1. INTRODUCTION
Poultry industries are increasing rapidly day by day. The growth of this sector is good as it provides career opportunities to the people, but problems regarding waste management also come with it. Poultry waste includes chicken feathers, chicken poop, bedding material, split feed, and hatchery material. Keratin comes primarily from the poultry industry. In actuality, the global poultry business processes about 400 million chickens/week, resulting in approximately 5 million tons of feathers [1]. These cause the problem regarding waste disposal. As a result of their wide range of uses, keratinase are earning a lot of interest. They are used in waste treatment, agricultural, pharmaceutical, and biomedical areas right now and in the future. Nanobiotechnology, leather, cosmetics, and detergent manufacturing industries are some of the other sectors. Several microbial strains with exceptional feather-degrading abilities could be valuable in the management of feather waste. There is also environmental pollution due to the recalcitrant character of keratinaceous substances [2].
Keratin, an insoluble protein macromolecule with exceptionally high stability and a low breakdown rate, is the main component of chicken feathers. Hair, feathers, nails, wool, and horns are the primary components that contain keratin [3]. Keratin waste contains a high amount of protein content, which can be utilized as a source of amino acids and proteins by recycling it. The feathers can be recapitulated, which may provide an alternative and inexpensive protein meal. Furthermore, this protein meal stuff can be used for animals’ fodder. Yet, keratin is not digested easily as it is difficult to recycle [4]. An extracellular enzyme, keratinase, is used for the biodegradation of keratin. Normally, microbial keratinase is generated in the presence of a keratin substrate. To degrade keratin substrate, this enzyme primarily destroys its disulfide bond. Microorganisms that produce keratinase are capable of destroying chicken feathers, hair, nails, wool, and other materials [5].
In soil collected from sites where poultry waste is dumped, keratinolytic bacteria were isolated and identified. Selvam et al. (2013) show that keratin is resistant to peptidases such as pepsin, papain, and trypsin [6]. The protein chains in keratin are tightly packed either into ?-helix or into β-sheet structures that fold into three-dimensional form [7]. One of the important characteristics of keratin is that it has high mechanical stability due to the presence of hydrogen and disulfide bonds [8]. Keratin is also a prime component of the epidermis and its appendages, such as nails, horns, hoofs, hair, feathers, scales, and wool. According to their sulfur content, keratins are classified into two types; one is hard keratins and the other is soft keratins. The hard keratins are found in appendages, while the soft keratins are found in skin and callus. In soft keratin, there is a low content of disulfide bonds [6]. Keratinolytic enzymes are generated by bacteria, fungi, and actinobacteria that have been commonly isolated from soils, where keratinous substances are accrued. Normally, thermophilic microorganisms are related with increased keratinolytic activity, microorganisms that decompose keratin wastes [9].
Feathers are generally composed of over 90% protein and are produced in large quantities as a waste product by poultry processing industries worldwide. Feathers accumulating in the environment may lead to wastage of feather protein [10]. Various keratinous substances such as feathers, wool, and hair can be used as substrates for the production of keratinase [5]. Bacterial keratinase has potential for the degradation of exclusive insoluble keratin substrates and on wide kinds of protein materials [11]. This enzymes function as pioneer biocatalysts which have various avail in different sectors such as in cosmetics industries, detergent industries, leather industries, and textile industries. This enzyme also works on drug delivery systems, production of biodegradable integument, or bio-processing of used X-ray integument and is also used in agroindustrial waste degradation [12,13].
In many ecological environments, a cluster of microbes that can produce keratinase enzymes and break down keratinous waste includes bacteria, saprophytic fungus, actinomycetes, and dermatophytes [14]. Therefore, the present research work was an attempt to isolate keratinase-producing microorganisms from the selected 30 poultry waste dumping soils, screen, and characterize keratinase-producing microbes from the poultry dumping sites of Himachal Pradesh.
2. MATERIALS AND METHODS
2.1. Collection of poultry waste soil samples
A total of 30 poultry waste soil samples were collected from local poultry processing industries in Himachal Pradesh, India. The rotted feathers and poultry waste soil samples were collected from regular poultry waste dumping sites. From a depth of 3–4 cm, samples of poultry soil with deteriorated feathers were taken and put into sterile plastic bags [15]. The samples of poultry waste soil were brought to a laboratory for further processing. The details of sample collection are shown in Table 1.
Table 1: List of poultry waste soil samples collected from different locations in Himachal Pradesh.
| S. No. | Sampling site | Soil Sample Name |
|---|---|---|
| 1. | Hills Poultry Farm, Dharwad Pater Talwara Chowk, Kuthera Distt.-Bilaspur H.P. 174026. | KP |
| 2. | Hemraj Poultry Farm, VPO- Hartlyalagar, Distt.- Bilaspur. | HT |
| 3. | Him Chicken Corner, Bus Stand, Ghumarwin, Bilaspur H.P. 174021 | HC |
| 4. | Jai Maa Kali, Kadaknath Poultry Farm Jahu, Mandi (H.P.) 177118 | JP |
| 5. | Chauhan Poultry and Hatchery, Vill- Lunapani, P.O. Bhangrotu, Mandi | CP |
| 6. | Him Hatchery Govt. Hatchery, Sundernagar, Mandi, (H.P.) | HH |
| 7. | The Happy Village Hens Farm and Hatchery, Khungan, P.O. – Kaphara, Bilaspur, (H.P.) | HV |
| 8. | Vishal’s Farm Near, Dakri Chowk, Ghumarwin Distt.- Bilaspur (H.P) | VP |
| 9. | Chauhan Poultry Farm, Township Dhanotu Chowk, Mandi (H.P.) 174402 | DP |
| 10. | Sandha Desi Murgi Poultry Farm, Hamirpur, (H.P.) | SD |
| 11. | Poultry Farm (Haroli) Chicken Hatchery, Palakwah (H.P.) | HR |
| 12. | Akib Poultry Farm Mahadev,(H.P.) 175019 | AP |
| 13. | Birbal Poultry Farm, Gagal, Mahadev Nagar, Mandi 175121 | BP |
| 14. | Naitre Singh Poultry Farm, Dugrain Teh. & P.O.- Chachiot 175020 | NP |
| 15. | Mahinder Singh Poultry Farm, Naugraon, Near Abhilashi University P.O.- Chailchowk, Mandi, (H.P.) | MP |
| 16. | Thakur Poultry Farm, Hamirpur H.P. 177301 | TP |
| 17. | VK Poultry Farm Vill.- Loharli P.O.- Jahallan Nadaun Hamirpur 177301 | VK |
| 18. | Pride dairy &Poultry Farm Galore, Hamirpur 177001 | PDP |
| 19. | Kaku Poultry Farm & Poultry House Palampur | KPF |
| 20. | Shiv G Poultry Farm Sundernagar (H.P.) | SP |
| 21. | Poultry Farm Haroli, Una Palakwah. 177220 | PH |
| 22. | Dr. Kuldeep’s desi Poultry and Kadaknath Poultry | DKP |
| 23. | Kadaknath Poultry Farm, Man Simble, Uparla, H.P. 176083 | MS |
| 24. | Chicken Neck Poultry Farm, Raakh, Baijnath, Kangra (H.P.) 176071 | CNP |
| 25. | Namay Desi Poultry Farm, Ladraur, Bari Chowk (H.P.) 174028 | LD |
| 26. | Rajput Abhishek Jamwal Poultry Farm, Vill. Samlohal Teh.- Ghumarwin, Bilaspur (H.P.) 174029 | SG |
| 27. | Patyal Poultry Farm, Vill. Bhallu P.O. Berthin Teh.- Jhandutta, Bilaspur (H.P.) 174029 | PP |
| 28. | Anurag Poultry and Fish Farm, Soyad Khad near Sandyar Androli, Chhat, H.P. 174029 | SA |
| 29. | AL Nadeem Kadknath Farm Danoh, Bilaspur H.P. 174001 | AB |
| 30. | AQSA Chicken Shop Ghumarwin, Bilaspur, (H.P.) 174021 | AQG |
2.2. Isolation of Microbes from Poultry Soils
Microorganisms were isolated from poultry waste soil samples by the serial dilution and plating method on skimmed milk agar. Ten grams of poultry waste soil samples were mixed thoroughly in 90 mL of sterilized feather basal medium. For 24 h, the flasks were placed in a rotator shaker incubator at 120 rpm. The sample was serially diluted up to a 10-7 dilution. The diluted sample in a 0.1 mL volume was inoculated in skimmed milk medium and spread evenly on petri plates using a spreader. Plates had a 24- to 48-h incubation period. At 37°C [16], the colonies were purified using the streak plate method. A pure culture was obtained by further subculturing the isolated colonies on nutrient agar [17].
2.3. Primary Screening of Proteolytic Bacteria
The skimmed milk agar medium was then inoculated with the microbial isolates. The inoculated plates were incubated in the incubator at 37°C for 24 h, and the observed positive appearance of a clear zone around the colony suggested the proteolytic activity [18]. The diameter of each zone was recorded. The microbial colonies that have produced large zones of hydrolysis were, further, selected for secondary screening. Pure bacterial isolates were kept for further research on nutrient agar medium slants at 4°C.
2.4. Secondary Screening of Keratinolytic Bacteria
The bacterial isolates with a clear zone around the inoculum were further used for secondary screening. The isolates were put in a base medium that also contained chicken feathers. The cultures were inoculated, the pH was adjusted to 8.0, and they were incubated at 37°C on a rotary shaker at a speed of 150 rpm [19].
2.5. Characterization of Isolates
The isolated bacteria were identified based on their molecular makeup as determined by 16S rRNA sequencing, Gram staining, biochemical assays, and culture features. The color, shape, size, kind of colony, and pigmentation of the colony were all noted [20]. The bacterial isolates were gram stained and examined in a light microscope with a high power magnifying lens [21]. The bacterial isolates were characterized biochemically by the Indole test, methyl red test, Voges–proskauer’s test, Citrate utilization test, motility test, catalase test, Oxidase test, Nitrate Reductase, Urease, H2S production, and Carbohydrate test [22]. The selective isolates with maximum keratinase producing activity were further characterized for 16S rRNA gene sequencing and phylogenetic analysis.
2.6. Isolation of Bacterial Genomic DNA
In 5 ml of sterile feather basal media, purified bacterial strains were inoculated and allowed to grow overnight. The next day, the cells were collected by centrifugation for 5 min at 10,000 rpm. In 1 mL of extraction buffer (100 mM Tris HCl pH 8.0, 50 mM EDTA pH 8.0, 500 mM NaCl, 0.07% -mercaptoethanol, and 20 mg/ml lysozyme), the bacterial cell pellets were once again re-dissolved. Following this, 50 μl of 10% SDS was added, carefully mixed, and then incubated for 30 min at 65°C. The samples were spun at 10,000 rpm for 20 min, and the supernatant was then poured into a fresh tube. Each sample was treated with RNase (1–2 μL of 1 mg/mL stock) and, then, incubated at 37°C for an hour. The samples were extracted 3 times using an equal volume of chloroform/tris-saturated phenol (1:1). DNA was precipitated by mixing 2.5 volumes of pure ethanol and 1/10 sample volume of 5 M NaCl. The samples were thoroughly mixed before being centrifuged for 20 min at 12,000 rpm after being incubated at −20°C for 2 h. The DNA pellet was cleaned with 1 mL of 70% ethanol, allowed to air dry, and then dissolved in TE (10 mM Tris HCl, 1mM EDTA) buffer. It was, then, stored at −20°C until it was needed again [23].
2.7. Sequence Analysis of the 16S rRNA Gene
Strain analysis was done through a comparative genomic method using 16S rRNA gene sequencing. Gene sequencing was performed by Shankarnarayan Life Sciences, Ltd. Bengaluru. The primers used for 16S rDNA amplification were fP1 (5’-GGGGGATCCAGAGTTTGATCCTGGCTCA-3’) and rP2 (3’- GGGGGATCCAGAGTTTGATCCTGGCTCA-5’) (Weisburg et al. 1991). The thermocycling settings included a preliminary denaturation stage at 94 °C for 5 min, 33 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min, and a concluding extension step at 72°C for 10 min. MEGA 11 used the Neighbor-Joining (N-J) method to build the phylogenetic tree [24,15].
2.8. Submission of gene sequences
The 16S rDNA sequence of potential keratinase producing bacterial isolates TP1, JP1, VP4, and PP1 were deposited in the GenBank database under the accession number OP748783, OP748782, OP420549, and OP748781 respectively.
3. RESULTS
3.1. Isolation of keratinase producing bacteria from poultry waste soil samples
The poultry waste soil samples were collected from 30 different local poultry farming sites in Himachal Pradesh [Table 2]. For isolation of microbes 10 g of poultry, waste soil sample was transferred to 90 mL of sterilized feather basal medium. 1 mL of soil suspension serially diluted up to 7 times. The diluted sample in 0.1 mL volume from each dilution was transferred to skimmed milk agar plates. After 24 h of incubation, the microbial diversity appears on the skimmed milk agar plates, as shown in Figure 1. A total of 64 well-formed single colonies were selected on the basis of their morphological differences. All the 64 isolates were subjected to primary screening on skimmed milk agar plates. Out of 64 isolates, 17 isolates named KP1, HT2, HT3, JP1, HH1, VP3, VP4, AP2, BP1, NP2, MP1, TP1, SP3, DKP1, LD1, and PP1 demonstrated a clear zone on skimmed milk agar plates after 24 h of incubation at 37°C. The zone size was recorded in mm after 48 h of incubation [Figure 2 and Table 3].
Table 2: Details of bacterial isolates isolated from different soil samples.
| S. No. | Soil Sample | Isolated bacteria on nutrient agar | Isolates showing keratinolytic activity on skimmed milk agar |
|---|---|---|---|
| 1. | KP | KP1, KP2, KP3 | KP1 |
| 2. | HT | HT1, HT2, HT3 | HT2, HT3 |
| 3. | HC | HC1 | Not Detected |
| 4. | JP | JP1, JP2 | JP1 |
| 5. | CP | CP1 | Not Detected |
| 6. | HH | HH1, HH2, HH3 | HH1 |
| 7. | HV | HV1 | Not Detected |
| 8. | VP | VP1, VP2, VP3, VP4 | VP3, VP4 |
| 9. | DP | DP1, DP2 | Not Detected |
| 10. | SD | SD1, SD2 | Not Detected |
| 11. | HR | HR1 | Not Detected |
| 12. | AP | AP1, AP2 | AP2 |
| 13. | BP | BP1, BP2, BP3 | BP1, BP3 |
| 14. | NP | NP1, NP2, NP3 | NP2 |
| 15. | MP | MP1, MP2, MP3 | MP1 |
| 16. | TP | TP1, TP2, TP3 | TP1 |
| 17. | VK | VK1, VK2 | Not Detected |
| 18. | PDP | PDP1, PDP2 | Not Detected |
| 19. | KPF | KPF1, KPF2 | Not Detected |
| 20. | SP | SP1, SP2, SP3 | SP3 |
| 21. | PH | PH 1, PH 2 | Not Detected |
| 22. | DKP | DKP1, DKP2, DK3 | DKP1 |
| 23. | MS | MS1, MS2 | Not Detected |
| 24. | CNP | CNP1 | Not Detected |
| 25. | LD | LD1, LD2 | LD1 |
| 26. | SG | SG1, SG2 | Not Detected |
| 27. | PP | PP1, PP2 | PP1 |
| 28. | SA | SA1 | Not Detected |
| 29. | AB | AB1 | Not Detected |
| 30. | AQG | AQG1, AQG2 | Not Detected |
| Total isolates | 64 | 17 | |
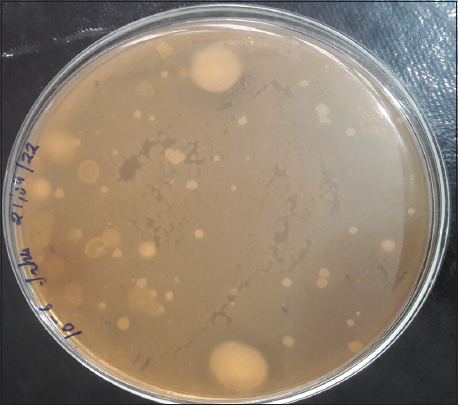 | Figure 1: Microbial diversity on skimmed milk agar plate. [Click here to view] |
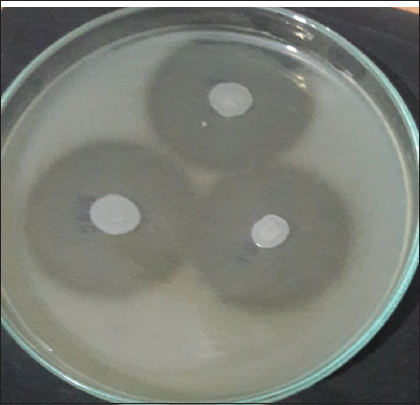 | Figure 2: Zone of clearance around the bacterial colonies after 48 h of incubation. [Click here to view] |
3.2. Secondary Screening
Secondary screening was done to find out the feather degradation capability of selected seventeen isolates. For secondary screening, basal media enriched with feathers were inoculated with KP1, HT2, HT3, JP1, HH1, VP3, VP4, AP2, BP1, BP3, NP2, MP1, TP1, SP3, DKP1, LD1, and PP1. The isolates use feathers as a single carbon and nitrogen source. After 15 days of incubation periods at 37°C, it was found that there was complete degradation of feathers by the isolates [Figure 3 and Table 3].
 | Figure 3: Feather degradation test for selected keratinolytic bacterial isolates (a) initial stage (b) after 15 days of incubation. [Click here to view] |
Table 3: Bacterial isolates showing keratinolytic activities on skimmed milk agar and feather basal media.
| S. No. | Isolates | Diameter of zone of clearance around the colonies after 48 h on skimmed milk agar (mm) | Rate of feather degradation on feather basal broth media (after 15 days)* |
|---|---|---|---|
| 1. | KP1 | 24 | +++++ |
| 2. | HT2 | 24 | +++++ |
| 3. | HT3 | 28 | +++++ |
| 4. | JP1 | 28 | +++++ |
| 5. | HH1 | 26 | ++++ |
| 6. | VP3 | 32 | ++++ |
| 7. | VP4 | 28 | +++++ |
| 8. | AP2 | 30 | ++++ |
| 9. | BP1 | 32 | ++++ |
| 10. | BP3 | 30 | ++++ |
| 11. | NP2 | 24 | ++++ |
| 12. | MP1 | 28 | ++++ |
| 13. | TP1 | 20 | +++++ |
| 14. | SP3 | 14 | + |
| 15. | DKP1 | 32 | ++++ |
| 16. | LD1 | 28 | +++++ |
| 17. | PP1 | 32 | +++++ |
* (+) low degradation; (++++) moderate degradation; (+++++) complete degradation
3.3. Biochemical and Morphological Identification of Keratinolytic Bacteria
The characterization of all seventeen keratinase producer microbes was done on the basis of morphological and biochemical tests. Seven isolates belong to Gram-positive, rod-shaped bacteria and ten isolates were identified as Gram-negative, rod-shaped bacteria Table 4. The morphological and biochemical characterization were compared with Bergey’s manual of determinative bacteriology [Table 5].
Table 4: Colony morphology and microscopic characterization of keratinolytic bacterial isolates.
| Colony morphology and microscopic characterization | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Isolates | Colony Characteristics | Gram staining | |||||||
| Size | Shape | Margin | Texture | Elevation | Opacity | Color | |||
| KP1 | Large | Irregular | Uneven | Smooth | Raised | Opaque | White | + | Rod Shaped |
| HT2 | Large | Irregular | Uneven | Rough | Raised | Opaque | White | + | Rod Shaped |
| HT3 | Small | Irregular | Even | Smooth | Raised | Opaque | White | + | Rod Shaped |
| JP1 | Medium | Round | Even | Smooth | Raised | Opaque | Cream | + | Rod Shaped |
| HH1 | Medium | Round | Uneven | Rough | Raised | Opaque | White | - | Rod Shaped |
| VP3 | Small | Irregular | Even | Smooth | Raised | Opaque | Cream | - | Rod Shaped |
| VP4 | Medium | Irregular | Entire | Smooth | Raised | Opaque | White | + | Rod Shaped |
| AP2 | Medium | Irregular | Uneven | Smooth | Raised | Opaque | Cream | - | Rod Shaped |
| BP1 | Medium | Round | Even | Rough | Flat | Opaque | White | - | Rod Shaped |
| BP3 | Large | Round | Uneven | Smooth | Raised | Opaque | White | - | Rod Shaped |
| NP2 | Medium | Round | Uneven | Smooth | Umbonate | Opaque | White | - | Rod Shaped |
| MP1 | Small | Irregular | Uneven | Rough | Umbonate | Opaque | White | - | Rod Shaped |
| TP1 | Large | Irregular | Uneven | Smooth | Umbonate | Opaque | White | + | Rod Shaped |
| SP3 | Medium | Round | Uneven | Smooth | Raised | Opaque | White | - | Rod Shaped |
| DKP1 | Large | Round | Uneven | Smooth | Raised | Opaque | White | - | Rod Shaped |
| LD1 | Medium | Round | Uneven | Smooth | Flat | Opaque | White | - | Rod Shaped |
| PP1 | Large | Round | Uneven | Smooth | Flat | Opaque | White | + | Rod Shaped |
+: Gram-positive; -: Gram-negative
Table 5: Biochemical characterization of keratinolytic bacteria.
| Isolates | Indole test | Methyl Red test | Vogas Proskaur test | Citrate Test | Motility Test | Catalase Test | Oxidase Test | Nitrate Reductase | Urease | H2S Production | Dextrose fermentation | Lactose fermentation | Sucrose fermentation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KP1 | - | + | - | - | + | - | - | + | - | - | + | - | - |
| HT2 | + | + | - | - | + | - | - | + | - | - | + | - | - |
| HT3 | + | + | - | - | + | - | + | + | + | - | + | - | - |
| JP1 | - | - | + | + | + | + | - | + | - | - | + | - | - |
| HH1 | + | + | - | - | + | - | - | + | - | - | + | - | - |
| VP3 | - | + | - | - | + | - | - | + | - | - | + | - | - |
| VP4 | + | + | - | + | + | - | + | + | - | - | - | - | - |
| AP2 | - | + | - | - | + | - | - | + | - | - | - | - | - |
| BP1 | + | + | - | - | + | - | - | + | - | - | + | - | - |
| BP3 | + | + | - | - | + | - | - | + | - | - | + | - | - |
| NP2 | - | + | - | - | + | - | + | + | + | - | + | - | - |
| MP1 | - | + | - | + | + | + | + | + | - | - | - | - | - |
| TP1 | - | - | + | + | + | + | - | + | + | - | - | - | - |
| SP3 | + | - | - | - | + | + | + | + | - | - | - | - | - |
| DKP1 | + | + | + | - | + | - | - | + | - | - | - | - | - |
| LD1 | - | + | - | - | + | - | - | + | - | - | - | - | - |
| PP1 | - | - | + | + | + | + | - | + | - | - | + | - | - |
+: Positive, -: Negative
3.4. Molecular Identification of Keratinase Producing Bacteria
The bacterial isolates, namely, JP1, TP1, VP4, and PP1 exhibited maximum keratinase producing activity and were selected for molecular characterization through 16S rRNA gene sequencing and phylogenetic analysis [Figures 4 and 5]. Using the Bioedit tool, the forward and reverse primer results were combined to extract the entire aligned sequences. The NCBI BLAST was performed to confirm the identity of specimens [25]. Utilizing BLAST, the 16S rRNA gene sequence was compared to the Gen-bank database. The isolates TP1, JP1, VP4, and PP1 displayed high levels of sequence similarity with members of the genus Bacillus in their 16S rRNA gene sequences. The NCBI database was queried using the “BLASTn” tool to assign sequences, and the highest-scoring hit from each query was used to identify the bacterial species. A phylogenetic tree was constructed using MEGA 11 [Figure 5]. The bacterial strains JP1, TP1, and PP1 were identified as Bacillus cereus, while strain VP4 was identified as Bacillus thuringiensis.
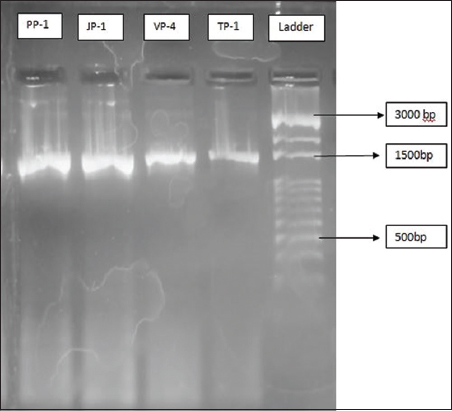 | Figure 4: Polymerase chain reaction amplification of 16S rDNA fragment from bacterial samples. [Click here to view] |
 | Figure 5: Phylogenic tree of strain VP4, JP1, TP1, and PP1 constructed by Neighbor-Joining method derived from analysis of the 16S rRNA gene sequences and related sequences obtained from NCBI. Scale bar, 0.050 substitutions per nucleotide. [Click here to view] |
4. DISCUSSION
Feathers have more than 90% protein. Due to insoluble nature of keratinous materials such as feather, hair, and wool, they are resistant to degradation by common microbial proteases. Thus, the annual production of several million tons of feathers by the chicken sector causes troublesome environmental contamination and waste of protein-rich reserve [10,26].
It has been observed that keratinase enzymes are a powerful enzyme for breaking down feathers, hair, and poultry waste. In this current research work, a total of 64 bacterial isolates were isolated from poultry waste soil samples from 30 different poultry farming industries in Himachal Pradesh, India. Another author covered the isolation of the Bacillus spp. from the wastewater of a poultry processing plant, as well as its growing conditions and keratinolytic activity [8].
Seventeen isolates from poultry waste has been shown to degrade feather keratin. Seventeen isolates exhibited the zone of clearance around the colonies on skimmed milk agar plates. On the basis of the zone of clearance, the isolates were grown in basal media, in which chicken feathers were used as a source of carbon and nitrogen. Although these 17 isolated strains have shown the significant property to degrade the feathers which are sophisticated to achieve. Isolates KP1, HT2, HT3, JP1, HH1, VP3, VP4, AP2, BP1, NP2, MP1, TP1, SP3, DKP1, LD1, and PP1 completely degraded the chicken feathers. However, in comparison to other isolates, strain SP3 has demonstrated dissimilar characteristics in the same feather basal medium as it has taken longer time for feather degradation. The degradation rate has been introduced by many authors as an indicator evidently correlated to the keratinase activity of a microbial strain [27].
After biochemical tests, these microorganisms were confirmed to belong to the Bacillus and Pseudomonas species. The previous studies have documented the isolation of keratinase-producing strains of Bacillus licheniformis, B. subtilis, B. pumilis, B. cereus, B. halodurans, B. pseudoformis [28], and Pseudomonas sp. [29]. In another study, Arthrobacter sp. was also reported to produce keratinase [30]. Arthrobacter creatinolyticus KP015744 was observed to be a better keratinase producer microorganism [31]. All biochemical tests for results are equivalent to those reported by various authors who proved that Gram-positive bacteria, inclusive of Bacillus, can hydrolyze keratin [5]. Likewise, species of Bacillus are considered the best producers of keratinase. Bacillus sp. has been identified as a potential keratinolytic organism in the previous research for the isolation of keratinolytic organisms from soil and other natural sources [32]. These studies suggest that this organism may be used in field studies for the biodegradation of feathers in feather processing units. Keratinases also have applications in personal care products to form skin whitening and bleaching creams [33].
The 16S rRNA sequencing results of these isolates demonstrate high levels of sequence similarity with genus Bacillus. For phylogenetic analysis, small subunit of rRNA is used that is extremely faithful method to determinate the taxonomic relationships between microorganisms [34]. According to both biochemical characterization and 16S rRNA gene sequence analysis, the JP1, TP1, PP1, and VP4 strains were identified as B. cereus and B. thuringenesis, respectively.
5. CONCLUSION
These keratinolytic isolates have the potential to degrade the feather keratin, which can be used to make animal feed proteins, can be used to make slow nitrogen-releasing fertilizers, and can also overcome the problems regarding waste management, because poultry waste causes environmental pollution. Therefore, the microorganisms isolated in this study exhibited potential for biotechnological uses.
6. ACKNOWLEDGMENT
I am grateful to Dr. Vijay Kumar, Assistant Professor, Career Point University, Hamirpur, for giving the possibility to complete this research work. I would like to give special thanks to Kehloor Biosciences and Research Center, Ghumarwin, India for their assistance in the practical work.
7. AUTHORS’ CONTRIBUTIONS
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Richa and Vijay Kumar. The first draft of the manuscript was written by Richa and second author commented on previous versions of the manuscript. All authors read and approved the final manuscript.
8. FUNDING
The authors received no financial support for this research work.
9. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
Most of the data supporting our findings included in the manuscript.
12. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Godheja J, Shekhar SK, Modi DR. Biodegradation of keratin from chicken feathers by fungal species as a means of sustainable development. Asian J Pharm Technol 2014;4:69-73. [CrossRef]
2. Lateef A, Oloke JK, Kana EG, Sobowale BO, Ajao SO, Bello BY. Keratinolytic activities of a new feather-degrading isolate of
3. Qiu J, Wilkens C, Barrett K, Meyer AS. Microbial enzymes catalyzing keratin degradation:Classification, structure, function. Biotechnol Adv 2020;44:107607. [CrossRef]
4. Jana P, Saha RP, Das A. A review on isolation of keratin protein from non-conventional resources and its application in daily diet to enhance hair quality. In:Advances in Medical Physics and Healthcare Engineering. Singapore:Springer;2021. 405-14. [CrossRef]
5. Gupta R, Ramnani P. Microbial keratinases and their prospective applications:An overview. Appl Microbiol Biotechnol 2006;70:21-33. [CrossRef]
6. Selvam K, Vishnupriya B, Yamuna M. Isolation and description of keratinase producing marine actinobacteria from South Indian coastal region. Afr J Biotechnol 2013;12:19-26. [CrossRef]
7. Sharma R, Devi S. Versatility and commercial status of microbial keratinases:A review. Rev Environ Sci Bio Technol 2018;17:19-45. [CrossRef]
8. Matikeviciene V, Masiliuniene D, Grigiskis S. Degradation of keratin containing wastes by bacteria with keratinolytic activity. Environ Technol Resour 2009;1:284-9. [CrossRef]
9. Nam GW, Lee DW, Lee HS, Lee NJ, Kim BC, Choe EA,
10. Gousterova A, Braikova D, Goshev I, Christov P, Tishinov K, Tonkova VE,
11. Lin X, Shih JC, Swaisgood HE. Hydrolysis of feather keratin by immobilized keratinase. Appl Environ Microbiol 1996;62:4273-5. [CrossRef]
12. Mohanapriya M, Parvathi L, Archana B, Suneetha VA. potential beta-keratin degrading bacteria from Vellore Emu feather dumped soil. Int J Pharm Sci Rev Res 2014;25:224-8.
13. Lucas FS, Broennimann O, Febbraro I, Heeb P. High diversity among feather-degrading bacteria from a dry meadow soil. Microb Ecol 2003;45:282-90. [CrossRef]
14. Tork S, Aly MM, Nawar L. Biochemical and molecular characterization of a new local keratinase producing
15. Prasad VH, Kumar G, Kartik L, Rao KV. Screening of extracellular keratinase producing bacteria from feather processing areas in Vellore, Tamil Nadu, India. J Sci Res 2010;2:559. [CrossRef]
16. Jayasudha J, Kumar G, Karthik L, Rao KV. Biological control of vibriosis by antagonistic actinobacteria-an
17. Sunita JV, Dolly PR, Bateja S, Vivek NU. Isolation and screening for hydrocarbon utilizing bacteria (HUB) from petroleum samples. In J Curr Microbiol Appl Sci 2013;2:48-60.
18. Jeong JH, Park KH, Oh DJ, Hwang DY, Kim HS, Lee CY,
19. Al Hasan MM. Isolation and Identification of Extracellular Keratinase Producing Bacteria from Soil. Bangladesh:BRAC University;2018.
20. Ashwini K, Gaurav K, Karthik L, Rao KV. Optimization, production and partial purification of extracellular a-amylase from
21. Sen SK, Raut S, Satpathy S, Rout PR, Bandyopadhyay B, Mohapatra PK. Characterizing novel thermophilic amylase producing bacteria from Taptapani hot spring, Odisha, India. Jundishapur J Microbiol 2014;7:e11800. [CrossRef]
22. Uddin ME, Maitra P, Faruquee HM, Alam MF. Isolation and characterization of proteases enzyme from locally isolated
23. Allpress JD, Mountain G, Gowland PC. Production, purification and characterization of an extracellular keratinase from
24. Saitou N, Nei M. The neighbor-joining method:A new method for reconstructing Phylogenetic trees. Mol Biol Evol 1987;4:406-25.
25. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215:403-10. [CrossRef]
26. Onifade AA, Al-Sane NA, Al-Musallam AA, Al-Zarban S. A review:Potentials for biotechnological applications of keratin-degrading microorganisms and their enzymes for nutritional improvement of feathers and other keratins as livestock feed resources. Bioresource Technol 1998;66:1-11. [CrossRef]
27. Veenayohini K, Sangeetha D. Isolation and identification of keratinolytic bacteria from poultry waste and assessment of its keratinase activity on chicken feathers. Int J Appl Res 2016;2:396-402.
28. Gessesse A, Hatti-Kaul R, Gashe BA, Mattiasson BO. Novel alkaline proteases from alkaliphilic bacteria grown on chicken feather. Enzyme Microb Technol 2003;32:519-24. [CrossRef]
29. Riffel A, Brandelli A. Keratinolytic bacteria isolated from feather waste. Braz J Microbiol 2006;37:395-9. [CrossRef]
30. Pereira JQ, Lopes FC, Petry MV, Medina LF, Brandelli A. Isolation of three novel Antarctic psychrotolerant feather-degrading bacteria and partial purifcation of keratinolytic enzyme from
31. Kate S, Pethe A. Study of efficiency of keratinase production by
32. Mazotto AM, de Melo AC, Macrae A, Rosado AS, Peixoto R, Cedrola SM,
33. Gupta R, Rajput R, Sharma R, Gupta N. Biotechnological applications and prospective market of microbial keratinases. Appl Microbiol Biotechnol 2013;97:9931-40. [CrossRef]
34. Iwamoto K, Yoshikawa T, Sakata T, Maeda H. Phylogenetic analysis of marine algicidal filamentous bacteria inferred from SSU rDNA and intergenic spacer regions. Microbes Environ 2001;16:213-21. https://doi.org/10.1264/jsme2.2001.213