1. INTRODUCTION
Plastic, one of the tenacious and indispensible materials in our lives and backbone of many industries, is ideally suited for a wide variety of applications to the modern generations. Plastics flaunt an unparalled and useful set of properties, namely, durability, strength, light weightedness, water and corrosion-resistance, cheapness, and relative ease in processing making it economical and superior over other materials [1]. Since its approbation, its consumption and production have escalated exponentially leading to beefing up in polymer fabrication and consequently in waste generation [2-4]. Worldwide annual plastic production scaled up from 2 Mt in 1950 to 367m tones in 2020, according to trade association Plastics Europe and is contemplated to further spike in the coming decades outweighing all the fishes in the sea by 2050.
Plastics and their associated compounds exert multitudinous biological impacts on environmental microbiomes. Manufacturing plastic takes heavy toll on fossil fuels extraction, with all the pollution risks that encompasses. Production and incineration of plastic vents greenhouse gases that significantly contribute to global warming [5]. In addition, the accumulation of such polymers which are non-biodegradable due to reprehensible waste management and disposal in the soil leads to low fertility.
Globally, out of total plastics produced, it has been estimated that only one-tenth of plastics undergo recycling, 14% incinerated and the remaining 76% finds their way to landfills or sneak into the natural environment [6]. This anthropogenic detritus have inimically affected life on the planet, as well as probity and sustainability [7]. Under the impact of solar UV radiation along with other natural factors, plastic disintegrates into smaller fragments called microplastics or nanoplastics. There are spectra of pernicious impacts that microplastics impinges on the environment [8]. Over the past decade, there have been series of efforts made to keep an eye on impacts of microplastics on the marine environment. According to an investigation, every year, 100,000 tons of plastic products have been jettisoned into the aquatic environment, affecting marine flora and fauna [9]. The accruing of waste spawns problems on all scales right from the ingestion of microscopic particles and the entanglement and throttling of unsuspected animals, benefacting the transport of invasive species across habitats on floating rubble posing significant hazards to marine life [10]. Choking of digestive tracts, deterioration of stomach mucosal lining, or diminishing appetite due to ingestion of plastics and injuries inflicted, starvation due to occlusion of esophagus, and suffocation because of respiratory tract strangulation, all may result due to entanglement of marine animals by plastic debris. Some microplastics have been shown to contain compounds that are known to be carcinogenic and mutagens that intermeddle with the body’s organ systems, causing various disorders in both humans and wild animals [11,12]. These chemicals may enter the food chain and results in bioaccumulation at multiple trophic levels [13].
In light of the growing concern about the deleterious repercussions of these non-biodegradable plastics on environmental and human health, microorganisms should be considered extensively for remediation of plastic pollution in the environment. Recently, a handful of microorganisms were reported to eat certain plastics, breaking them down into their constituent molecules. These microorganisms can form biofilms on the periphery of pollutants resulting in a zone called plastisphere where they interact and engender acids or enzymes for the ruin of plastics. These miniature organisms represent a great vision to cope waste plastic materials with no uninvited impacts and could quickly play a key role in building a greener economy. Hence, this review article has been prepared to put in the picture the role of microbes in the biodegradation of plastic waste.
2. IMPACT OF PLASTICS ON ENVIRONMENT
From the Arctic to the Antarctic, plastic garbage can be found all over the planet. It clogs municipal sewers, pollutes campgrounds and national parks, and even builds up on Mount Everest. However, due to runoff and our habit of throwing rubbish into the nearby river or lake, plastic is becoming more prevalent in the world’s oceans. The amount of plastic debris in the oceans is so large that it is referred to as the “seventh continent.” If present trends persist, the oceans will have more plastic than fish by 2050. Floating plastic even generates vast “trash patches” in the Pacific Ocean middle, thousands of kilometers from land, as that flotilla of freed rubber duckies proved [14]. Mountains of plastic debris have been found all across the world’s oceans, from Henderson Island, a small deserted coral island in the Pacific Ocean’s middle, to the Mariana Trench, which reaches a depth of 36,070 ft [15,16].
The most visible and painful consequences of marine plastics include suffocation, tangling, and ingestion of thousands of marine animals. Plastic trash is mistaken for food by whales, turtles, seabirds, and fish and the majority of them starve to death as their tummies fill with trash. Infections, lacerations, internal injuries, and reduced swimming abilities are also present. Invasive marine species and bacteria are also propagated by floating trash, damaging ecosystems. Invasive species are transported by plastics. Plastics serve as a way for long-distance dispersal; bring species to deserted places where they compete with local species. For example, insect eggs were found on 24% of the plastic pellets analyzed in a research in the Western Atlantic [17].
Microplastic has been discovered in beer, tap water, and salt, as well as in all ocean specimens examined across the world, including the Arctic. Numerous chemicals used in the production of plastic items are classified as carcinogenic and alter the endocrine system, causing reproductive, developmental, immunological, and neurological problems in humans and wildlife. Toxic contaminants grow up on the surface of plastic products after prolonged contact with seawater. Plastic waste consumed by marine animals enters their digestive tracts, where it builds in the food chain over time. Although it is yet to be completely researched, the transmission of contaminants from marine organisms to humans through seafood consumption has been highlighted as a health risk [18]. Plastic pollution can impair not only the waterways and land, but also human health, by harming wildlife and habitats. Plastic, a petroleum-based product, contributes to global warming as well. When plastic garbage is burned, carbon dioxide is released into the air, raising carbon emissions [17]. Plastic waste reduces the visual appeal of tourist destinations, leading to the lower tourism-related income and hefty cleaning and maintenance costs.
3. BIODEGRADATION OF PLASTICS
Plastic waste can be degraded through biotic and abiotic means which are referred to as biodegradation and physicochemical processes, respectively. The first step in any degradation process is usually considered the breakdown of the polymeric material by mechanical forces [19,20]. Microorganisms convert biochemical into compounds and this process is called biodegradation [21]. Depending on the degree of biodegradability and microbial assimilation, fossil-based and bio-based polymers can be included in biodegradable plastics. Plastics degrade due to a variety of factors, including mobility, crystalline structure, functional groups, molecular weight, and chemical additions to the polymers. During the biodegradation of plastics, microorganisms on the surface firstly decrease the molecular weight of the plastics, followed by the transformation of the polymer to its monomers, which are then broken down in a process of mineralization with the release carbon dioxide, water, and methane [21,22]. In case of big polymers, the depolymerization of into smaller monomers is important before being taken by microbes as big polymers pose difficulty in entering through the cellular membrane. Plastics can be degraded in a variety of ways, one of which is enzymatic degradation, in which enzymes attack the polymer substrate after hydrolytic breakage. Bacteria and fungi aid in the degradation of both natural and manmade polymers. Another type of degradation is clear zone development, which occurs when a polymer enters the synthetic medium agar as small particles. Agar plates containing emulsified polymers are often used procedures for the degradation of plastics, which result in the creation of halo zones around the plastics when treated with the microbe.
4. BIODIVERSITY OF PLASTIC DEGRADING MICROBES
The production of the synthetic plastic is one of the fastest growing fields of global industry. It has been estimated that approximately 80% of the total global plastic usage constitutes petrochemical plastic, which includes polyvinyl chloride (PVC), polyethylene (PE), polypropylene (PP), polystyrene (PS), and polyethylene terephthalate (PET). No doubt, plastic materials form an important part of global economy still the challenges and issues in association with their wide ranging applications and utilization cannot be ignored [23]. The major challenge associated with the plastics is the environmental pollution. Microbial communities are the most versatile organisms which play a major role in biodegradation in which microbes either break down the polymers into or utilize these compounds and convert them into simpler waste compounds while others are still able to utilize the excreted wastes [24]. Biodegradation involves four major steps [25]. The first step is biodeterioration in which the metabolic activities of microbes provoke plastic cracks. In the process, the physical properties of the plastics worsen or the microstructure of the matrix is changed. This step is followed by biofragmentation and release of the oligomers. Biofragmentation is followed by degradation and conversion of the oligomers to monomeric units in which oligomers enter inside the cells and secondary degraders assimilate them as a source of carbon. Assimilation of oligomers occurs in the last step in which excretion of completely oxidized metabolites to H2O, CO2, N2, and CH4. This section will discuss plastic degradation by different groups of microbes [Figure 1].
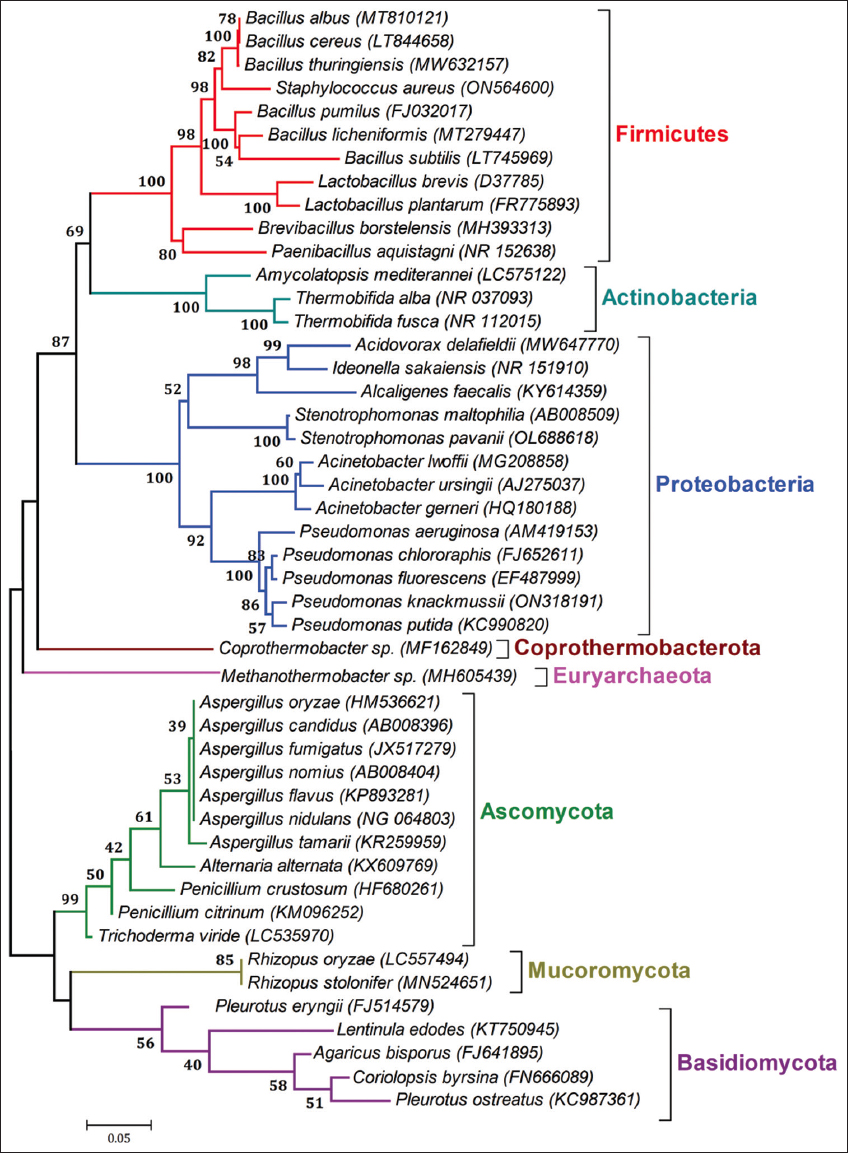 | Figure 1: Phylogenetic tree showing relationships among diverse phyla of plastic degrading microbes. [Click here to view] |
4.1. Archaea
Archaea are one of the most versatile groups of the microbes thriving in the extreme environmental conditions which even define physical barrier or too severe for existence of life including deep-sea hydrothermal vents, hypersaline ponds, or strictly anoxic ecosystems [26]. They may be extreme halophiles, hyperthermophiles, methanogens, and sulfur-metabolizing thermophiles. These extremophilic microbial communities possess unusual characteristics which make them a valuable bioresource for the development of innovative biotechnological processes. These extremophiles have been known though for more than 40 years but the research has increased in the last two decades as the conditions under which they survive have now become much broader than it was previously thought which has led to exposure of many unexplored habitats. Second, the increasing research and screening of extremophiles have unlocked various novel potential features of these organisms which can be utilized in diverse industrial sectors [27]. The role of the archaea in the biodegradation of the plastics is scarce and needs to be explored. The study of Jin et al. [28] recently reported the biodegradation of biodegradable plastics by species of Methanothermobacter.
4.2. Bacteria
Plastic wastes are dangerous for the natural environment as they accumulate in the rivers and oceans [23,29,30]. Fishing, industries, and coastal tourism are the major sources of plastics into the marine environment and pose a direct impact on seas and oceans [31]. The released plastics in the marine environment are colonized by the microbes [32-34]. It has been known fact that in sea water, colonization of the plastics by the bacterial communities starts immediately as they are introduced. Within a few hours, microbial assemblages are formed and surface of the plastics is covered in a step called as attachment. Microbial assemblages during this step may catalyze series of metabolic reactions that lead to the adsorption, desorption, and fragmentation of micro plastic-associated compounds [35].
Bacteria are emerging as an excellent agent for degradation of the plastics. Studies have reported that bacteria with capability to degrade plastics such as Moritella sp., Pseudomonas sp., Psychrobacter sp., and Shewanella sp. have been reported to degrade PCL [36]. Vibrio alginolyticus and Vibrio parahemolyticus have been reported for PVA-LLDPE degradation [37]. Singh et al. [38] reported Bacillus sp., Pseudomonas sp., and Staphylococcus sp. for polythene degradation. Bacillus sp., Klebsiella sp., and Pseudomonas sp. have been reported to degrade polyethylene synthetic plastic. Clonostachys rosea, Pseudomonas sp., Rhodococcus sp., and Trichoderma sp. from the arctic soil have been reported for degradation of PCL and commercial available bag based on potato and corn starch [39].
The use of the thermophiles for degradation of the plastics in the biological treatment of polluted thermal habitats is highly beneficial. The thermophilic microbes greatly improve the substrate bioavailability and solubility due to the changes in polymer properties including physical and optical [40]. PET majorly used in textile industry is available for hydrolysis by the activity of enzymes at about 65–75°C temperature due to the enhanced mobility of the amorphous sectors of the polymer chains [41,42]. The degradation of PET by Thermobifida alba, Thermomonospora curvata and Thermobifida halotolerans has been reported [43-45]. Recently, Jin et al. [28] reported Coprothermobacter sp. belonging to new phylum of anaerobic and thermophilic bacteria, Coprothermobacterota as degrader of biodegradable plastics.
4.3. Fungi
With the increasing research on the sustainable environment, fungal communities are getting greater attention as a potential bioresource in biodegradation of the plastics. Usman et al. [46] Alternaria alternate, Aspergillus candidus, Aspergillus flavus, Aspergillus nidulans, Aspergillus niger, Aspergillus ornatus, Aspergillus terreus, and Rhizopus stolonifera as potential degraders of polythene bags and bottom of plastic bottle. Cosgrove et al. [47] for the first time reported fungal communities from the surface of plastics such as polyester polyurethane during burial in situ in soil. In the study, Geomyces pannorum and Phoma sp. were reported as the potential candidates for PU degradation. Many studies have focused on plastic degradation by mushroom species. Da Luz et al. [48] reported the oxobiodegradable plastic biodegrading ability of Pleurotus ostreatus. Hock et al. [49] reported Agaricus bisporus, Lentinula edodes, Pleurotus eryngii, and Pleurotus ostreatus as di(2- ethylhexyl) phthalate degraders [Table 1].
Table 1: Diversity of plastic degrading microbes.
| Plastic degrading microbes | Type of plastic degraded | References |
|---|---|---|
| Achromobacter sp. | Low-density Polyethylene | Dey et al. [81] |
| Acinetobacter ursingii | Low-density Polyethylene | Hussein et al. [82] |
| Agaricus bisporus | di (2-ethylhexyl) phthalate | Hock et al. [49] |
| Aspergillus flavus | Polyethylene | Deepika and Jaya [83] |
| Aspergillus fumigatus | Polypropylene | OLIYA et al. [84] |
| Aspergillus niger | Polyethylene | Deepika and Jaya [83] |
| Aspergillus niger | Low-density Polyethylene | Ogunbayo et al. [85] |
| Aspergillus niger | Low-density Polyethylene | Mohamad [86] |
| Aspergillus nomius | Low-density Polyethylene | Munir et al. [87] |
| Bacillus licheniformis | Polyethylene | Ibrahim et al. [88] |
| Bacillus sp. | Low-density Polyethylene | Gupta et al. [89] |
| Bacillus sp. | Low-density Polyethylene | Joshi et al. [90] |
| Bacillus sp. | Polyethylene | Biki et al. [91] |
| Bacillus subtilis | Polycaprolactone | Widyananto et al. [92] |
| Bacillus subtilis | Polyethylene | Ibrahim et al. [88] |
| Bacillus subtilis | High-density Polyethylene | Tadimeti [93] |
| Bacillus subtilis | Low-density Polyethylene | Tadimeti [93] |
| Coriolopsis byrsina | Synthetic plastic | Kuswytasari et al. [94] |
| Enterobacter sp. | Polyethylene | Ren et al. [95] |
| Geomyces pannorum | Polyester polyurethane | Cosgrove et al. [47] |
| Ideonella sakaiensis | Polyethylene terephathalate | Juliana et al. [96] |
| Lentinula edodes | di (2-ethylhexyl) phthalate | Hock et al. [49] |
| Micrococcus sp. | Low-density Polyethylene | Gupta et al. [89] |
| Oceanimonas sp. | Low-density Polyethylene | Joshi et al. [90] |
| Paenibacillus sp. | Low-density Polyethylene | Bardají et al. [97] |
| Paenibacillus sp. | Low-density Polyethylene | Joshi et al. [90] |
| Penicillium citrinum | Low-density Polyethylene | Khan et al. [98] |
| Phoma sp. | Polyester polyurethane | Cosgrove et al. [47] |
| Pleurotus eryngii | di (2-ethylhexyl) phthalate | Hock et al. [49] |
| Pleurotus ostreatus | Oxo-biodegradable (D2W) plastic | da Luz et al. [48] |
| Pleurotus ostreatus | di (2-ethylhexyl) phthalate | Hock et al. [49] |
| Pseudomonas aeruginosa | Low-density Polyethylene | Hussein et al. [82] |
| Pseudomonas aeruginosa | Polyethylene | Hou et al. [99] |
| Pseudomonas aeruginosa | Polyethylene | Shahreza et al. [100] |
| Pseudomonas fluorescens | Low-density Polyethylene | Hussein et al. [82] |
| Pseudomonas knackmussii | Polyethylene | Hou et al. [99] |
| Pseudomonas putida | Polyethylene | Ibrahim et al. [88] |
| Pseudomonas sp. | Polyethylene | Deepika and Jaya [83] |
| Pseudomonas sp. | Low-density Polyethylene | Gupta et al. [89] |
| Pseudomonas sp. | Low-density Polyethylene | Ogunbayo et al. [85] |
| Pseudoxanthomonas sp. | Bisphenol-A polycarbonate | Yue et al. [101] |
| Ralstonia sp. | Polyethylene | Biki et al. [91] |
| Rheinheimera sp. | Low-density Polyethylene | Joshi et al. [90] |
| Rhizopus oryzae | Low-density Polyethylene | Mohamad [86] |
| Shewanella sp. | Low-density Polyethylene | Joshi et al. [90] |
| Staphylococcus sp. | Polypropylene | OLIYA et al. [84] |
| Stenotrophomonas pavanii | Polyethylene terephathalate | Huang et al. [102] |
| Stenotrophomonas sp. | Low-density Polyethylene | Dey et al. [81] |
| Streptomyces sp. | Polyethylene | Deepika and Jaya [83] |
| Streptomyces sp. | Low-density Polyethylene | Soud [103] |
| Thermobifida fusca | Polyethylene terephathalate | Huang et al. [102] |
| Trichoderma viride | Low-density Polyethylene | Munir et al. [87] |
| Trichoderma sp. | Low-density Polyethylene | Hikmah et al. [104] |
| Vibrio sp. | Low-density Polyethylene | Joshi et al. [90] |
5. FACTORS AFFECTING PLASTIC DEGRADATION
The properties of polymers, as well as their response to diverse biotic and abiotic stimuli, influence synthetic plastic degradation [50]. The physicochemical properties influence breakdown as its susceptibility to biotic or abiotic degradation is determined by their backbone composition and chain length. Polymeric materials possess long carbon chains such as those found in polypropylene which make them resistant to biodegradation. However, the insertion of heteroatoms in the carbon chain, as in the case of oxygen-containing polymers, renders them prone to heat and biodegradation. Furthermore, hydrophobicity is another factor which affects the efficiency of breakdown of these polymers. The rate of degradation rises as hydrophilicity rises [51].
The crystallinity impacts rate of plastic degradation. The more the crystallinity of the polymer, the greater is the need for oxygen and water, which may trigger the degradation process. The amorphous portions of the polymers are also known to have some role in degradation process as they are thought to be more sensitive to thermal oxidation. Furthermore, the molecular weight of a polymer might also impact its breakdown rate. Polymers with a high MW, according to this idea, degrade more slowly due to their smaller relative surface area [19].
Plastic polymer manufacturing involves additives that might affect the rate of degradation. Nanoadditives, for instance, can improve the polymeric properties for industrial applications. Nanoscale reinforcements increase the surface area of the interface for improved performance. The integration of these nanoparticles into the polymers intends to improve mechanical, rheological, electrical, and thermal properties. This further makes the recycling and degradation process easier. The use of nanoparticle additions in polymer manufacturing may potentially help to overcome biodegradation issues [52]. The degradability of the polymer may be influenced by the fabrication process. Copolymerized PP (polypropylene), for example, is less photodegradable than PP made by bulk (mass) polymerization or using the ZieglerNatta catalyst.
Stabilizers are frequently employed as additives in plastic manufacture to reduce the rate of deterioration. The chemical functionalization type in a polymeric structure irreversibly modifies the physicochemical qualities and its degradation rate. The morphological properties of the polymer have an impact on its degradability. In reality, the rate of degradation will rise as the surface roughness increases, because greater surface area promotes biofilm growth more than smooth surfaces [53].
The plastic degradation process mechanisms and rate may be affected by geographical location, air pollution, and prevailing meteorological conditions. For example, PET bottles may survive on the seabed for more than 15 years [54]. Water availability is a key factor in the degradation process because hydrolytic cleavage of functional groups that are vulnerable to hydrolysis results in polymeric chain separation [55]. The rate of degradation of plastics is influenced by oxygen availability. The availability of high levels of oxygen leads to accelerated polymeric degradation due to reactivity of oxygen with carbon-centered radicals which are created during the early degradation steps [56].
6. ROLE OF MICROBIAL ENZYMES IN PLASTIC DEGRADATION
Microbial enzymes are the most important environmental agents contributing to the biodegradation process. The process of biodegradation results in the conversion of the carbon in the polymer chains into smaller biomolecules or into carbon dioxide and water [57]. Thus, adding to the soil fertility and decreased accumulation of the plastics in turn reducing the cost of the waste management. The biodegradation of polyethylene through microbial enzymes consists of two steps. In the first step, there is adhesion of enzyme to the polyethylene substrate followed by the hydrolytic cleavage. Intracellular and extracellular depolymerases produced by fungi and bacteria are involved in the biodegradation of the polyethylene [58]. Microbial enzymes involved in lignin degradation have been reported to play a role in the biodegradation of polyethylene [59-62]. These include laccases, lignin peroxidases, and manganese peroxidases. Manganese peroxidase from Phanerochaete chrysosporium has been reported to play a chief role in degradation of a high molecular weight PE membrane [63]. The purified protease from Pseudomonas fluorescens, an esterase from Comamonas acidovorans, three esterases from Pseudomonas chlororaphis, and a lipase from Bacillus subtilis have been shown to exhibit hydrolytic capacity to emulsify polyester PUR [64-68]. Lipase and polyurethane esterase from Rhizopus delemer and Comamonas acidovorans have been reported to degrade PLA of low molecular weight and Amycalotopsis sp. has been reported for degradation of high molecular weight PLA [69]. Pestalotiopsis microspora containing serine hydrolase has been revealed to utilize the PU as a substrate, carbon source, and degrade it [70]. A thermostable laccase from Rhodococcus ruber has been investigated to degrade UV-irradiated films of PE [71]. Carboxylesterases from Thermobifida fusca, Bacillus subtilis, and Bacillus licheniformis have been shown to hydrolyze PET fibers partially as well as demonstrated a high activity against PET oligomers [72-76]. Thus, microbial enzymes play an important role in plastic waste management. Novel strategies and innovative solutions are important for reducing the production of non-biodegradable and non-reusable plastics and improvement of target enzymes or microbiomes in plastic biodegradation as tools for plastic management [77] [Table 2, Figures 2 and 3].
Table 2: Microbial enzymes involved in the degradation of different types of plastics.
| Plastic degrading microbes | Plastic used | Enzyme | References |
|---|---|---|---|
| Acidovorax delafieldii | poly (tetramethylene succinate)-co-adipate | Lipase | Uchida et al. [105] |
| Acinetobacter gerneri | Polyurethane | Polyurethanase | Howard et al. [106] |
| Acinetobacter lwoffii | Polyhydroxyalkanoates | Lipase | Sharma et al. [107] |
| Actinomadura sp. | Polyester | Depolymerases | Sriyapai et al. [108] |
| Alcaligens faecalis | Polyethylene | Lipase | Nag et al. [109] |
| Alcaligens faecalis | Polyethylene | CMCase | Nag et al. [109] |
| Alcaligens faecalis | Polyethylene | Xylanases | Nag et al. [109] |
| Alcaligens faecalis | Polyethylene | Protease | Nag et al. [109] |
| Amycolatopsis mediterannei | Poly(e-caprolactone) | Cutinase | Tan et al. [110] |
| Aspergillus melleus | Poly(e-caprolactone) | Lipase | Amin et al. [111] |
| Aspergillus niger | Poly (lactic acid) | Lipase | Nakajima-Kambe et al. [112] |
| Aspergillus niger | Poly(e-caprolactone) | Lipase | Nakajima-Kambe et al. [112] |
| Aspergillus tamarii | Polyethylene terephathalate | Lipase | Anbalagan et al. [113] |
| Aspergillus tamarii | Polyethylene terephathalate | Cutinase | Anbalagan et al. [113] |
| Bacillus pumilus | Polyurethane | Lipase | Nair and Kumar [114] |
| Bacillus sp. | Poly (butylene adipate co terephthalate) | Lipase | Zhang et al. [115] |
| Bacillus subtilis | Polyurethane | Polyurethanase-lipase | Rowe and Howard [66] |
| Bacillus sp. | Cellulose acetate plastic | Lipase | Ishigaki et al. [116] |
| Bacillus sp. | Cellulose acetate plastic | Cellulase | Ishigaki et al. [116] |
| Candida antarctica | Poly (butylene succinate) | Cutinase | Shi et al. [117] |
| Fusarium sp. | Poly (butylene succinate) | Lipase | Shi et al. [117] |
| Ideonella sakaiensis | Polyethylene terephthalate | PETase | Alfieri et al. [118] |
| Laceyella sp. | Polyester | Depolymerases | Sriyapai et al. [108] |
| Lactobacillus brevis | Poly(e-caprolactone) | Lipase | Khan et al. [119] |
| Lactobacillus plantarum | Poly(e-caprolactone) | Lipase | Khan et al. [119] |
| Moraxella sp. | Synthetic polymers | Esterase | Nikolaivits et al. [120] |
| Penicillium crustosum | Polyethylene terephathalate | Lipase | Anbalagan et al. [113] |
| Penicillium crustosum | Polyethylene terephathalate | Cutinase | Anbalagan et al. [113] |
| Pseudomonas chlororaphis | Polyhydroxyalkanoates | Lipase | Sharma et al. [107] |
| Pseudomonas chlororaphis | - | Lipase | Mohanan et al. [121] |
| Saccharothrix sp. | Poly (butylene succinate) | Depolymerases | Sriyapai et al. [122] |
| Stenotrophomonas sp. | Poly (butylene adipate-co-terephthalate) | Lipase | Jia et al. [123] |
| Streptomyces sp. | Polyester | Depolymerases | Sriyapai et al. [108] |
| Thermobifida alba | Aliphatic-aromatic copolyester film | Esterase | Hu et al. [43] |
 | Figure 2: Mechanism of biodegradation of plastics through microbial enzymes. [Click here to view] |
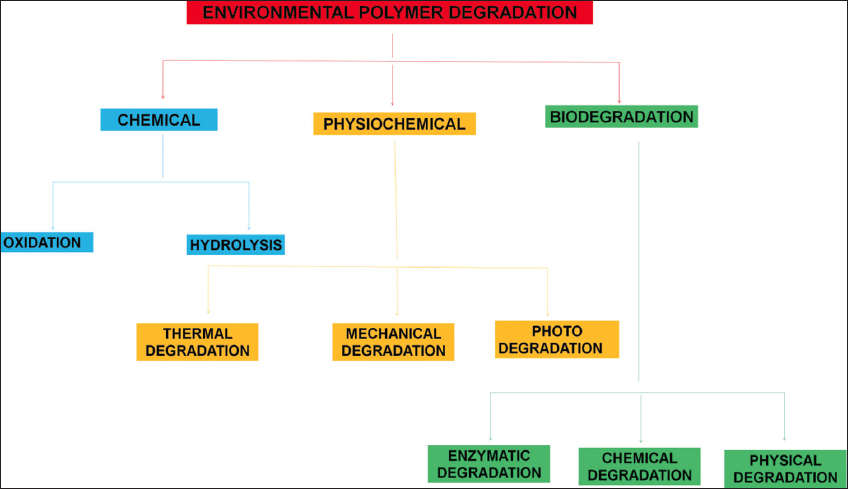 | Figure 3: Different methods used for polymer degradation. [Click here to view] |
7. DRAFT GENOME
The role of the microbes in the environmental cleaning especially the degradation of the plastics is an emerging field. The studies of the genomes of plastic degrading microbes are limited. To understand the metabolic pathways and genes associated with biodegradation is of major importance. A few researchers have reported the draft genomes of the plastic degrading microbes [78-80] [Table 3].
Table 3: Genome sequencing of plastic degrading microbes.
| Plastic degraders | Size (Mb) | GC% | Protein | Gene | References |
|---|---|---|---|---|---|
| Azoarcus sp. PA01T | 3.9 | 66.08 | 3625 | 3712 | Junghare et al. [124] |
| Bacillus albus PFYN01 | 6.4 | 34.90 | 6566 | - | León-Zayas et al. [78] |
| Bacillus sp. AIIW2 | 4.4 | 45.70 | - | 4714 | Kumari et al. [125] |
| Bacillus thuringiensis C15 | 5.2 | 35.0 | 5275 | - | León-Zayas et al. [78] |
| Bacillus sp. Y-01 | 5.8 | 38.24 | 4996 | - | Wang et al. [126] |
| Paenibacillus aquistagni DK1 | 5.4 | 47.40 | 4754 | 4950 | Furlan et al. [127] |
| Pseudomonas aeruginosa S3 | 6.6 | 66.17 | 6437 | Satti et al. [79] | |
| Pseudomonas putida CA-3 | 6.1 | 61.89 | 5608 | - | Almeida et al. [128] |
| Pseudomonas sp. B10 | 6.2 | 60.70 | 5565 | - | León-Zayas et al. [78] |
| Pseudomonas sp. SWI36 | 5.7 | 61.90 | 5186 | - | León-Zayas et al. [78] |
| Pseudomonas sp. SWI36 | 5.7 | 61.90 | 5193 | - | León-Zayas et al. [78] |
| Sphinogobacterium sp. S2 | 5.6 | 43.50 | 5385 | - | Satti et al. [79] |
| Stenotrophomonas maltophilia PE591 | 4.7 | 66.50 | 4432 | Frederico et al. [80] |
8. CONCLUSION AND FUTURE PERSPECTIVES
Plastic is the most utile fabricated polymer that has become an imminent part of human life, spanning countless sectors, all with inimitable attributes. However, unpredicted use of synthetic polymers, our throw-away culture, and widespread mismanagement has become ubiquitous in all the natural habitats, leading to rampant ingress of plastics into the environment and has become a global disquiet. The pernicious effects of plastics are apparent and intimidate food safety and quality, human, and wildlife health and contribute to climate change. Given the overwhelming impediment of dealing with worldwide plastic pollution, the novel microbial approach could be a pivotal component of the solution. Microbial communities are proficient in degrading inorganic and organic materials. The interest has aroused to study microbiomes for their capability to degrade plastic polymers. However, the multeity of known enzymes and microbes acting on synthetic polymers is still rather restricted. Hence, further exploration and screening of effectual microbial strains, suitable in-situ and ex-situ remediation techniques, and apposite maintenance of microbial growth and physicochemical conditions are imperative to curtail polymer hazards for the surroundings. Besides, at molecular level, identification of genes accountable for producing enzymes with potential of plastic degradation and recombinant DNA technology can perk up and expedite remediation of plastic waste. Another bonafide, effectual, and sustainable approach is to bolster the use of bio-based and biodegradable plastics. In fact, the replacing conventional plastics with bioplastics can lead to considerable energy and GHGs emissions savings. Furthermore, plastic pollution being a serious issue across the globe solicits a clamant and international rejoinder involving all germane participants at different levels. Unless waste management practices are revitalized, the flux of plastics to the oceans could amplify by an order of magnitude in the ensuing decade. Hence, awareness about plastic pollution and its negative and undesirable effects on living organisms and environment is important. The awareness should be spawned at the school level. Furthermore, students must be guided on biodegradable and non-biodegradable plastic waste and their separation before disposal. To knuckle down, the predicament of plastic debris in the oceans is a herculean task and a catholic approach and collated action is urgently obligated that combines technology, public/policy initiatives and advocacy to circumvent further plastic pollution, and the subsequent affliction to aquatic ecosystems and human health.
9. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
10. FUNDING
This work is supported by Eternal University, Baru Sahib, Himachal Pradesh, India
11. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
12. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
13. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
14. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Muthukumar A, Veerappapillai S. Biodegradation of plastics-a brief review. Int J Pharm Sci Rev Res 2015;31:204-9.
2. Andrady AL, Neal MA. Applications and societal benefits of plastics. Philos Trans R Soc Lond B Biol Sci 2009;364:1977-84. [CrossRef]
3. Geyer R. Production, use, and fate of synthetic polymers. In:Plastic Waste and Recycling. London:Elsevier Inc.;2020. 13-32. [CrossRef]
4. Miller L, Soulliere K, Sawyer-Beaulieu S, Tseng S, Tam E. Challenges and alternatives to plastics recycling in the automotive sector. In:Waste Management and Valorization. United States:Academic Press;2017. 237-66. [CrossRef]
5. Kale SK, Deshmukh AG, Dudhare MS, Patil VB. Microbial degradation of plastic:A review. J BiochemTechnol 2015;6:952-61.
6. Geyer R. Production, use, and fate of synthetic polymers. In:Letcher TM, editor. Plastic Waste and Recycling. United States:Academic Press;2020. 13-32. [CrossRef]
7. Lewis J, Hayes M. Reduce, Reuse, Recycle, Rejected:Why Canada's Recycling Industry is in Crisis Mode. Toronto, Ontario:The Globe and Mail;2019. 22.
8. de Souza Machado AA, Lau CW, Kloas W, Bergmann J, Bachelier JB, Faltin E,
9. Rutkowska M, Heimowska A, Krasowska K, Janik HZ. Biodegradability of polyethylene starch blends in sea water. Pol J Environ Stud 2002;11:267-72.
10. Kiessling T, Gutow L, Thiel M. Marine litter as habitat and dispersal vector. In:Marine Anthropogenic Litter. Cham:Springer;2015. 141-81. [CrossRef]
11. Wright SL, Kelly FJ. Plastic and human health:A micro issue?Environ Sci Technol 2017;51:6634-47. [CrossRef]
12. Wong JK, Lee KK, Tang KH, Yap PS. Microplastics in the freshwater and terrestrial environments:Prevalence, fates, impacts and sustainable solutions. Sci Total Environ 2020;719:137512. [CrossRef]
13. Farrell P, Nelson K. Trophic level transfer of microplastic:
14. Nelson B. What can 28,000 Rubber Duckies Lost at Sea Teach Us about Our Oceans. Vol. 3. Atlanta:Mother Nature Network;2011. 1.
15. Lavers JL, Bond AL. Exceptional and rapid accumulation of anthropogenic debris on one of the world's most remote and pristine islands. Proc Natl Acad Sci 2017;114:6052-5. [CrossRef]
16. Jamieson AJ, Malkocs T, Piertney SB, Fujii T, Zhang Z. Bioaccumulation of persistent organic pollutants in the deepest ocean fauna. Nat Ecol Evol 2017;1:51. [CrossRef]
17. Bender M. An Earth Law Solution to Ocean Plastic Pollution. New York City:Earth Law Center;2018. 9.
18. Boucher J, Friot D. Primary Microplastics in the Oceans:A Global Evaluation of Sources. Vol. 10. Switzerland:IUCN Gland;2017. [CrossRef]
19. Li J, Wang Y, Wang X, Wu D. Crystalline characteristics, mechanical properties, thermal degradation kinetics and hydration behavior of biodegradable fibers melt-spun from polyoxymethylene/poly (l-lactic acid) blends. Polymers 2019;11:1753. [CrossRef]
20. Ali SS, Elsamahy T, Koutra E, Kornaros M, El-Sheekh M, Abdelkarim E,
21. Zheng Y, Yanful EK, Bassi AS. A review of plastic waste biodegradation. Crit Rev Biotechnol 2005;25:243-50. [CrossRef]
22. Shah AA, Hasan F, Hameed A, Ahmed S. Biological degradation of plastics:A comprehensive review. Biotechnol Adv 2008;26:246-65. [CrossRef]
23. Urbanek AK, Rymowicz W, Miro?czuk AM. Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl Microbiol Biotechnol 2018;102):7669-78. [CrossRef]
24. Atanasova N, Stoitsova S, Paunova-Krasteva T, Kambourova M. Plastic degradation by extremophilic bacteria. Int J Mol Sci 2021;22:5610. [CrossRef]
25. Dussud C, Ghiglione JF. Bacterial degradation of synthetic plastics. CIESM Workshop Monogr 2014;46:49-54.
26. Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nat Rev Microbiol 2005;3:479-88. [CrossRef]
27. Schiraldi C, Giuliano M, de Rosa M. Perspectives on biotechnological applications of archaea. Archaea 2002;1:75-86. [CrossRef]
28. Jin Y, Cai F, Song C, Liu G, Chen C. Degradation of biodegradable plastics by anaerobic digestion:Morphological, micro-structural changes and microbial community dynamics. Sci Total Environ 2022;834:155167. [CrossRef]
29. Eriksen M, Lebreton LC, Carson HS, Thiel M, Moore CJ, Borerro JC,
30. Lebreton L, Slat B, Ferrari F, Sainte-Rose B, Aitken J, Marthouse R,
31. Cole M, Lindeque P, Halsband C, Galloway TS. Microplastics as contaminants in the marine environment:A review. Mar Pollut Bull 2011;62:2588-97. [CrossRef]
32. De Tender CA, Devriese LI, Haegeman A, Maes S, Ruttink T, Dawyndt P. Bacterial community profiling of plastic litter in the Belgian part of the North Sea. Environ Sci Technol 2015;49:9629-38. [CrossRef]
33. Pauli NC, Petermann JS, Lott C, Weber M. Macrofouling communities and the degradation of plastic bags in the sea:An
34. Rummel CD, Jahnke A, Gorokhova E, Kühnel D, Schmitt-Jansen M. Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environ Sci Technol Lett 2017;4:258-67. [CrossRef]
35. Harrison JP, Sapp M, Schratzberger M, Osborn AM. Interactions between microorganisms and marine microplastics:A call for research. Mar Technol Soc J 2011;45:12-20. [CrossRef]
36. Sekiguchi T, Sato T, Enoki M, Kanehiro H, Uematsu K, Kato C. Isolation and characterization of biodegradable plastic degrading bacteria from deep-sea environments. JAMSTEC Rep Res Develop 2011;11:33-41. [CrossRef]
37. Raghul S, Bhat S, Chandrasekaran M, Francis V, Thachil E. Biodegradation of polyvinyl alcohol-low linear density polyethylene-blended plastic film by consortium of marine benthic vibrios. Int J Environ Scia Technol 2014;11:1827-34. [CrossRef]
38. Singh G, Singh AK, Bhatt K. Biodegradation of polythenes by bacteria isolated from soil. Int J Res Dev Pharm Life Sci 2016;5:2056-62.
39. Urbanek AK, Rymowicz W, Strzelecki MC, Kociuba W, Franczak ?, Miro?czuk AM. Isolation and characterization of Arctic microorganisms decomposing bioplastics. AMB Express 2017;7:148. [CrossRef]
40. Ahmed T, Shahid M, Azeem F, Rasul I, Shah AA, Noman M,
41. Geyer R, Jambeck JR, Law KL. Production, use, and fate of all plastics ever made. Sci Adv 2017;3:e1700782. [CrossRef]
42. Ru J, Huo Y, Yang Y. Microbial degradation and valorization of plastic wastes. Front Microbiol 2020;11:442. [CrossRef]
43. Hu X, Thumarat U, Zhang X, Tang M, Kawai F. Diversity of polyester-degrading bacteria in compost and molecular analysis of a thermoactive esterase from
44. Ribitsch D, Acero EH, Greimel K, Dellacher A, Zitzenbacher S, Marold A,
45. Wei R, Oeser T, Then J, Kühn N, Barth M, Schmidt J,
46. Usman L, Yerima R, Haruna M, Adamu S, Nafiu M, Lawal N,
47. Cosgrove L, McGeechan PL, Robson GD, Handley PS. Fungal communities associated with degradation of polyester polyurethane in soil. Appl Environ Microbiol 2007;73:5817-24. [CrossRef]
48. da Luz JM, Paes SA, Nunes MD, da Silva MD, Kasuya MC. Degradation of oxo-biodegradable plastic by
49. Hock OG, De Qin D, Lum HW, Hee CW, Shing WL. Evaluation of the plastic degradation ability of edible mushroom species based on their growth and manganese peroxidase activity. Curr Top Toxicol 2020;16:65-72.
50. Artham T, Doble M. Biodegradation of aliphatic and aromatic polycarbonates. Macromol Biosci 2008;8:14-24. [CrossRef]
51. Ali SS, Elsamahy T, Al-Tohamy R, Zhu D, Mahmoud Y, Koutra E,
52. de Dicastillo CL, Velásquez E, Rojas A, Guarda A, Galotto MJ. The use of nanoadditives within recycled polymers for food packaging:Properties, recyclability, and safety. Compr Rev Food Sci Food Saf 2020;19:1760-76. [CrossRef]
53. Booth AM, Kubowicz S, Beegle-Krause CJ, Skancke J, Nordam T, Landsem E,
54. Fotopoulou KN, Karapanagioti HK. Degradation of various plastics in the environment. In:Hazardous Chemicals Associated with Plastics in the Marine Environment. Germany:Springer;2017. 71-92. [CrossRef]
55. Pitt CG. Non-microbial degradation of polyesters. In:Mechanisms and Modifications. Vol. 180. Cambridge:Royal Society of Chemistry;1992. 7-12.
56. Price D, Horrocks A. Combustion processes of textile fibres. In:Handbook of Fire Resistant Textiles. United Kingdom:Woodhead Publishing;2013. 3-25. [CrossRef]
57. Mir S, Asghar B, Khan AK, Rashid R, Shaikh AJ, Khan RA,
58. Bhardwaj H, Gupta R, Tiwari A. Communities of microbial enzymes associated with biodegradation of plastics. J Pol Environ 2013;21:575-9. [CrossRef]
59. Carrott PJ, Carrott MR. Lignin-from natural adsorbent to activated carbon:A review. Bioresour Technol 2007;98:2301-2. [CrossRef]
60. Wei R, Zimmermann W. Microbial enzymes for the recycling of recalcitrant petroleum-based plastics:How far are we?Microb Biotechnol 2017;10:1308-22. [CrossRef]
61. Restrepo-Flórez JM, Bassi A, Thompson MR. Microbial degradation and deterioration of polyethylene-a review. Int Biodeterior Biodegrad 2014;88:83-90. [CrossRef]
62. Krueger MC, Harms H, Schlosser D. Prospects for microbiological solutions to environmental pollution with plastics. Appl Microbiol Biotechnol 2015;99:8857-74. [CrossRef]
63. Iiyoshi Y, Tsutsumi Y, Nishida T. Polyethylene degradation by lignin-degrading fungi and manganese peroxidase. J Wood Sci 1998;44:222-9. [CrossRef]
64. Allen AB, Hilliard NP, Howard GT. Purification and characterization of a solublepolyurethane degrading enzyme from
65. Howard GT, Ruiz C, Hilliard NP. Growth of
66. Rowe L, Howard GT. Growth of
67. Ruiz C, Howard GT. Nucleotide sequencing of a polyurethanase gene (pulA) from
68. Vega RE, Main T, Howard GT. Cloning and expression in
69. Masaki K, Kamini NR, Ikeda H, Iefuji H. Cutinase-like enzyme from the yeast
70. Russell JR, Huang J, Anand P, Kucera K, Sandoval AG, Dantzler KW,
71. Santo M, Weitsman R, Sivan A. The role of the copper-binding enzyme-laccase-in the biodegradation of polyethylene by the actinomycete
72. Billig S, Oeser T, Birkemeyer C, Zimmermann W. Hydrolysis of cyclic poly (ethylene terephthalate) trimers by a carboxylesterase from
73. Barth M, Honak A, Oeser T, Wei R, Belisário-Ferrari MR, Then J,
74. Lülsdorf N, Vojcic L, Hellmuth H, Weber TT, Mußmann N, Martinez R,
75. Oeser T, Wei R, Baumgarten T, Billig S, Föllner C, Zimmermann W. High level expression of a hydrophobic poly (ethylene terephthalate)-hydrolyzing carboxylesterase from
76. Ribitsch D, Heumann S, Trotscha E, Acero EH, Greimel K, Leber R,
77. Gricajeva A, Nadda AK, Gudiukaite R. Insights into polyester plastic biodegradation by carboxyl ester hydrolases. J Chem Technol Biotechnol 2022;97:359-80. [CrossRef]
78. León-Zayas R, Roberts C, Vague M, Mellies JL. Draft genome sequences of five environmental bacterial isolates that degrade polyethylene terephthalate plastic. Microbiol Resour Announc 2019;8:e00237-19. [CrossRef]
79. Satti SM, Shah AA, Auras R, Marsh TL. Genome annotation of Poly (lactic acid) degrading
80. Frederico TD, Peixoto J, de Sousa JF, Vizzotto CS, Steindorff AS, Pinto OH,
81. Dey AS, Bose H, Mohapatra B, Sar P. Biodegradation of unpretreated low-density polyethylene (LDPE) by
82. Hussein AA, Al-Mayaly IK, Khudeir SH. Isolation, Screening and Identification of Low Density Polyethylene (LDPE) degrading bacteria from contaminated soil with plastic wastes. Mesopotamia Environ J 2015;1:1-14.
83. Deepika S, Jaya M. Biodegradation of low density polyethylene by microorganisms from garbage soil. J Exp Biol Agric Sci 2015;3:1-5.
84. Oliya P, Singh S, Goel N, Singh UP, Srivastava AK. Polypropylene degradation potential of microbes isolated from solid waste dumping site. Pollut Res Pap 2020;39:268-77.
85. Ogunbayo A, Olanipekun O, Adamu I. Preliminary studies on the microbial degradation of plastic waste using
86. Mohamad NN. Biodegradation of Low-Density Polyethylene (LDPE) Mixed with Corn Strach by
87. Munir E, Harefa R, Priyani N, Suryanto D. Plastic degrading fungi
88. Ibrahim S, Gupta RK, War AR, Hussain B, Kumar A, Sofi T,
89. Gupta KK, Devi D, Rana D. Isolation and screening of Low Density Polyethylene (Ldpe) degrading bacterial strains from waste disposal sites. World J Pharma Res 2016;5:1633-43.
90. Joshi G, Goswami P, Verma P, Prakash G, Simon P, Vinithkumar NV,
91. Biki SP, Mahmud S, Akhter S, Rahman MJ, Rix JJ, Al Bachchu MA,
92. Widyananto PA, Muchlissin S, Radjasa O, Sabdono A. Aliphatic polyester biodegradation by coral-associated bacteria from Karimunjawa Marine National Park, Java Sea. IOP Conf Ser Earth Environ Sci 2022;967:012045. [CrossRef]
93. Tadimeti A. The effects of different aquatic environments on the rate of HDPE and LDPE degradation by
94. Kuswytasari ND, Kurniawati AR, Alami NH, Zulaika E, Shovitri M,
95. Ren L, Men L, Zhang Z, Guan F, Tian J, Wang B,
96. Juliana S, Parhusip M, Simanullang A, Tita E, Irawati W. Potential of
97. BardajíDK, Furlan JP, Stehling EG. Isolation of a polyethylene degrading
98. Khan S, Ali SA, Ali AS. Biodegradation of Low Density Polyethylene (LDPE) by Mesophilic fungus “
99. Hou L, Xi J, Liu J, Wang P, Xu T, Liu T,
100. Shahreza H, Sepahy AA, Hosseini F, Nejad RK. Molecular identification of pseudomonas strains with polyethylene degradation ability from soil and cloning of alkB gene. Arch Pharm Pract 2019;10:3-48.
101. Yue W, Yin CF, Sun L, Zhang J, Xu Y, Zhou NY. Biodegradation of bisphenol-A polycarbonate plastic by
102. Huang QS, Yan ZF, Chen XQ, Du YY, Li J, Liu ZZ,
103. Soud SA. Biodegradation of polyethylene LDPE plastic waste using locally isolated
104. Hikmah M, Setyaningsih R, Pangastuti A. The potential of lignolytic trichoderma isolates in LDPE (Low Density Polyethylene) plastic biodegradation. IOP Conf Ser Mater Sci Eng 2018;333:012076. [CrossRef]
105. Uchida H, Nakajima-Kambe T, Shigeno-Akutsu Y, Nomura N, Tokiwa Y, Nakahara T. Properties of a bacterium which degrades solid poly (tetramethylene succinate)-co-adipate, a biodegradable plastic. FEMS Microbiol Lett 2000;189:25-9. [CrossRef]
106. Howard GT, Norton WN, Burks T. Growth of
107. Sharma PK, Mohanan N, Sidhu R, Levin DB. Colonization and degradation of polyhydroxyalkanoates by lipase-producing bacteria. Can J Microbiol 2019;65:461-75. [CrossRef]
108. Sriyapai P, Chansiri K, Sriyapai T. Isolation and characterization of polyester-based plastics-degrading bacteria from compost soils. Microbiology 2018;87:290-300. [CrossRef]
109. Nag M, Lahiri D, Dutta B, Jadav G, Ray RR. Biodegradation of used polyethylene bags by a new marine strain of
110. Tan Y, Henehan GT, Kinsella GK, Ryan BJ. An extracellular lipase from
111. Amin M, Bhatti HN, Bilal M. Kinetic and thermodynamic characterization of lipase from
112. Nakajima-Kambe T, Edwinoliver N, Maeda H, Thirunavukarasu K, Gowthaman M, Masaki K,
113. Anbalagan S, Venkatakrishnan HR, Ravindran J, Sathyamoorthy J, Rangabashyam KA, Ragini YP,
114. Nair S, Kumar P. Molecular characterization of a lipase-producing
115. Zhang M, Sharaf F, Chengtao L. Screening and characterization of novel lipase producing
116. Ishigaki T, Sugano W, Ike M, Fujita M. Enzymatic degradation of cellulose acetate plastic by novel degrading bacterium
117. Shi K, Su T, Wang Z. Comparison of poly (butylene succinate) biodegradation by
118. Alfieri B, Alfieri M, Kelly M, Kilcoyne S, Poprik L, Sanyal A,
119. Khan I, Dutta JR, Ganesan R.
120. Nikolaivits E, Taxeidis G, Gkountela C, Vouyiouka S, Maslak V, Nikodinovic-Runic J,
121. Mohanan N, Wong CH, Budisa N, Levin DB. Characterization of polymer degrading lipases, LIP1 and LIP2 from
122. Sriyapai P, Sriyapai T, Sukrakanchana L. Optimization of polybutylene succinate (PBS)-degrading enzyme production from
123. Jia H, Zhang M, Weng Y, Zhao Y, Li C, Kanwal A. Degradation of poly (butylene adipate-co-terephthalate) by
124. Junghare M, Patil Y, Schink B. Draft genome sequence of a nitrate-reducing, o-phthalate degrading bacterium,
125. Kumari A, Bano N, Chaudhary DR, Jha B. Draft genome sequence of plastic degrading
126. Wang X, Qu C, Wang W, Zheng Z, Liu F, An M,
127. Furlan JP, Lopes R, Stehling EG. Whole-genome sequence-based analysis of the
128. Almeida EL, Margassery LM, O'Leary N, Dobson AD. Draft genome sequence of pseudomonas putida CA-3, a bacterium capable of styrene degradation and medium-chain-length polyhydroxyalkanoate synthesis. Gen Announc 2018;6:e01534-17. https://doi.org/10.1128/genomeA.01534-17 PMid:29371359 PMCid:PMC5786685