy to enhance fungal lipid and gamma-linolenic acid (GLA) production. Sci Rep. 2022;12(1):13111. [CrossRef]
40. Effiom H, Lennox J. Optimization of independent variables for the production of extracellular alpha amylase by Bacillus subtilis IMD34 using Plackett-Burman design. Sultan Qaboos Univ J Sci. 2022;27(2):77-83. [CrossRef]
41. Yue H, Zhong J, Li Z, Zhou J, Yang J, Wei H, et al. Optimization of iturin A production from Bacillus subtilis ZK-H2 in submerge fermentation by response surface methodology. 3 Biotech. 2021;11(2). [CrossRef]
42. Zhong H, Wang L, Zhao J-y, Xiao Z. Fermentative production of chiral acetoin by wild-type Bacillus strains. Prep Biochem Biotechnol. 2019;50(2):116-22. [CrossRef]
43. Ullah A, Sun B, Wang F, Yin X, Xu B, Ali N, et al. Isolation of selenium-resistant bacteria and advancement under enrichment conditions for selected probiotic Bacillus subtilis (BSN313). J Food Biochem. 2020;44(6). [CrossRef]
44. Wang H, Sun X, Wang L, Wu H, Zhao G, Liu H, et al. Coproduction of menaquinone-7 and nattokinase by Bacillus subtilis using soybean curd residue as a renewable substrate combined with a dissolved oxygen control strategy. Ann Microbiol. 2018;68(10):655-65. [CrossRef]
45. Yánez-Mendizábal V, Falconí CE, Kanaley K. Production optimization of antifungal lipopeptides by Bacillus subtilis CtpxS2-1 using low-cost optimized medium. Biol Control. 2023;185:105306. [CrossRef]
46. Kanwal M, Wattoo AG, Khushnood RA, Liaqat A, Iqbal R, Song Z. Advancements and challenges in production of biosurfactants. In: Inamuddin, Charles Oluwaseun Adetunji, editors. Applications of Next Generation Biosurfactants in the Food Sector, Academic Press; 2023:239-59. [CrossRef]
47. Huang J, Pan Y, Zheng X, Lin H, Lin W, Liu J. Enhanced the biomass of Bacillus subtilis cultured in industrial wastewater through statistical methodologies. J Biotech Res. 2020;11:155-68.
48. Park MK, Lee S, Kim YS. Effects of pH and osmotic changes on the metabolic expressions of Bacillus subtilis strain 168 in metabolite pathways including leucine metabolism. Metabolites. 2022;12(2):112. [CrossRef]
49. Petrackova D, Vecer J, Svobodova J, Herman P. Long-term adaptation of Bacillus subtilis 168 to extreme pH affects chemical and physical properties of the cellular membrane. J Membr Biol. 2010;233(1-3):73-83. [CrossRef]
50. Sánchez-Clemente R, Guijo MI, Nogales J, Blasco R. Carbon source influence on extracellular pH changes along bacterial cell-growth. Genes. 2020;11(11):1292. [CrossRef]
51. Singh V, Haque S, Niwas R, Srivastava A, Pasupuleti M, Tripathi C. Strategies for fermentation medium optimization: an in-depth review. Front Microbiol. 2017;7:2087. [CrossRef]
52. Odds FC, Hall CA, Abbott AB. Peptones and mycological reproducibility. Med Mycol. 1978;16(4):237-46. [CrossRef]
53. Yadav J, Balabantaray S, Patra N. Statistical optimization of fermentation conditions for the improved production of poly-β-hydroxybutyrate from Bacillus subtilis. Chem Eng Commun. 2017;204(10):1122-8. [CrossRef]
54. Ghasemi S, Ahmadzadeh M. Optimization of a cost-effective culture medium for the large-scale production of Bacillus subtilis UTB96. Arch Phytopathol Pflanzenschutz. 2013;46(13):1552-63. [CrossRef]
55. Uzeh RE, Akinola SO, Olatope SO. Production of peptone from soya beans (Glycine max L merr) and African locust beans (Parkia biglobosa). Afr J Biotechnol. 2006;5(18).
56. Rochmaningsih, Harismah K, Mulyaningtyas A. Optimization of peptone production from soybeans. Proceedings of the 4th International Conference Current Breakthrough in Pharmacy (ICB-Pharma 2022); 2022;295-305. Atlantis Press. [CrossRef]