1. INTRODUCTION
Cancer is the second-leading cause of death in developing countries. This is mostly due to cancer-causing gene mutations. Tumor suppressor genes, DNA repair genes, and genes involved in cell formation increase, and differentiating enzymes such as L-asparaginase, L-glutaminase, and L-methionase remove essential amino acids from metabolic sources. By inhibiting the growth of cancer cells. Although it has high specificity, it can cause fever, immune, and allergic reactions. [1,2]. L-methionase is a carbon-sulfur lyase that converts L-methionine to α-ketobutyrate, methanethiol, and ammonia through an elimination reaction [3]. The study thoroughly examined the enhancements in cancer cell dependence on methionine and the biochemical findings of L-methionase as a therapeutic drug. Therefore, using L-methionase to remove L-methionine-dependent cancer cells is a potent, acceptable therapeutic technique [4,5].
L-methionase, a pyridoxal-5-phosphate-dependent enzyme [6], has shown efficacy as an anticancer drug against various tumor cells [7]. These comprise neuroblastoma, lung, kidney, and breast cancer, along with glioblastoma, and could have applications in the food and pharmaceutical industries [8]. Methionine is required for the production of proteins and the growth of cancer cells. However, it differs from normal cells, which stay unaffected as they can synthesize amino acids from external sources. Targeting methionine can inhibit cancer cell growth and cause apoptosis [9]. The therapeutic preparation of L-Methionase has been conjugated with polyethylene glycol to decrease its immunogenicity [10].
However, some studies have shown that tumor cells lack the enzyme methionine synthase, which normal cells require to synthesize methionine from the precursor homocysteine [4]. Normal cells are independent of methionine. While tumor cells require methionine for protein synthesis and DNA regulation. Homocysteine, which is formed from the demethylation of L-methionine, is associated with cardiovascular disease and Alzheimer’s disease [11]. L-methionase is used pharmacologically to reduce homocysteine levels, reduce histone methylation, and control weight in breast and prostate cancer cells [12]. L-methionase has potential therapeutic uses in the treatment of cancer, drug resistance, infections, aging, and heart disease [13]. It can also be employed as a biotechnology-relevant treatment for dangerous microorganisms [14].
Enzyme production and activity by isolated microorganisms is affected by culture conditions and the composition of the culture medium. This is different from traditional optimization approaches that adjust variables one at a time. This is also called one factor at a time (OFAT), by keeping other parameters constant. Statistical methods use tools such as response surface methods (RSM) to analyze relationships between variables and optimize culture parameters. The statistical RSM optimizes culture parameters by establishing relationships between responses and independent variables. This technique analyzes individual or combined variables, enhancing the optimal culture parameter through efficient statistical experimental design [10].
The main objective of this study was to screen L-methionase-producing fungi and increase L-methionase production from Aspergillus fumigatus MF 13 media components. The nutrient source and the culture conditions of this fungus were optimized using statistical methods.
2. MATERIALS AND METHODS
2.1. Materials
The chemicals used in this study were L-methionine (SRL, Maharashtra, India), phenol red, dextrose, sodium hydroxide, pyridoxal-5-phosphate, Nessler’s reagent, Folin reagent, yeast extract, sucrose, magnesium sulfate, potassium chloride, iron, potassium nitrate, ammonium sulfate, maltose, fructose, lactose, potato dextrose agar (PDA) (potato infusion: 4 g/l, dextrose: 20 g/l, and agar-agar: 20 g/l) and C’zapek Dox broth (sucrose: 30 g/l, sodium nitrate: 2 g/l, dipotassium phosphate: 1 g/l, magnesium sulfate: 0.50 g/l, potassium chloride: 0.50 g/l, ferrous sulfate: 0.010 g/l, and agar-agar: 20 g/l), all of which were purchased from Himedia, Mumbai, India.
2.2. Isolation of Fungi
Soil samples were collected at a depth of 5 cm from different locations. Including garbage dump sites (Rajkot, India), agricultural land (Rajkot, India), marine soil (Porbandar, India), pond soil (Morbi, India), and river soil (Aji River, Rajkot, India) after the sample was placed in a sterile zip-lock plastic bag and transported to the laboratory. Soil samples were stored at room temperature until further inoculated on PDA plates and incubated at 28°C for 5 to 6 days, and then the pure cultures were stored at 4°C for use further [15].
2.3. Screening of Fungal Isolates for L-methionase Production
This study used an agar-well diffusion technique to qualitatively investigate L-methionase production. Fungal isolates were inoculated into a modified C’Zapec-Dox medium and incubated for 5 to 7 days after the incubation period. Centrifugation was used to extract fungal cells free of filters. To determine L-methionase activity, 100 µl of fungal cell-free culture was introduced to the wells [16].
2.4. L-methionase Enzyme Assay
L-methionase production was examined by measuring the ammonia released from L-methionine. 1 ml of 1% methionine in 0.5 M potassium phosphate buffer along with 1 ml of crude enzyme made up the optimal reaction system. After adding 0.1 ml of 1.5 M trichloroacetic acid and centrifuging it, enzyme activity was inhibited. 3.7 ml of distilled water was mixed with 0.1 ml of the supernatant. Nessler’s reagent was used to identify the released ammonia, and 480 nm was used to analyze the color compounds that were produced [17].
2.5. Protein Determination
Using bovine serum albumin as the standard, the Folin–Lowry method was used to determine the protein concentration [14].
2.6. Morphological and Molecular Characterization
Macroscopic and microscopic examination of fungal morphology was carried out, with particular focus given to characteristics such as color, size, shape, and hyphae staining with lactophenol cotton blue [18].
Molecular identification of fungal isolates was performed by SLS Research Pvt. Co., Ltd. Genomic DNA was extracted from fungal isolate on PDA plates using a DNeasy® Fungal Mini Kit (QIAGEN, Hilden, Germany.) According to the manufacturer’s instructions, an assessment of the concentration and quality of the extracted DNA using a spectrophotometer (LABMAN, Chennai, Tamil Nadu) was performed using a total of 20 ng of DNA. The ITS region of the ribosome was amplified using the forward primer ITS1-F (5′-CTACCTGATCCGAGGTCAAC-3′) and the reverse primer ITS4-R (5′-AGGTGGACCCAGGGCCCTCA-3′) in a Bio-Rad thermocycler (Hercules, CA, USA). 25 µl of polymerase chain reaction (PCR) reaction mix contained 2.5 µl of 10× PCR buffer, 1.5 µl of MgCl2 (50 mM), 5.0 µl of dNTPs (10 mM), 1.0 µl of each primer, and 0.9 µl of Taq DNA polymerase (10 U/μl) (Amid Bioscience, USA), and 19.6 µl of molecular biology water. Amplification was achieved with the following PCR conditions: initial denaturation at 94°C for 5 minutes, followed by 33 cycles of denaturation at 94°C for 30 seconds, and annealing at 55°C for 30 seconds, extension at 72°C for 2 minutes, each time at 72°C, ending with a final extension for 5 minutes, PCR amplicons were purified using the PCR Clean-up Kit (Sigma-Aldrich, St. Louis, MO). They carried out the sequencing reaction with sequencing 27F (TACGTCCCTGCCCTTTGTAC) using the BigDye Terminator v3.1 cycle sequencing kit (Thermo Fisher Scientific, USA) on an ABI 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA). They performed similarity searches for the nucleotide sequences by BLAST (http://www.ncbi.nlm.nih.gov/blast) against the GenBank database to identify the isolate.
The 18S rRNA gene sequence of strain MF13 was used to search for similar sequences in the nucleotide sequence database by running the BLAST program. The highest-scoring sequences similar to the 18S rRNA gene sequence were identified from the results and retrieved. From the GenBank database, the identified sequences were aligned using MEGA 6.0 software, and the phylogenetic tree was estimated using neighbor-joining bootstrap analysis with the help of MEGA 6.0 software.
2.7. Media Optimization Using OFAT
The effects of culture conditions and media composition on L-methionase production were studied using the OFAT method with different parameters such as pH (from 6 to 10), temperature (25°C, 30°C, 35°C, and 40°C), substrate concentration (0.2% to 0.5%), inoculum size (1% to 3%), and incubation period (4 to 8 days) Nitrogen source (peptone, yeast extract, ammonium sulfate, and potassium nitrate), and carbon sources (glucose, maltose, fructose, and lactose).
2.8. Statistical Modeling and Optimization of L-methionase Production
Statistical modeling and optimization techniques were used to increase L-methionase production in A. fumigatus MF 13 fungal strains using the RSM and Plackett-Burman design (PBD). All experiments, data analysis, and verification of the results were carried out using Design Expert 13 software (Stat-Ease).
2.9. Plackett Berman Design
The experiment aimed to optimize the design of Plackett–Berman (PB) and identify the effect of fermentation factors on the production of the L-methionase enzyme. Eight parameters were examined at the highest (+) and lowest (-) levels. These parameters include temperature, pH, incubation time, potassium phosphate, magnesium sulfate, glucose, yeast extract, potassium chloride, and potassium nitrate Flask culture tests were performed, and response was measured in U/ml/minute and calculated as enzyme yield.
2.10. Optimization of Selected Parameters Using RSM
Pareto chart from the experimental design of PB It was used to analyze medium-sized components that affect enzyme production. This was then optimized using a central composite design (CCD). The experiment involved 20 trials and 6 central points, with the L-methionase activity averaged and denoted as response R1.
2.11. Validation of the Model
Conduct experiments to validate the statistical model at its optimal level of the most significant variables under a predicted set of conditions.
3. RESULTS
3.1. Screening of Fungal Isolates for L-methionase Production
For the production of L-methionase, we screened 50 fungal isolates obtained from soil samples. When 10 strains were grown on C’zapex dox agar, a yellow zone formed around their colonies. Seven isolates with larger zone dimensions, yellow appearances, and stronger intensity were found during the qualitative screening. Fungal isolate MF 13 showed the highest zone diameter, measuring 35 mm (Fig. 1), and L-methionase activity, releasing 4.31 U/ml/minute of ammonia under the assay condition (Fig. 2).
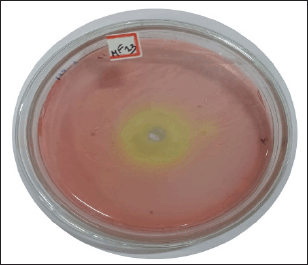 | Figure 1. Qualitative assay of L-methionase by rapid plate method. [Click here to view] |
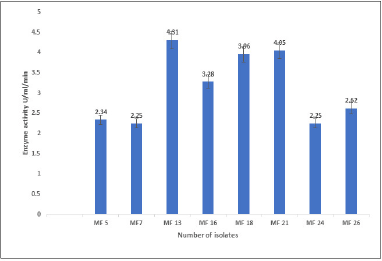 | Figure 2. Quantitative assay of L-methionase activity produce by the selected fungal isolates by Nessler’s method. [Click here to view] |
3.2. Morphological and Molecular Characterization
The morphological features of the fungal isolate MF 13 are typical. On PDA agar, colonies develop into gray-colored colonies in 7 days, with a maximum diameter of 70 mm. Under a microscope, the MF 13 conidia head on PDA agar revealed a columnar structure, ovate to flask-shaped vesicles, hyaline conidiophores, and uniseriate sterigmata. The shape of the conidia ranged from globose to prolate, with sizes between 1.5 and 2.5 µm (Fig. 3).
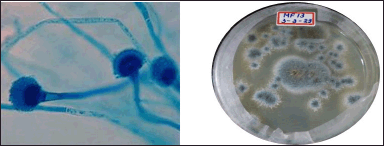 | Figure 3. Micro morphological and macro morphological identification of MF13 by lactophenol cotton blue method. [Click here to view] |
Using the neighbor-joining method in MEGA 6.0 software and the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the analysis of the 18S rRNA sequence showed 99.82% similarity to A. fumigatus. With A. fumigatus, a phylogenetic tree was constructed with a bootstrap value of 98%. The NCBI database has A. fumigatus strain MF13 with accession number OQ690549 associated with the 18S rRNA gene sequence (Fig. 4).
 | Figure 4. Phylogenetic tree analysis of A. fumigatus MF 13 by the neighbor-joining method. [Click here to view] |
3.3. Media Optimization Using OFAT
The study found that there was a significant relationship between pH and the amount of enzyme produced, with pH 8 (2.8 U/ml/minute) showing the highest level of L-methionase production after being examined at several temperatures. It was found that at 30°C the highest result for enzyme production was U/ml/minute. Substrate concentration ranged from 0.2% to 0.5%, with a maximum enzyme yield of 0.882 U/ml/minute, observed at a concentration of 0.2%. This study examined enzyme production at different inoculum sizes, with a maximum production of 0.476 U/ml/minute at an inoculum dose of 3%. Measurement of the production of enzymes took place from day 4 to 8 of incubation, with a peak of 1.75 ml/minute on day 7 and a subsequent decline to 0.683 ml/minute. When the impact of carbon sources on the production of L-methionase was evaluated, glucose produced the highest yield of the enzyme. The production of enzymes is also influenced by nitrogen sources like potassium nitrate and yeast extract (Fig. 5).
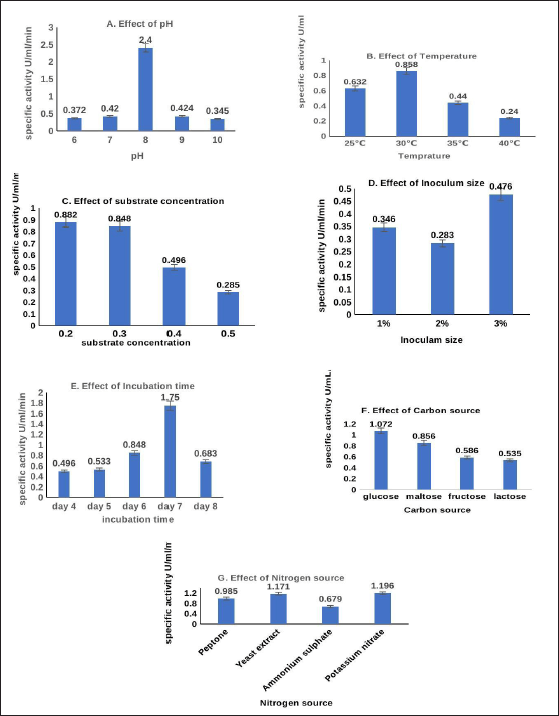 | Figure 5. Optimization of A. pH, B. temperature C. substrate concentration D. inoculum size E. incubation time F. carbon sources G. nitrogen sources on L-Methionase production by A. fumigatus MF 13 through OFAT method. [Click here to view] |
3.4. Plackett Burman design
The determination of important physicochemical parameters of PB (pH, temperature, incubation time, glucose, yeast extract, magnesium sulfate, potassium chloride, potassium nitrate, and potassium phosphate) to increase L-methionase production is an effective selection method. The study involved nine variables and 12 run trials, which were analyzed at two levels and coded as (−1) for low level and (+1) for high level (Table 1). L-methionase activity in A. fumigatus MF13 showed a wide variation, from 0.613 to 1.655 U/ml/minute, as shown in Table 2.
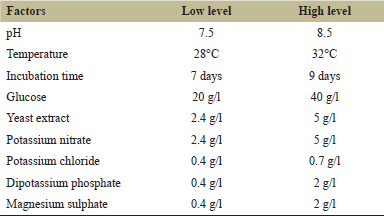 | Table 1. Experimental variable at two levels used for the production of L-methionase by A. fumigatus MF 13 using PBD. [Click here to view] |
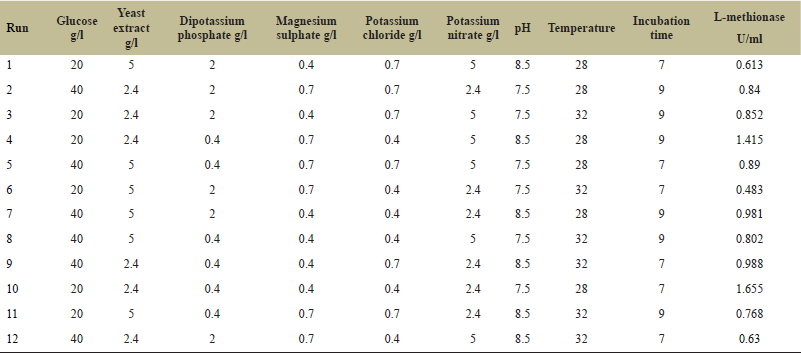 | Table 2. PBD for optimization of parameters influencing L-methionase production by A. fumigatus MF 13. [Click here to view] |
The Pareto chart indicates the most important factors affecting L-methionase production, mainly temperature, yeast extract, and dipotassium phosphate, as shown in Figure 6. These factors were selected for the CCD with their p values < 0.05 (Table 3), indicating their significance in enzyme production.
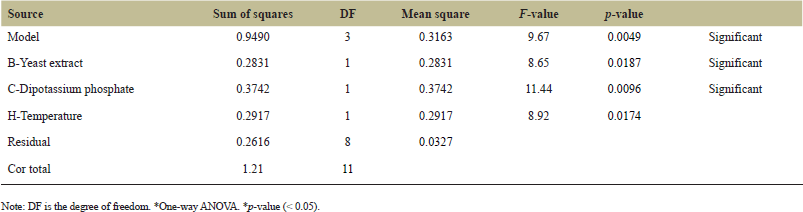 | Table 3. ANOVA for the experimental result of PBD. [Click here to view] |
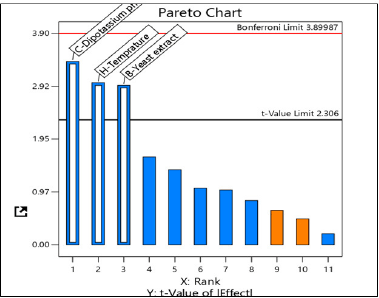 | Figure 6. Pareto chart of the standardized effects of nine medium factors on L-methionase production by A. fumigatus MF13, temperature, yeast extract, and dipotassium phosphate the significant factors positively affecting enzyme production. [Click here to view] |
3.5. Central Composite Design
Studies indicate that temperature, yeast extract, and dipotassium phosphate are key factors in increasing L-methionase production, which is further optimized using the CCD. The experimental design and response obtained for L-methionase production are shown in Table 4. The study included 20 experimental designs using CCD, and the highest L-methionase activity of 2.57 U/ml/minute was detected. In run number 15, the lowest L-methionase activity of 0.644 U/ml/minute was detected in run number 7. Regression analysis examined both the positive and negative effects of the significant variables on L-methionase production, where the results were interpreted in terms of probability values (p values) as shown in Table 5. The high F value of the model is 146.6 and the p-value is less than 0.05, with a poor fit, indicating the accuracy of predicting L-methionase production. The study found no significant effect of other interacting conditions on L-methionase production. Through regression analysis, using Design Expert software yields the following equations:
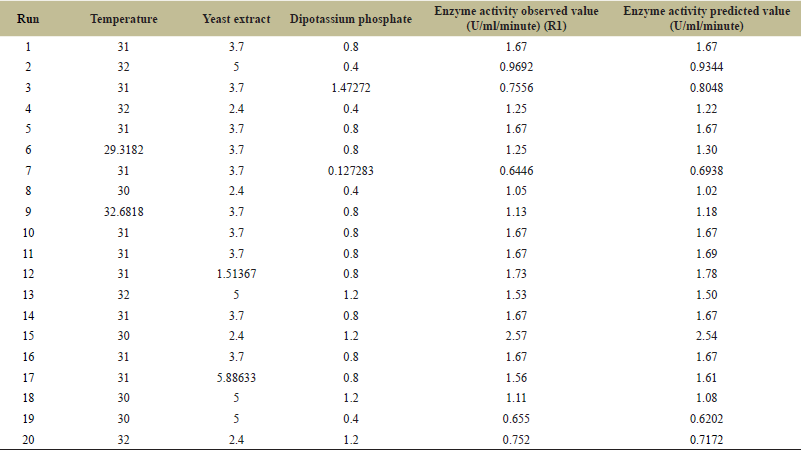 | Table 4. CCD for optimization experiment for L-methionase by A. fumigatus MF 13. [Click here to view] |
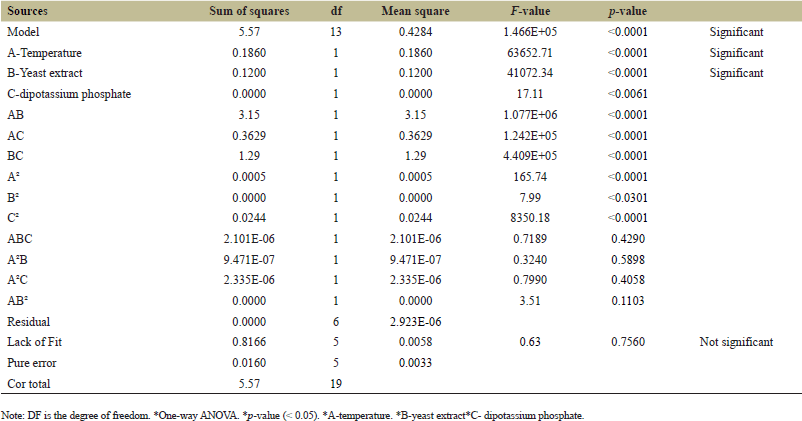 | Table 5. Analysis of ANOVA and significance level of the response surface of the full quadratic model for the L-methionase production. [Click here to view] |
Yi = β0 + ? βi xj + ? βii xj2 + ? βij xi
Y = 1.67 + 0.0795 temperature + 0.1204 yeast extract + 0.1629 dipotassium phosphate + 0.2940 temperature *Yeast extract: 0.2390 temperature * dipotassium phosphate = 0.00083 yeast extract* dipotassium phosphate = 0.1513 (temperature) 2 + 0.0096 (yeast extract)2 – 0.3245 (dipotassium phosphate)2.
In the above equation, βi is the linear effect coefficient, βii is the quadratic effect coefficient, βij is the interaction effect coefficient, and Y shows the response of the model to L-methionase production, in good agreement with the indicated R2 value of 95.4%, while the predicted R2 value was 0.943 (94.8%).
Design experts have created RSM 3D surface plots to graphically display regression equations of key parameters in the activities of 13 enzymes. It shows the effect and relationship of the variables on L-methionase production (Fig. 7).
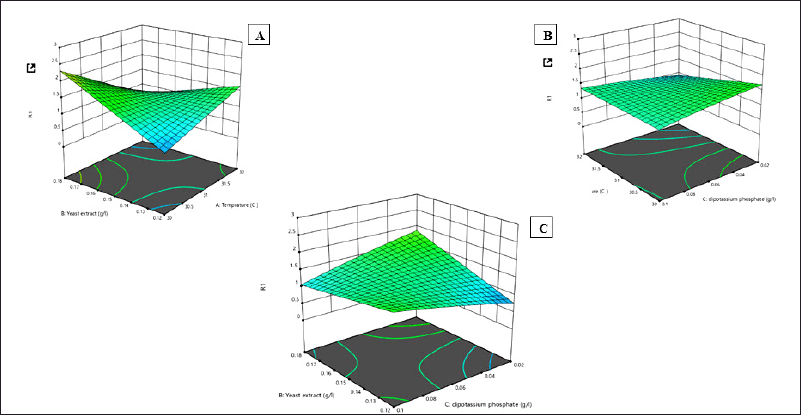 | Figure 7. (A–C) RSM 3D surface plots obtain by design expert 13 representing the effect and relationship between different variables in L-methionase production A) Temperature versus yeast extract, B) Temperature versus Dipotassium phosphate C) Yeast extract versus Dipotassium phosphate. [Click here to view] |
3.6. Data Validation
The obtained parameters of temperature (30°C), yeast extract (2.4 g/l), and dipotassium phosphate (1.2 g/l) were validated for their accuracy. These parameters indicated that the experimental enzyme activity was 2.565 U/ml/minute (Fig. 8). The model was found to be effective in increasing enzyme production considering all factors.
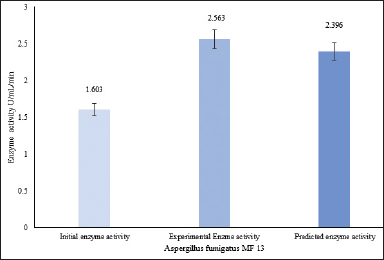 | Figure 8. Validation of the ideal conditions for A. fumigatus MF 13 to produce L-methionase enzyme. [Click here to view] |
4. DISCUSSION
This study presents A. fumigatus ability to secrete the L-methionase enzyme, a bioactive compound widely observed in various microorganisms, including bacteria, yeast, and fungi. L-methionase is known for its potential therapeutic and industrial applications, making its production by different organisms of significant interest.
L-methionase production levels by A. fumigatus were found to be comparable to other well-studied organisms such as A. flavipes, Trichoderma harzianum, Candida tropicalis, Serratia marcescens, Pseudomonas putida and Chaetomium globosum [4,9,11,19,20]. This finding expands the possible sources of L-methionase and emphasizes the versatility of A. fumigatus in biotechnology applications. Research results indicate that A. fumigatus could be a valuable organism for further research and development of L-methionase production.
The agar-well diffusion technique was used for quantitative screening by isolating L-methionase on agar plates, which was considered a potential producer of Selim et al. [15]. A previous study by Hendy et al. [14] reported that Aspergillus oryzae exhibited a 20-mm zone of diameter and L-methionase activity, releasing 2.13 U/ml/minute of ammonia under the assay condition. In this study, the isolate A. fumigatus MF 13 demonstrated the highest zone diameter, measuring 35 mm, and L-methionase activity, releasing 4.31 U/ml/minute of ammonia under the assay condition.
The challenge lies in finding new manufacturers for the large-scale production of L-methionase and in developing new growth media. This study also highlights the importance of culture parameters and diet composition in influencing L-methionase production in a one-time, single-factor approach. The physical conditions and chemical composition of the culture medium significantly affect cell growth and metabolism production.
In the present study of L-methionase from A. fumigatus MF 13, we observed higher enzyme production at 30°C, while a sharp decrease in enzyme production was detected at 40°C. Another thing is pH. We found that pH 8.0 was effective in increasing L-methionase production. The carbon source in the culture medium is important for microbial growth and metabolism production. Aspergillus flavipes [21] was identified as the best carbon source of glucose for L-methionase production. In our study, glucose also results in higher L-methionase production by A. fumigatus. Yeast extract has also been shown to be highly effective in increasing L-methionase production as a nitrogen source. According to a previous report [22], peptone was identified as the most suitable organic nitrogen supplement for L-methionase production.
PBD experiments were used in the present work to test different media components for maximum L-methionase production by A. fumigatus MF 13. Previous studies by Selim et al. [23] showed that PBD was used to optimize the media components produced successfully by Candida specie. This experiment identified three factors, namely, temperature, yeast extract, and dipotassium phosphate, as statistically significant using the PBD. It revealed that yeast extracts are an important source of nitrogen for the production of PBD L-methionase. A report by Salim et al. [22] illustrates the significance of yeast extract in the production of L-methionase from T. harzianum. Ions are essential for building cell mass and activating biosynthetic enzymes. This study identified dipotassium phosphate as an important ion source for increasing L-methionase production.
Other statistical methods include artificial neural networks that consist of neurons. It learns complex relationships between independent and dependent variables using a multi-layer feedforward architecture with a backpropagation algorithm for nonlinear mapping. According to Salim et al. [22], neural networks in an implant with hidden neurons have the minimum mean square error and provide good predictions of the results for training and validation. The overall R2 of the model is 0.993, which indicates a good fit. Activity of L-methionase. The predicted maximum was 30.17 U/ml/minute, which was close to the experimental value of 30.2 U/ml/minute [22]. A full qualitative fractional of the Box–Behnken design, a fraction of the full factorial, to evaluate quadratic effects and two-way interactions between variables, determining the nonlinear nature of the response. According to Mohkam et al. [24] yeast extract 0.2%; lactose 0.75%; pH 6; temperature 35°C. Additionally, this model predicted 13.1 U/ml/minute of L-methionase activity [24].
The study optimized factors for maximizing L-methionase activity using the CCD of RSM. The model’s strength was confirmed by an R2 value of 0.96, and the lack of fit F value (0.63) confirmed its significance. The model was generally significant, contrasting with previously reported data by Kavya and Nadumane [25], which indicated a high F value (145.31) and a lower F value (1.59), suggesting non-significance in the optimization of L-methionase production.
The main objective of this research is to find naturally occurring microorganisms for large-scale production of the enzyme L-methionase, avoiding the use of genetically modified organisms (GMOs). If GMOs are used to produce L-methionase, there is a chance that will be released accidentally. To the environment: This can destroy local ecosystems and have unexpected environmental impacts. Our research therefore has the potential to support biodiversity conservation efforts.
5. CONCLUSION
The study’s outcomes show how well fungal isolates were isolated, screened, and characterized to produce L-methionase. To find possible producers of L-methionase, several qualitative and quantitative testing techniques were performed. The fungal isolate MF13 was chosen to check its molecular identity out of the 50 isolates based on their highest enzymatic activity. Moreover, the fungus A. fumigatus was detected. L-methionase production from A. fumigatus was optimized by a OFAT and reached 7.69 µg/ml/minute. Statistical optimization was then completed using the PBD and RSM. This study included a total of seven parameters, and the most important parameter identified was the temperature (30°C), yeast extract (2.4 g/l), and dipotassium phosphate (1.2 g/l). RSM showed an optimal response to the production of L-methionase. RSM depicts the optimum response for L-methionase production. To the best of our knowledge, it is the first time that RSM and its representatives have been employed and demonstrated significant results for optimizing the medium components for L-Methionase production by A. fumigatus MF 13. Optimum culture conditions enhanced L-Methionase production by 4.4-fold compared to that under natural conditions. These optimized experimental designs can provide a quick and meaningful approach to improving the productivity of L-methionase.
6. ACKNOWLEDGMENT
The author would like to thank the management of Atmiya University, Rajkot, India, for continuous support in carrying out this study. The research leading to these results received funding from SHODH—Scheme of Developing High-quality Research, Education Department, Government of Gujarat, India (Grant Agreement Ref No. 202201241).
7. AUTHOR CONTRIBUTION
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVAL
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Dhankhar R, Gupta V, Kumar S, Kapoor RK, Gulati P. Microbial enzymes for deprivation of amino acid metabolism in malignant cells: biological strategy for cancer treatment. Appl Microbiol Biotechnol 2020;104(7):2857–69; CrossRef
2. Liu J, Huang J, Xin P, Liu G, Wu J. Biomedical applications of methionine-based systems. Biomater Sci 2021;9(6):1961–73; CrossRef
3. Al-Zahrani NH, Bukhari KA. Molecular identification, gene detection, and improving L-methioninase production of Serratia Sp. isolate. Pharmacophore 2019;10(6):14–25.
4. Kaiser P. Methionine dependence of cancer. Biomolecules 2020;10(4):7–9; CrossRef
5. Jonsson WO, Margolies NS, Anthony TG. Dietary sulfur amino acid restriction and the integrated stress response: mechanistic insights. Nutrients 2019;11(6):1349; CrossRef
6. Hassabo A, Mostafa E, Saad M, Selim M. Utilization of orange pulp and corn steep liquor for L-methioninase production by Wickerhamomyces subpelliculosus. Egypt Pharm J 2021;20(1):8–16.
7. Hoffman RM. Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: a 40-year odyssey. Expert Opin Biol Ther 2015;15(1):21–31.
8. Tan Y, Xu M, Hoffman RM. Broad selective efficacy of recombinant methioninase and polyethylene glycol-modified recombinant methioninase on cancer cells in vitro. Anticancer Res 2010;30(4):1041–6.
9. Teege VSL, Kondapalli K. Characterization and optimization of fermentation conditions for increased production of L-asparaginase from marine fungi. Eur J Mol Clin Med 2021;08(03):1758–69.
10. Alzaeemi SA, Noman EA, Al-shaibani MM, Al-Gheethi A, Mohamed RMSR, Almoheer R, et al. Improvement of L-asparaginase, an anticancer agent of Aspergillus arenarioides EAN603 in submerged fermentation using a radial basis function neural network with a specific genetic algorithm (RBFNN-GA). Fermentation 2023;9(3):200.
11. Alshehri WA. Bacterium Hafnia alvei secretes L-methioninase enzyme: optimization of the enzyme secretion conditions. Saudi J Biol Sci 2020;27(5):1222–7; CrossRef
12. Inagaki K. Structure of the antitumour enzyme L-methionine -Lyase from Pseudomonas putida at 1. 8 A resolution. J Biochem 2007 Apr;141(4):535–44; CrossRef
13. Pokrovsky VS, Anisimova NY, Davydov DZ, Bazhenov SV, Bulushova NV, Zavilgelsky GB. Methionine gamma lyase from Clostridium sporogenes increases the anticancer effect of doxorubicin in A549 cells and human cancer xenografts. Invest New Drugs 2018 Apr;37(2):201–9.
14. Hendy M, Hashem A, Sultan M, El-Ghamery A, Abdelraof M. L-methionine γ-lyase from thermo-tolerant fungi: isolation, identification of the potent producers, and statistical optimization of production via response surface methodology. Egypt J Chem 2021; CrossRef
15. Selim MH, Elshikh HH, El-Hadedy DE, Saad MM, Eliwa E, Abdelraof M. L-methioninase from some Streptomyces isolates I: isolation, identification of best producers and some properties of the crude enzyme produced. J Genet Eng Biotechnol 2015;13(2):129–37; CrossRef
16. Bopaiah BBK, Kumar DAN, Balan K, Dehingia L, Reddy MKRV, Suresh AB, et al. Purification, characterization, and antiproliferative activity of L-methioninase from a new isolate of Bacillus haynesii JUB2. J Appl Pharm Sci 2020;10(10):54–61; CrossRef
17. Khalaf SA, El-Sayed ASA. L-methioninase production by filamentous fungi: I-screening and optimization under submerged conditions. Curr Microbiol 2009;58(3):219–26; CrossRef
18. El-Sayed ASA, Ruff LE, Ghany SEA, Ali GS, Esener S. Molecular and spectroscopic characterization of Aspergillus flavipes and Pseudomonas putida L-methionine γ-Lyase in vitro. Appl Biochem Biotechnol 2017;181(4):1513–32; CrossRef
19. Sundar WA, Nellaiah H. Production of methioninase from Serratia marcescens isolated from soil and its anti-cancer activity against Dalton’s lymphoma ascitic (DLA) and Ehrlich ascitic carcinoma (EAC) in Swiss albino mice. Trop J Pharm Res 2013;12(5):699–704; CrossRef
20. Ito S, Nakamura T, Eguchi Y. Purification and characterization of methioninase from Pseudomonas putida. J Biochem 1976 Jun;79(6):1263–72.
21. El-Sayed ASA. L-methioninase production by Aspergillus flavipes under solid-state fermentation. J Basic Microbiol 2009;49(4):331–41; CrossRef
22. Salim N, Santhiagu A, Joji K. Process modeling and optimization of high yielding L-methioninase from a newly isolated Trichoderma harzianum using response surface methodology and artificial neural network coupled genetic algorithm. Biocatal Agric Biotechnol 2019;17:299–308.
23. Selim MH, Karm Eldin EZ, Saad MM, Mostafa ESE, Shetia YH, Anise AAH. Purification, characterization of L-methioninase from Candida tropicalis, and its application as an anticancer. Biotechnol Res Int 2015;2015:1–10.
24. Mohkam M, Taleban Y, Golkar N, Berenjian A, Dehshahri A, Mobasher MA, et al. Isolation and identification of novel L-Methioninase producing bacteria and optimization of its production by experimental design method. Biocatal Agric Biotechnol 2020;26:101566.
25. Kavya D, Nadumane VK. Identification of highest l-methioninase enzyme producers among soil microbial isolates, with potential antioxidant and anticancer properties. J Appl Biol Biotechnol 2020;8(6):21–7.